Ubiquitin Conjugating Enzyme E2 H (UBE2H) Is Linked to Poor Outcomes and Metastasis in Lung Adenocarcinoma
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. RNA Sequencing
2.3. Bioinformatic Analysis
2.4. Cell Culture and Transfection
2.5. qPCR
2.6. Western Blot
2.7. Wound-Healing Assay
2.8. Transwell Migration Assay
2.9. Statistics
3. Results
3.1. Identification of Ubiquitin Conjugating Enzyme E2 H (UBE2H) in the Malignant Pleural of Lung Adenocarcinoma
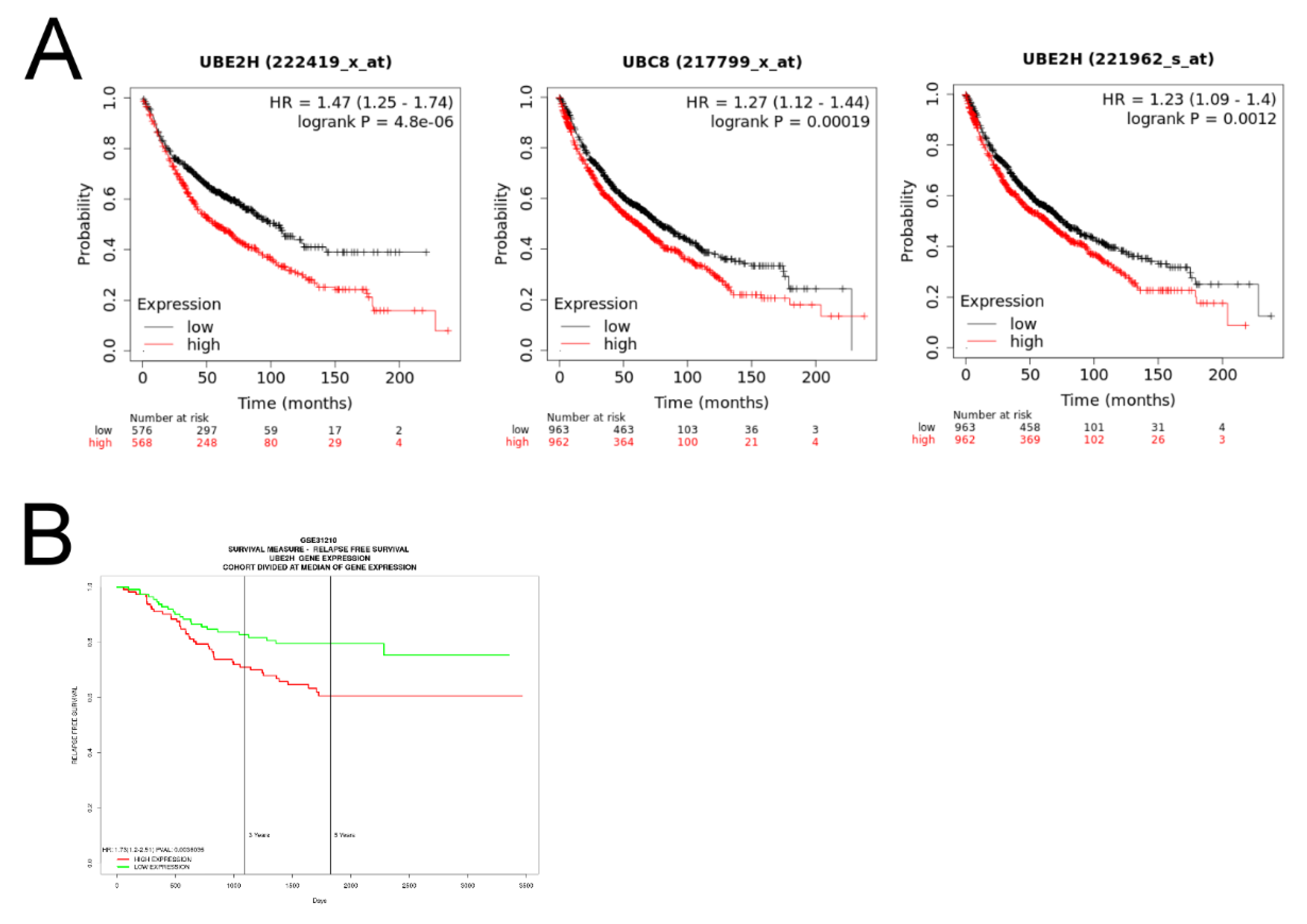
3.2. Investigation of the Association between UBE2H Expression and Survival in Patients with LUAD
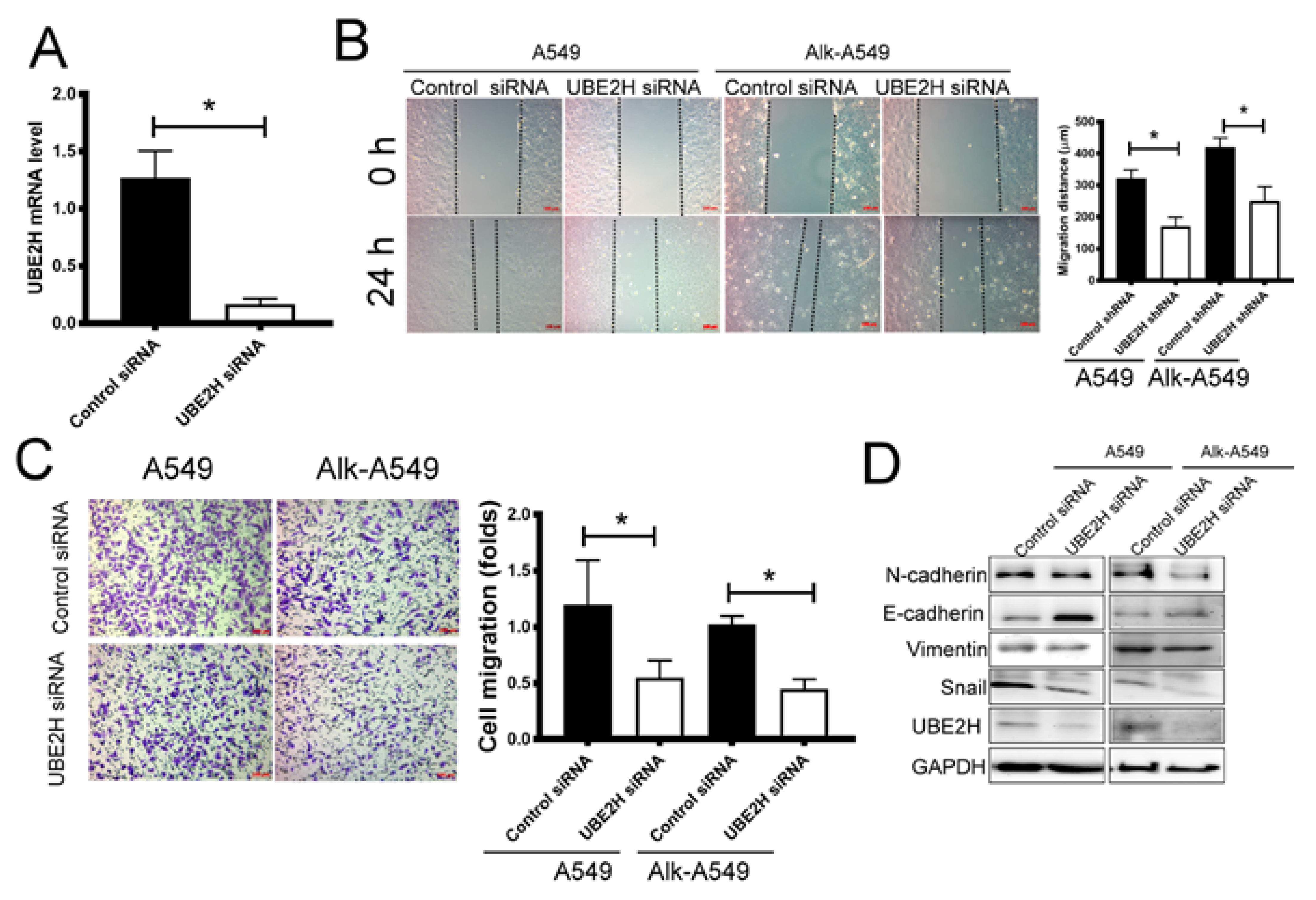
3.3. Investigating the Role of UBE2H in Migration Capacity of LUAD Cell Lines
3.4. The Potential Regulatory Mechanisms for UBE2H Expression in LUAD
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef]
- Duma, N.; Santana-Davila, R.; Molina, J.R. Non-Small Cell Lung Cancer: Epidemiology, Screening, Diagnosis, and Treatment. Mayo Clin. Proc. 2019, 94, 1623–1640. [Google Scholar] [CrossRef]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. Introduction to The 2015 World Health Organization Classification of Tumors of the Lung, Pleura, Thymus, and Heart. J. Thorac. Oncol. 2015, 10, 1240–1242. [Google Scholar] [CrossRef] [PubMed]
- Arbour, K.C.; Riely, G.J. Systemic Therapy for Locally Advanced and Metastatic Non-Small Cell Lung Cancer: A Review. JAMA 2019, 322, 764–774. [Google Scholar] [CrossRef] [PubMed]
- Elkrief, A.; Joubert, P.; Florescu, M.; Tehfe, M.; Blais, N.; Routy, B. Therapeutic landscape of metastatic non-small-cell lung cancer in Canada in 2020. Curr. Oncol. 2020, 27, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Reckamp, K.L. Targeted Therapy for Patients With Metastatic Non-Small Cell Lung Cancer. J. Natl. Compr. Cancer Netw. 2018, 16, 601–604. [Google Scholar] [CrossRef] [PubMed]
- Tarbe, N.; Losch, S.; Burtscher, H.; Jarsch, M.; Weidle, U.H. Identification of rat pancreatic carcinoma genes associated with lymphogenous metastasis. Anticancer Res. 2002, 22, 2015–2027. [Google Scholar] [PubMed]
- Kitamura, T.; Qian, B.Z.; Pollard, J.W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 2015, 15, 73–86. [Google Scholar] [CrossRef]
- Rankin, E.B.; Giaccia, A.J. Hypoxic control of metastasis. Science 2016, 352, 175–180. [Google Scholar] [CrossRef] [PubMed]
- Rankin, E.B.; Nam, J.M.; Giaccia, A.J. Hypoxia: Signaling the Metastatic Cascade. Trends Cancer 2016, 2, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Fares, J.; Fares, M.Y.; Khachfe, H.H.; Salhab, H.A.; Fares, Y. Molecular principles of metastasis: A hallmark of cancer revisited. Signal. Transduct. Target. Ther. 2020, 5, 28. [Google Scholar] [CrossRef]
- Jany, B.; Welte, T. Pleural Effusion in Adults-Etiology, Diagnosis, and Treatment. Dtsch. Arztebl. Int. 2019, 116, 377–386. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.W.; Chang, W.A.; Liu, K.T.; Yen, M.C.; Kuo, P.L. Vascular endothelial growth factor and protein level in pleural effusion for differentiating malignant from benign pleural effusion. Oncol. Lett. 2017, 14, 3657–3662. [Google Scholar] [CrossRef]
- Epelbaum, O.; Rahman, N.M. Contemporary approach to the patient with malignant pleural effusion complicating lung cancer. Ann. Transl Med. 2019, 7, 352. [Google Scholar] [CrossRef] [PubMed]
- Agalioti, T.; Giannou, A.D.; Stathopoulos, G.T. Pleural involvement in lung cancer. J. Thorac. Dis. 2015, 7, 1021–1030. [Google Scholar] [CrossRef]
- Roberts, M.E.; Neville, E.; Berrisford, R.G.; Antunes, G.; Ali, N.J.; Group BTSPDG. Management of a malignant pleural effusion: British Thoracic Society Pleural Disease Guideline 2010. Thorax 2010, 65 (Suppl. 2), ii32–ii40. [Google Scholar] [CrossRef]
- Liu, L.; Shao, D.; Deng, Q.; Tang, H.; Wang, J.; Liu, J.; Guo, F.; Lin, Y.; Peng, Z.; Mao, M.; et al. Next generation sequencing-based molecular profiling of lung adenocarcinoma using pleural effusion specimens. J. Thorac. Dis. 2018, 10, 2631–2637. [Google Scholar] [CrossRef]
- Grigoriadou, G.; Esagian, S.M.; Ryu, H.S.; Nikas, I.P. Molecular Profiling of Malignant Pleural Effusions with Next Generation Sequencing (NGS): Evidence that Supports Its Role in Cancer Management. J. Pers. Med. 2020, 10, 206. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef]
- Mootha, V.K.; Lindgren, C.M.; Eriksson, K.F.; Subramanian, A.; Sihag, S.; Lehar, J.; Puigserver, P.; Carlsson, E.; Ridderstrale, M.; Laurila, E.; et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003, 34, 267–273. [Google Scholar] [CrossRef]
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Ponce-Rodriguez, I.; Chakravarthi, B.; Varambally, S. UALCAN: A Portal for Facilitating Tumor Subgroup Gene Expression and Survival Analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef] [PubMed]
- Gyorffy, B.; Surowiak, P.; Budczies, J.; Lanczky, A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE 2013, 8, e82241. [Google Scholar] [CrossRef] [PubMed]
- Aguirre-Gamboa, R.; Gomez-Rueda, H.; Martinez-Ledesma, E.; Martinez-Torteya, A.; Chacolla-Huaringa, R.; Rodriguez-Barrientos, A.; Tamez-Pena, J.G.; Trevino, V. SurvExpress: An online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 2013, 8, e74250. [Google Scholar] [CrossRef]
- Dweep, H.; Gretz, N. miRWalk2.0: A comprehensive atlas of microRNA-target interactions. Nat. Methods 2015, 12, 697. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Okayama, H.; Kohno, T.; Ishii, Y.; Shimada, Y.; Shiraishi, K.; Iwakawa, R.; Furuta, K.; Tsuta, K.; Shibata, T.; Yamamoto, S.; et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012, 72, 100–111. [Google Scholar] [CrossRef] [PubMed]
- Bayliss, R.; Choi, J.; Fennell, D.A.; Fry, A.M.; Richards, M.W. Molecular mechanisms that underpin EML4-ALK driven cancers and their response to targeted drugs. Cell. Mol. Life Sci. 2016, 73, 1209–1224. [Google Scholar] [CrossRef]
- Xiao, D.; He, J. Epithelial mesenchymal transition and lung cancer. J. Thorac. Dis. 2010, 2, 154–159. [Google Scholar] [CrossRef]
- Clague, M.J.; Heride, C.; Urbe, S. The demographics of the ubiquitin system. Trends Cell Biol. 2015, 25, 417–426. [Google Scholar] [CrossRef]
- Ayesha, A.K.; Hyodo, T.; Asano, E.; Sato, N.; Mansour, M.A.; Ito, S.; Hamaguchi, M.; Senga, T. UBE2S is associated with malignant characteristics of breast cancer cells. Tumour Biol. 2016, 37, 763–772. [Google Scholar] [CrossRef]
- Liu, Z.; Xu, L. UBE2S promotes the proliferation and survival of human lung adenocarcinoma cells. BMB Rep. 2018, 51, 642–647. [Google Scholar] [CrossRef]
- Tang, X.K.; Wang, K.J.; Tang, Y.K.; Chen, L. Effects of ubiquitin-conjugating enzyme 2C on invasion, proliferation and cell cycling of lung cancer cells. Asian Pac. J. Cancer Prev. 2014, 15, 3005–3009. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Huang, Y.; Du, Y.; Zhao, Y.; Ren, J.; Ma, S.; Wu, C. Identification of Prognostic Genes and Pathways in Lung Adenocarcinoma Using a Bayesian Approach. Cancer Inform. 2017, 16, 1176935116684825. [Google Scholar] [CrossRef] [PubMed]
- Martin, I.; Vourc’h, P.; Mahe, M.; Thepault, R.A.; Antar, C.; Vedrine, S.; Praline, J.; Camu, W.; Andres, C.R.; Corcia, P.; et al. Association study of the ubiquitin conjugating enzyme gene UBE2H in sporadic ALS. Amyotroph. Lateral Scler. 2009, 10, 432–435. [Google Scholar] [CrossRef]
- Lim, K.H.; Joo, J.Y. Predictive Potential of Circulating Ube2h mRNA as an E2 Ubiquitin-Conjugating Enzyme for Diagnosis or Treatment of Alzheimer’s Disease. Int. J. Mol. Sci 2020, 21, 3398. [Google Scholar] [CrossRef] [PubMed]
- Zuo, H.; Chen, L.; Li, N.; Song, Q. Identification of a Ubiquitination-Related Gene Risk Model for Predicting Survival in Patients With Pancreatic Cancer. Front. Genet. 2020, 11, 612196. [Google Scholar] [CrossRef] [PubMed]
- Keng, V.W.; Villanueva, A.; Chiang, D.Y.; Dupuy, A.J.; Ryan, B.J.; Matise, I.; Silverstein, K.A.; Sarver, A.; Starr, T.K.; Akagi, K.; et al. A conditional transposon-based insertional mutagenesis screen for genes associated with mouse hepatocellular carcinoma. Nat. Biotechnol. 2009, 27, 264–274. [Google Scholar] [CrossRef]
- Zhu, Y.C.; Wang, W.X.; Song, Z.B.; Zhang, Q.X.; Xu, C.W.; Chen, G.; Zhuang, W.; Lv, T.; Song, Y. MET-UBE2H Fusion as a Novel Mechanism of Acquired EGFR Resistance in Lung Adenocarcinoma. J. Thorac. Oncol. 2018, 13, e202–e204. [Google Scholar] [CrossRef]
- Inamura, K.; Shimoji, T.; Ninomiya, H.; Hiramatsu, M.; Okui, M.; Satoh, Y.; Okumura, S.; Nakagawa, K.; Noda, T.; Fukayama, M.; et al. A metastatic signature in entire lung adenocarcinomas irrespective of morphological heterogeneity. Hum. Pathol. 2007, 38, 702–709. [Google Scholar] [CrossRef]
- Li, X.; Peng, S. Identification of metastasis-associated genes in colorectal cancer through an integrated genomic and transcriptomic analysis. Chin. J. Cancer Res. 2013, 25, 623–636. [Google Scholar] [CrossRef]
- Voutsadakis, I.A. The ubiquitin-proteasome system and signal transduction pathways regulating Epithelial Mesenchymal transition of cancer. J. Biomed. Sci 2012, 19, 67. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Yao, F.; Xiao, Z.; Sun, Y.; Ma, L. MicroRNAs and metastasis: Small RNAs play big roles. Cancer Metastasis Rev. 2018, 37, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.G.; Chang, T.H.; Liu, Y.N.; Shih, J.Y. MicroRNA in Lung Cancer Metastasis. Cancers (Basel) 2019, 11, 265. [Google Scholar] [CrossRef] [PubMed]
- Han, L.; Chen, W.; Xia, Y.; Song, Y.; Zhao, Z.; Cheng, H.; Jiang, T. MiR-101 inhibits the proliferation and metastasis of lung cancer by targeting zinc finger E-box binding homeobox 1. Am. J. Transl. Res. 2018, 10, 1172–1183. [Google Scholar]
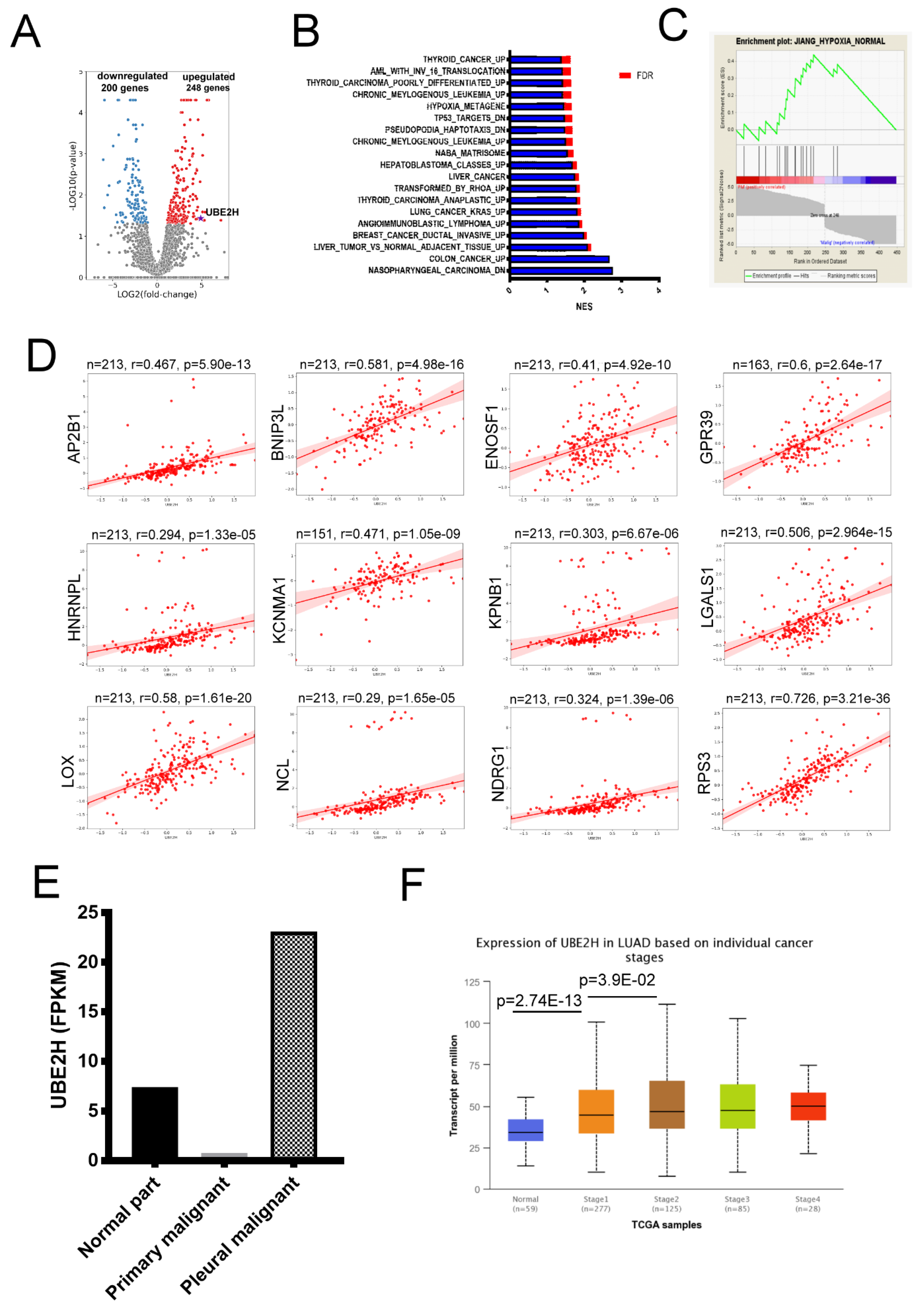


| Gene Symbol | Log2 (Fold) (Pleural Malignancy/Primary Malignancy) | Gene Symbol | Log2 (Fold) (Pleural Malignancy/Primary Malignancy) |
|---|---|---|---|
| LGALS1 | 18.337 | UBE2H | 4.889 |
| AP2B1 | 4.034 | ENOSF1 | 3.074 |
| NCL | 2.996 | KPNB1 | 2.816 |
| RPS3 | 2.768 | KCNMA1 | 2.708 |
| LOX | 2.304 | HNRNPC | 2.286 |
| HIF1A | 2.114 | BNIP3L | 2.087 |
| NDRG1 | 1.963 | GPR39 | 1.770 |
| miRNA | Fold (Pleural Malignant/Primary Malignant) | miRNA | Fold (Pleural Malignant/Primary Malignant) |
|---|---|---|---|
| hsa-miR-101-3p | 0.442752 | hsa-miR-122-5p | 0.000752 |
| hsa-miR-1275 | 0.486842 | hsa-miR-135a-5p | 0.035668 |
| hsa-miR-143-3p | 0.323242 | hsa-miR-30a-3p | 0.167051 |
| hsa-miR-30b-3p | 0.410777 | hsa-miR-3150b-5p | 0.000195 |
| hsa-miR-497-3p | 0.142195 | hsa-miR-328-3p | 0.477685 |
| hsa-miR-6724-5p | 0.413534 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yen, M.-C.; Wu, K.-L.; Liu, Y.-W.; Chang, Y.-Y.; Chang, C.-Y.; Hung, J.-Y.; Tsai, Y.-M.; Hsu, Y.-L. Ubiquitin Conjugating Enzyme E2 H (UBE2H) Is Linked to Poor Outcomes and Metastasis in Lung Adenocarcinoma. Biology 2021, 10, 378. https://doi.org/10.3390/biology10050378
Yen M-C, Wu K-L, Liu Y-W, Chang Y-Y, Chang C-Y, Hung J-Y, Tsai Y-M, Hsu Y-L. Ubiquitin Conjugating Enzyme E2 H (UBE2H) Is Linked to Poor Outcomes and Metastasis in Lung Adenocarcinoma. Biology. 2021; 10(5):378. https://doi.org/10.3390/biology10050378
Chicago/Turabian StyleYen, Meng-Chi, Kuan-Li Wu, Yu-Wei Liu, Yung-Yun Chang, Chao-Yuan Chang, Jen-Yu Hung, Ying-Ming Tsai, and Ya-Ling Hsu. 2021. "Ubiquitin Conjugating Enzyme E2 H (UBE2H) Is Linked to Poor Outcomes and Metastasis in Lung Adenocarcinoma" Biology 10, no. 5: 378. https://doi.org/10.3390/biology10050378
APA StyleYen, M.-C., Wu, K.-L., Liu, Y.-W., Chang, Y.-Y., Chang, C.-Y., Hung, J.-Y., Tsai, Y.-M., & Hsu, Y.-L. (2021). Ubiquitin Conjugating Enzyme E2 H (UBE2H) Is Linked to Poor Outcomes and Metastasis in Lung Adenocarcinoma. Biology, 10(5), 378. https://doi.org/10.3390/biology10050378






