Microarray and qPCR Analysis of Mitochondrial Metabolism Activation during Prenatal and Early Postnatal Development in Rats and Humans with Emphasis on CoQ10 Biosynthesis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. RNA Preparation and cDNA Reverse Transcription
2.2. Quantitative PCR
2.3. Protein Concentration
2.4. Spectrophotometric Analysis of Mitochondrial Enzyme Activities
2.5. CoQ9 and CoQ10 Content
2.6. cDNA Microarray Performance and its Subsequent Data Analysis
2.7. Data Analysis and Statistics
3. Results
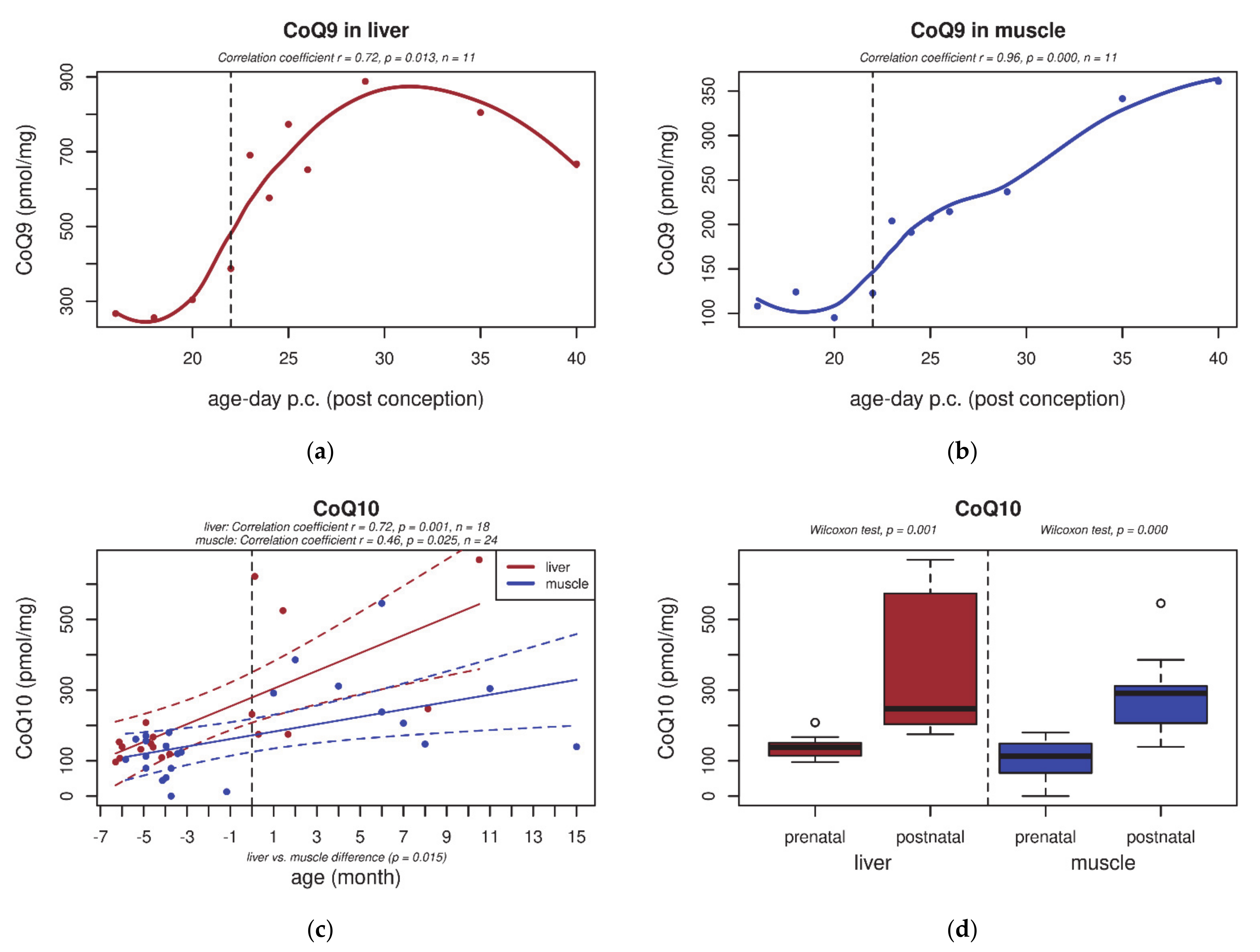
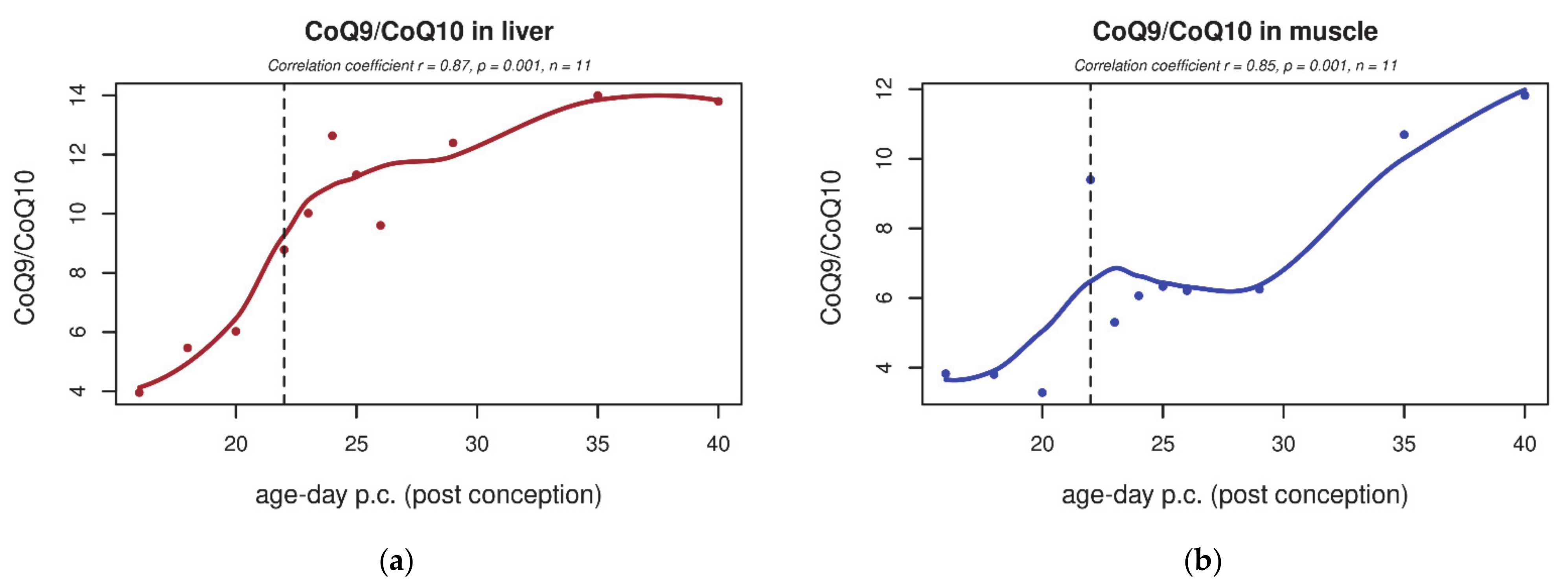
3.1. CoQ9 and CoQ10 Content
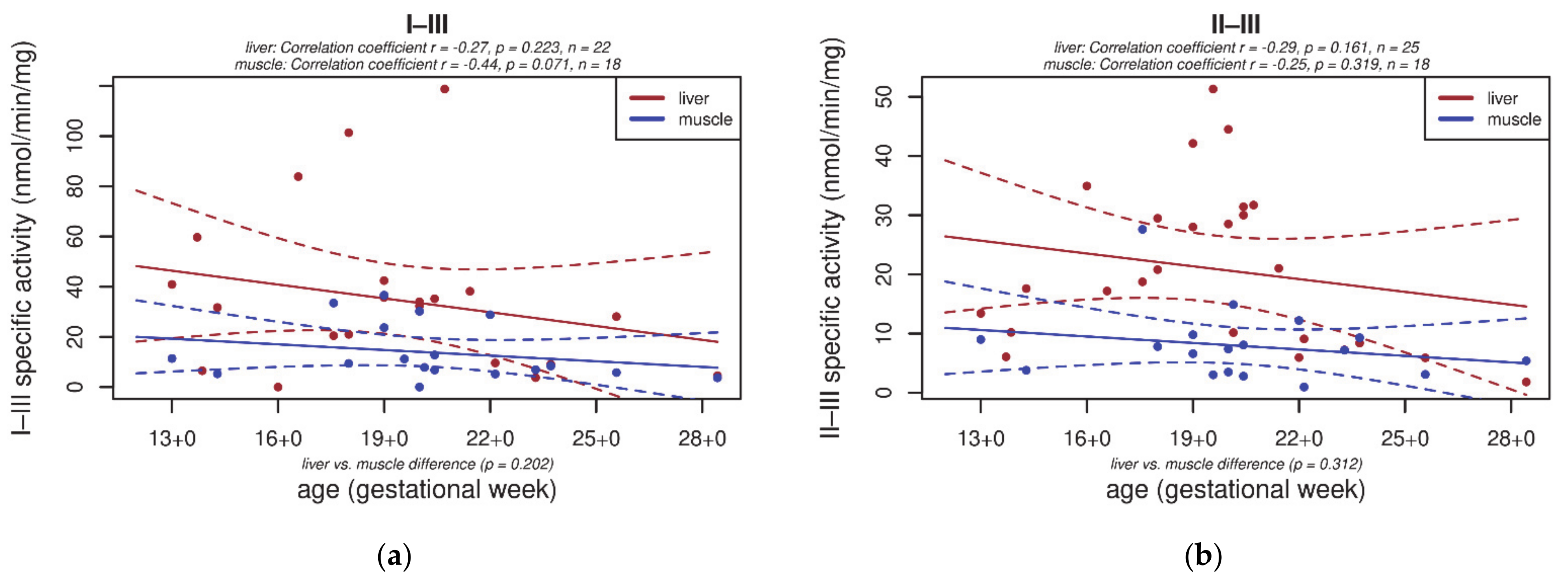
3.2. Electron Transport Chain (ETC) Enzyme Activities
3.3. Microarray Analysis
3.4. qPCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
Appendix A

| Cluster Number | Liver | Skeletal Muscle |
|---|---|---|
| Cluster 0 | 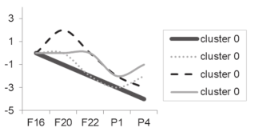 |  |
| Cluster 1 | 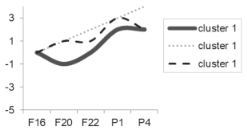 | 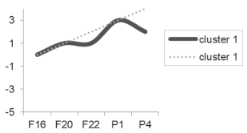 |
| Cluster 2 Cluster 3 | 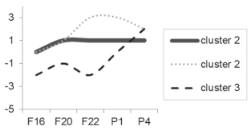 |  |
| Cluster 4 Cluster 5 | Not determined. | 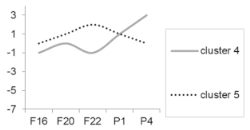 |
References
- Benard, G.; Faustin, B.; Passerieux, E.; Galinier, A.; Rocher, C.; Bellance, N.; Delage, J.P.; Casteilla, L.; Letellier, T.; Rossignol, R. Physiological diversity of mitochondrial oxidative phosphorylation. Am. J. Physiol. Cell Physiol. 2006, 291, C1172–C1182. [Google Scholar] [CrossRef]
- Burch, H.B.; Lowry, O.H.; Kuhlman, A.M.; Skerjance, J.; Diamant, E.J.; Lowry, S.R.; Von Dippe, P. Changes in patterns of enzymes of carbohydrate metabolism in the developing rat liver. J. Biol. Chem. 1963, 238, 2267–2273. [Google Scholar] [CrossRef]
- Prystowsky, H. Fetal blood studies. VII. The oxygen pressure gradient between the maternal and foetal bloods of the human in normal and abnormal pregnancy. Bull. Johns Hopkins Hosp. 1957, 101, 48–56. [Google Scholar]
- Sutton, R.; Pollak, J.K. The increasing adenine nucleotide concentration and the maturation of rat liver mitochondria during neonatal development. Differentiation 1978, 12, 15–21. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, J.M.; Cuezva, J.M. Control of the translational efficiency of beta-F1-ATPase mRNA depends on the regulation of a protein that binds the 3’ untranslated region of the mRNA. Mol. Cell Biol. 1997, 17, 5255–5268. [Google Scholar] [CrossRef] [Green Version]
- Izquierdo, J.M.; Jiménez, E.; Cuezva, J.M. Hypothyroidism affects the expression of the beta-F1-ATPase gene and limits mitochondrial proliferation in rat liver at all stages of development. Eur. J. Biochem. 1995, 232, 344–350. [Google Scholar] [CrossRef] [PubMed]
- Minai, L.; Martinovic, J.; Chretien, D.; Dumez, F.; Razavi, F.; Munnich, A.; Rötig, A. Mitochondrial respiratory chain complex assembly and function during human foetal development. Mol. Genet. Metab. 2008, 94, 120–126. [Google Scholar] [CrossRef]
- Pejznochová, M.; Tesarová, M.; Honzík, T.; Hansíková, H.; Magner, M.; Zeman, J. The developmental changes in mitochondrial DNA content per cell in human cord blood leukocytes during gestation. Physiol. Res. 2008, 57, 947–955. [Google Scholar]
- Pejznochova, M.; Tesarova, M.; Hansikova, H.; Magner, M.; Honzik, T.; Vinsova, K.; Hajkova, Z.; Havlickova, V.; Zeman, J. Mitochondrial DNA content and expression of genes involved in mtDNA transcription, regulation and maintenance during human foetal development. Mitochondrion 2010, 10, 321–329. [Google Scholar] [CrossRef] [PubMed]
- Lenaz, G.; Genova, M.L. Structural and functional organization of the mitochondrial respiratory chain: A dynamic super-assembly. Int. J. Biochem. Cell Biol. 2009, 41, 1750–1772. [Google Scholar] [CrossRef] [PubMed]
- Schägger, H.; Pfeiffer, K. The ratio of oxidative phosphorylation complexes I-V in bovine heart mitochondria and the composition of respiratory chain supercomplexes. J. Biol. Chem. 2001, 276, 37861–37867. [Google Scholar] [CrossRef] [PubMed]
- Lenaz, G.; Genova, M.L. Mobility and function of coenzyme Q (ubiquinone) in the mitochondrial respiratory chain. Biochim. Biophys. Acta 2009, 1787, 563–573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bentinger, M.; Brismar, K.; Dallner, G. The antioxidant role of coenzyme Q. Mitochondrion 2007, 7, S41–S50. [Google Scholar] [CrossRef]
- Hermans, N.; Cos, P.; De Meyer, G.R.; Maes, L.; Pieters, L.; Vanden Berghe, D.; Vlietinck, A.J.; De Bruyne, T. Study of potential systemic oxidative stress animal models for the evaluation of antioxidant activity: Status of lipid peroxidation and fat-soluble antioxidants. J. Pharm. Pharmacol. 2007, 59, 131–136. [Google Scholar] [CrossRef]
- Bravo, E.; Palleschi, S.; Rossi, B.; Napolitano, M.; Tiano, L.; D’Amore, E.; Botham, K.M. Coenzyme Q metabolism is disturbed in high fat diet-induced non-alcoholic fatty liver disease in rats. Int. J. Mol. Sci. 2012, 13, 1644–1657. [Google Scholar] [CrossRef] [Green Version]
- Turunen, M.; Olsson, J.; Dallner, G. Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 2004, 1660, 171–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guaras, A.; Perales-Clemente, E.; Calvo, E.; Acin-Perez, R.; Loureiro-Lopez, M.; Pujol, C.; Martinez-Carrascoso, I.; Nunez, E.; Garcia-Marques, F.; Rodriguez-Hernandez, M.A.; et al. The CoQH2/CoQ Ratio Serves as a Sensor of Respiratory Chain Efficiency. Cell Rep. 2016, 15, 197–209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, S.; Tsukahara, H.; Ohshima, Y.; Ohta, N.; Uchiyama, M.; Sekine, K.; Uetake, T.; Mayumi, M. Changes of plasma coenzyme Q10 levels in early infancy. Redox Rep. 2004, 9, 289–290. [Google Scholar] [CrossRef]
- Ochoa, J.J.; Ramirez-Tortosa, M.C.; Quiles, J.L.; Palomino, N.; Robles, R.; Mataix, J.; Huertas, J.R. Oxidative stress in erythrocytes from premature and full-term infants during their first 72 h of life. Free Radic. Res. 2003, 37, 317–322. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Navas, P.; Villalba, J.M.; de Cabo, R. The importance of plasma membrane coenzyme Q in aging and stress responses. Mitochondrion 2007, 7, S34–S40. [Google Scholar] [CrossRef]
- Martin, A.; Faes, C.; Debevec, T.; Rytz, C.; Millet, G.; Pialoux, V. Preterm birth and oxidative stress: Effects of acute physical exercise and hypoxia physiological responses. Redox Biol. 2018, 17, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Perez, M.; Robbins, M.E.; Revhaug, C.; Saugstad, O.D. Oxygen radical disease in the newborn, revisited: Oxidative stress and disease in the newborn period. Free Radic Biol Med. 2019, 142, 61–72. [Google Scholar] [CrossRef]
- Perrone, S.; Laschi, E.; Buonocore, G. Biomarkers of oxidative stress in the foetus and in the newborn. Free Radic. Biol. Med. 2019, 142, 23–31. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, G.; Carota, G.; Li Volti, G.; Giuffre, M. Biomarkers of Oxidative Stress for Neonatal Lung Disease. Front. Pediatr. 2021, 9, 618867. [Google Scholar] [CrossRef] [PubMed]
- Ozsurekci, Y.; Aykac, K. Oxidative Stress Related Diseases in Newborns. Oxid. Med. Cell Longev. 2016, 2016, 2768365. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torres-Cuevas, I.; Parra-Llorca, A.; Sanchez-Illana, A.; Nunez-Ramiro, A.; Kuligowski, J.; Chafer-Pericas, C.; Cernada, M.; Escobar, J.; Vento, M. Oxygen and oxidative stress in the perinatal period. Redox Biol. 2017, 12, 674–681. [Google Scholar] [CrossRef]
- Qin, X.; Cheng, J.; Zhong, Y.; Mahgoub, O.K.; Akter, F.; Fan, Y.; Aldughaim, M.; Xie, Q.; Qin, L.; Gu, L.; et al. Mechanism and Treatment Related to Oxidative Stress in Neonatal Hypoxic-Ischemic Encephalopathy. Front. Mol. Neurosci. 2019, 12, 88. [Google Scholar] [CrossRef]
- Serrenho, I.; Rosado, M.; Dinis, A.; Cardoso, M.C.; Graos, M.; Manadas, B.; Baltazar, G. Stem Cell Therapy for Neonatal Hypoxic-Ischemic Encephalopathy: A Systematic Review of Preclinical Studies. Int. J. Mol. Sci. 2021, 22, 3142. [Google Scholar] [CrossRef] [PubMed]
- Izquierdo, J.M.; Ricart, J.; Ostronoff, L.K.; Egea, G.; Cuezva, J.M. Changing patterns of transcriptional and post-transcriptional control of beta-F1-ATPase gene expression during mitochondrial biogenesis in liver. J. Biol. Chem. 1995, 270, 10342–10350. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Lecordier, A.; Bowman, L.H. Both nuclear and mitochondrial cytochrome c oxidase mRNA levels increase dramatically during mouse postnatal development. Biochem. J. 1995, 306, 353–358. [Google Scholar] [CrossRef] [Green Version]
- Cuezva, J.M.; Ostronoff, L.K.; Ricart, J.; López de Heredia, M.; Di Liegro, C.M.; Izquierdo, J.M. Mitochondrial biogenesis in the liver during development and oncogenesis. J. Bioenerg. Biomembr. 1997, 29, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Willers, I.M.; Martínez-Reyes, I.; Martínez-Diez, M.; Cuezva, J.M. miR-127-5p targets the 3’UTR of human β-F1-ATPase mRNA and inhibits its translation. Biochim. Biophys. Acta 2012, 1817, 838–848. [Google Scholar] [CrossRef] [PubMed]
- Spacilova, J.; Hulkova, M.; Hrustincova, A.; Capek, V.; Tesarova, M.; Hansikova, H.; Zeman, J. Analysis of Expression Profiles of Genes Involved in F0F1-ATP Synthase Biogenesis During Perinatal Development in Rat Liver and Skeletal Muscle. Physiol. Res. 2016, 65, 597–608. [Google Scholar] [CrossRef]
- Chapple, R.H.; Tizioto, P.C.; Wells, K.D.; Givan, S.A.; Kim, J.; McKay, S.D.; Schnabel, R.D.; Taylor, J.F. Characterization of the rat developmental liver transcriptome. Physiol. Genomics 2013, 45, 301–311. [Google Scholar] [CrossRef] [PubMed]
- Kolarova, H.; Krizova, J.; Hulkova, M.; Hansikova, H.; Hulkova, H.; Smid, V.; Zeman, J.; Honzik, T.; Tesarova, M. Changes in transcription pattern lead to a marked decrease in COX, CS and SQR activity after the developmental point of the 22(nd) gestational week. Physiol. Res. 2018, 67, 79–91. [Google Scholar] [CrossRef]
- R: A language and Environment for Statistical Computing, 2014: “R Core Team”. Available online: http://www.r-project.org/ (accessed on 3 May 2021).
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [CrossRef]
- Rustin, P.; Chretien, D.; Bourgeron, T.; Gérard, B.; Rötig, A.; Saudubray, J.M.; Munnich, A. Biochemical and molecular investigations in respiratory chain deficiencies. Clin. Chim. Acta 1994, 228, 35–51. [Google Scholar] [CrossRef]
- Wharton, D.; Tzagoloff, A. Cytochrome oxidase from beef heart mitochondria. Methods Enzymol. 1967, 10, 245–250. [Google Scholar] [CrossRef]
- Mosca, F.; Fattorini, D.; Bompadre, S.; Littarru, G.P. Assay of coenzyme Q(10) in plasma by a single dilution step. Anal. Biochem. 2002, 305, 49–54. [Google Scholar] [CrossRef]
- Lang, J.K.; Gohil, K.; Packer, L. Simultaneous determination of tocopherols, ubiquinols, and ubiquinones in blood, plasma, tissue homogenates, and subcellular fractions. Anal. Biochem. 1986, 157, 106–116. [Google Scholar] [CrossRef]
- The Gene Ontology, C. Expansion of the Gene Ontology knowledgebase and resources. Nucleic Acids Res. 2017, 45, D331–D338. [Google Scholar] [CrossRef] [Green Version]
- Mi, H.; Huang, X.; Muruganujan, A.; Tang, H.; Mills, C.; Kang, D.; Thomas, P.D. PANTHER version 11: Expanded annotation data from Gene Ontology and Reactome pathways, and data analysis tool enhancements. Nucleic Acids Res. 2017, 45, D183–D189. [Google Scholar] [CrossRef] [Green Version]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Davis, A.P.; Dolinski, K.; Dwight, S.S.; Eppig, J.T.; et al. Gene ontology: Tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edgar, R.; Domrachev, M.; Lash, A.E. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002, 30, 207–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vijay, V.; Han, T.; Moland, C.L.; Kwekel, J.C.; Fuscoe, J.C.; Desai, V.G. Sexual dimorphism in the expression of mitochondria-related genes in rat heart at different ages. PLoS ONE 2015, 10, e0117047. [Google Scholar] [CrossRef]
- Ernst, J.; Bar-Joseph, Z. STEM: A tool for the analysis of short time series gene expression data. BMC Bioinform. 2006, 7, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, A.I.; Sharov, V.; White, J.; Li, J.; Liang, W.; Bhagabati, N.; Braisted, J.; Klapa, M.; Currier, T.; Thiagarajan, M.; et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques 2003, 34, 374–378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, Y.; Yang, D.S.; Katti, P.; Glancy, B. Protein composition of the muscle mitochondrial reticulum during postnatal development. J. Physiol. 2019, 597, 2707–2727. [Google Scholar] [CrossRef]
- Ouyang, H.; Wang, Z.; Chen, X.; Yu, J.; Li, Z.; Nie, Q. Proteomic Analysis of Chicken Skeletal Muscle during Embryonic Development. Front. Physiol. 2017, 8, 281. [Google Scholar] [CrossRef] [Green Version]
- Piquereau, J.; Ventura-Clapier, R. Maturation of Cardiac Energy Metabolism During Perinatal Development. Front. Physiol. 2018, 9, 959. [Google Scholar] [CrossRef]
- Cardoso-Moreira, M.; Halbert, J.; Valloton, D.; Velten, B.; Chen, C.; Shao, Y.; Liechti, A.; Ascencao, K.; Rummel, C.; Ovchinnikova, S.; et al. Gene expression across mammalian organ development. Nature 2019, 571, 505–509. [Google Scholar] [CrossRef]
- Hurley, E.; Zabala, V.; Boylan, J.M.; Gruppuso, P.A.; Sanders, J.A. Hepatic Gene Expression During the Perinatal Transition in the Rat. Gene Expr. 2018, 19, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-Ayala, D.J.; Guerra, I.; Jimenez-Gancedo, S.; Cascajo, M.V.; Gavilan, A.; Dimauro, S.; Hirano, M.; Briones, P.; Artuch, R.; De Cabo, R.; et al. Survival transcriptome in the coenzyme Q10 deficiency syndrome is acquired by epigenetic modifications: A modelling study for human coenzyme Q10 deficiencies. BMJ Open 2013, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stefely, J.A.; Reidenbach, A.G.; Ulbrich, A.; Oruganty, K.; Floyd, B.J.; Jochem, A.; Saunders, J.M.; Johnson, I.E.; Minogue, C.E.; Wrobel, R.L.; et al. Mitochondrial ADCK3 employs an atypical protein kinase-like fold to enable coenzyme Q biosynthesis. Mol. Cell 2015, 57, 83–94. [Google Scholar] [CrossRef] [Green Version]
- Vazquez Fonseca, L.; Doimo, M.; Calderan, C.; Desbats, M.A.; Acosta, M.J.; Cerqua, C.; Cassina, M.; Ashraf, S.; Hildebrandt, F.; Sartori, G.; et al. Mutations in COQ8B (ADCK4) found in patients with steroid-resistant nephrotic syndrome alter COQ8B function. Hum. Mutat. 2018, 39, 406–414. [Google Scholar] [CrossRef]
- Cullen, J.K.; Abdul Murad, N.; Yeo, A.; McKenzie, M.; Ward, M.; Chong, K.L.; Schieber, N.L.; Parton, R.G.; Lim, Y.C.; Wolvetang, E.; et al. AarF Domain Containing Kinase 3 (ADCK3) Mutant Cells Display Signs of Oxidative Stress, Defects in Mitochondrial Homeostasis and Lysosomal Accumulation. PLoS ONE 2016, 11, e0148213. [Google Scholar] [CrossRef] [Green Version]
- Cullen, J.K.; Abdul Murad, N.; Yeo, A.; McKenzie, M.; Ward, M.; Chong, K.L.; Schieber, N.L.; Parton, R.G.; Lim, Y.C.; Wolvetang, E.; et al. Correction: AarF Domain Containing Kinase 3 (ADCK3) Mutant Cells Display Signs of Oxidative Stress, Defects in Mitochondrial Homeostasis and Lysosomal Accumulation. PLoS ONE 2016, 11, e0160162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzalez-Mariscal, I.; Martin-Montalvo, A.; Vazquez-Fonseca, L.; Pomares-Viciana, T.; Sanchez-Cuesta, A.; Fernandez-Ayala, D.J.; Navas, P.; Santos-Ocana, C. The mitochondrial phosphatase PPTC7 orchestrates mitochondrial metabolism regulating coenzyme Q10 biosynthesis. Biochim. Biophys. Acta Bioenerg. 2018, 1859, 1235–1248. [Google Scholar] [CrossRef] [PubMed]
- Lapointe, C.P.; Stefely, J.A.; Jochem, A.; Hutchins, P.D.; Wilson, G.M.; Kwiecien, N.W.; Coon, J.J.; Wickens, M.; Pagliarini, D.J. Multi-omics Reveal Specific Targets of the RNA-Binding Protein Puf3p and Its Orchestration of Mitochondrial Biogenesis. Cell Syst. 2018, 6, 125–135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kershaw, C.J.; Costello, J.L.; Talavera, D.; Rowe, W.; Castelli, L.M.; Sims, P.F.; Grant, C.M.; Ashe, M.P.; Hubbard, S.J.; Pavitt, G.D. Integrated multi-omics analyses reveal the pleiotropic nature of the control of gene expression by Puf3p. Sci Rep. 2015, 5, 15518. [Google Scholar] [CrossRef]
- Lin, K.; Qiang, W.; Zhu, M.; Ding, Y.; Shi, Q.; Chen, X.; Zsiros, E.; Wang, K.; Yang, X.; Kurita, T.; et al. Mammalian Pum1 and Pum2 Control Body Size via Translational Regulation of the Cell Cycle Inhibitor Cdkn1b. Cell Rep. 2019, 26, 2434–2450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veling, M.T.; Reidenbach, A.G.; Freiberger, E.C.; Kwiecien, N.W.; Hutchins, P.D.; Drahnak, M.J.; Jochem, A.; Ulbrich, A.; Rush, M.J.P.; Russell, J.D.; et al. Multi-omic Mitoprotease Profiling Defines a Role for Oct1p in Coenzyme Q Production. Mol. Cell 2017, 68, 970–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eldomery, M.K.; Akdemir, Z.C.; Vogtle, F.N.; Charng, W.L.; Mulica, P.; Rosenfeld, J.A.; Gambin, T.; Gu, S.; Burrage, L.C.; Al Shamsi, A.; et al. MIPEP recessive variants cause a syndrome of left ventricular non-compaction, hypotonia, and infantile death. Genome Med. 2016, 8, 106. [Google Scholar] [CrossRef] [Green Version]
- Gennarino, V.A.; Palmer, E.E.; McDonell, L.M.; Wang, L.; Adamski, C.J.; Koire, A.; See, L.; Chen, C.A.; Schaaf, C.P.; Rosenfeld, J.A.; et al. A Mild PUM1 Mutation Is Associated with Adult-Onset Ataxia, whereas Haploinsufficiency Causes Developmental Delay and Seizures. Cell 2018, 172, 924–936. [Google Scholar] [CrossRef] [Green Version]
- Shalata, A.; Edery, M.; Habib, C.; Genizi, J.; Mahroum, M.; Khalaily, L.; Assaf, N.; Segal, I.; Abed El Rahim, H.; Shapira, H.; et al. Primary Coenzyme Q deficiency Due to Novel ADCK3 Variants, Studies in Fibroblasts and Review of Literature. Neurochem. Res. 2019, 44, 2372–2384. [Google Scholar] [CrossRef]
- Traschutz, A.; Schirinzi, T.; Laugwitz, L.; Murray, N.H.; Bingman, C.A.; Reich, S.; Kern, J.; Heinzmann, A.; Vasco, G.; Bertini, E.; et al. Clinico-Genetic, Imaging and Molecular Delineation of COQ8A-Ataxia: A Multicenter Study of 59 Patients. Ann. Neurol. 2020. [Google Scholar] [CrossRef] [PubMed]
- Alcazar-Fabra, M.; Trevisson, E.; Brea-Calvo, G. Clinical syndromes associated with Coenzyme Q10 deficiency. Essays Biochem. 2018, 62, 377–398. [Google Scholar] [CrossRef]
- Hughes, B.G.; Harrison, P.M.; Hekimi, S. Estimating the occurrence of primary ubiquinone deficiency by analysis of large-scale sequencing data. Sci. Rep. 2017, 7, 17744. [Google Scholar] [CrossRef] [Green Version]
- Levavasseur, F.; Miyadera, H.; Sirois, J.; Tremblay, M.L.; Kita, K.; Shoubridge, E.; Hekimi, S. Ubiquinone is necessary for mouse embryonic development but is not essential for mitochondrial respiration. J. Biol. Chem. 2001, 276, 46160–46164. [Google Scholar] [CrossRef] [Green Version]
- Quinzii, C.M.; Garone, C.; Emmanuele, V.; Tadesse, S.; Krishna, S.; Dorado, B.; Hirano, M. Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 2013, 27, 612–621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia-Corzo, L.; Luna-Sanchez, M.; Doerrier, C.; Garcia, J.A.; Guaras, A.; Acin-Perez, R.; Bullejos-Peregrin, J.; Lopez, A.; Escames, G.; Enriquez, J.A.; et al. Dysfunctional Coq9 protein causes predominant encephalomyopathy associated with CoQ deficiency. Hum. Mol. Genet. 2013, 22, 1233–1248. [Google Scholar] [CrossRef] [PubMed] [Green Version]
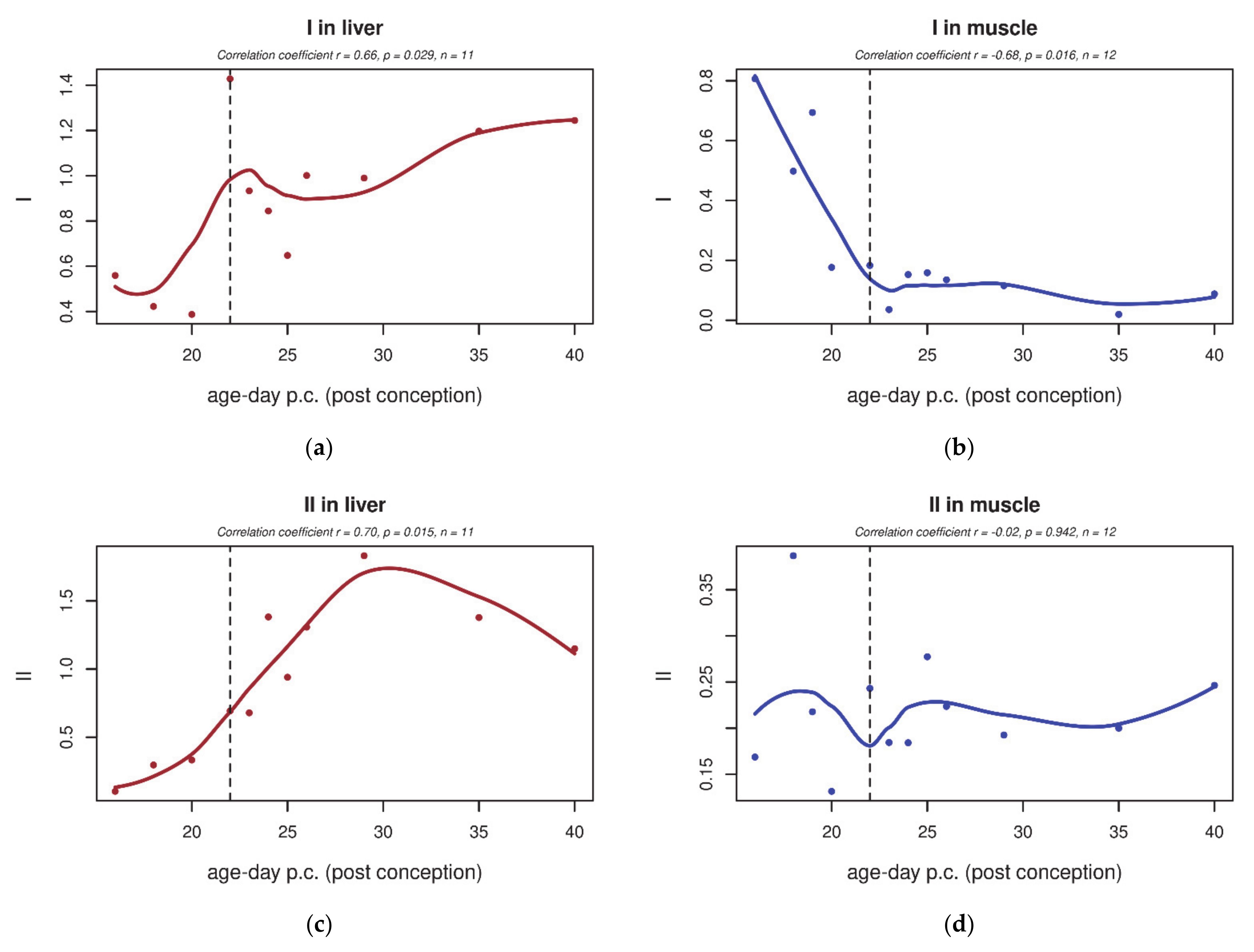

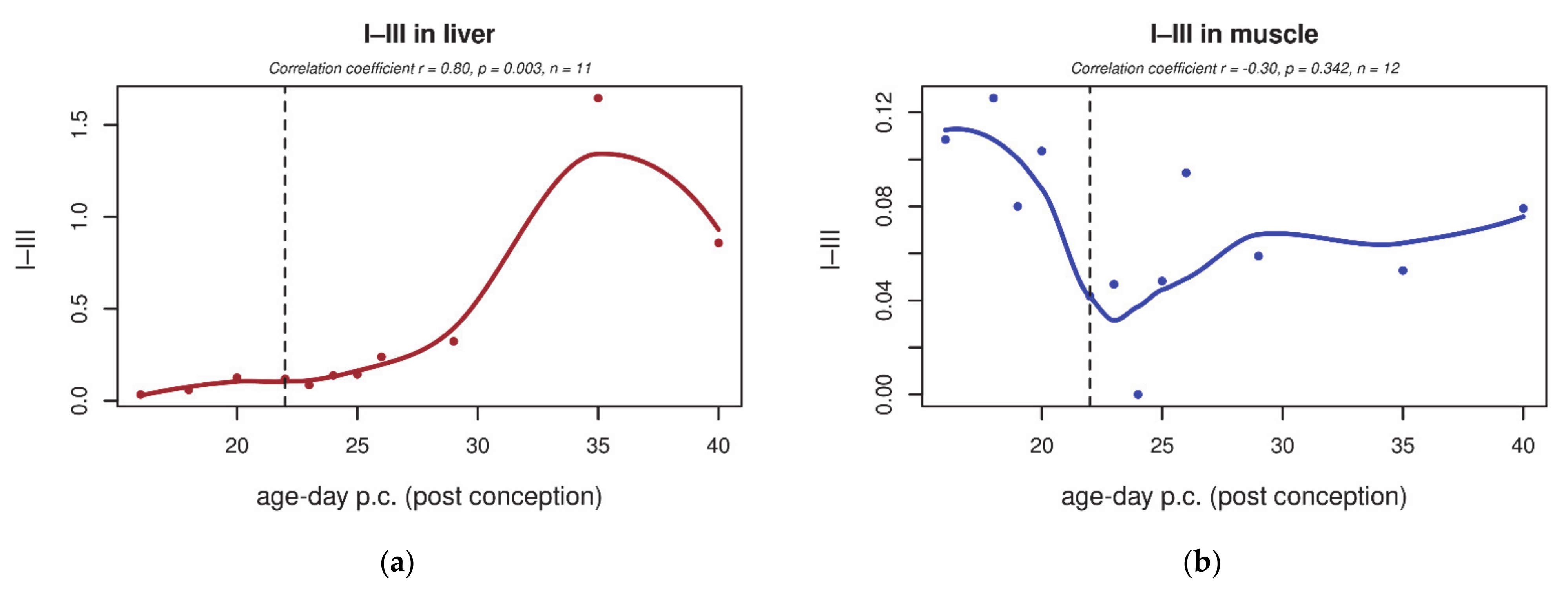
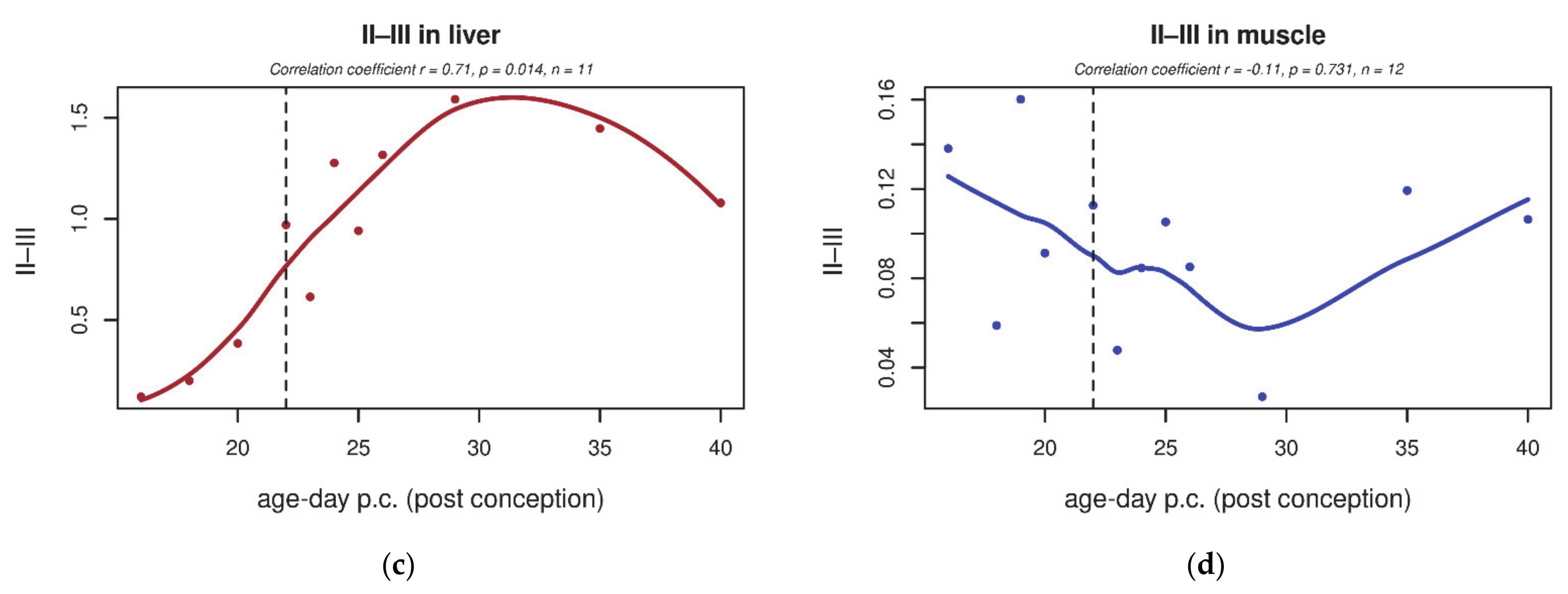
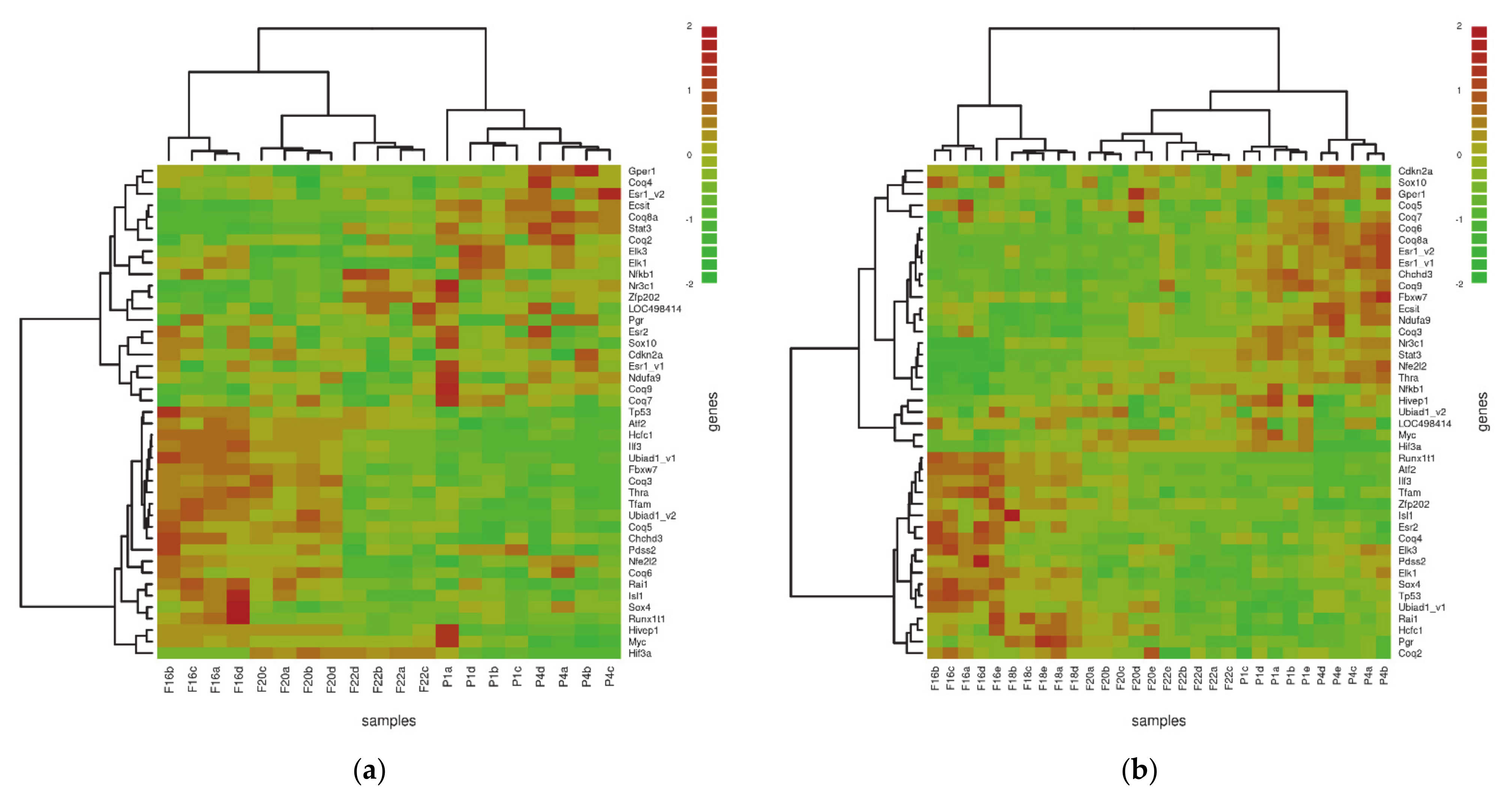
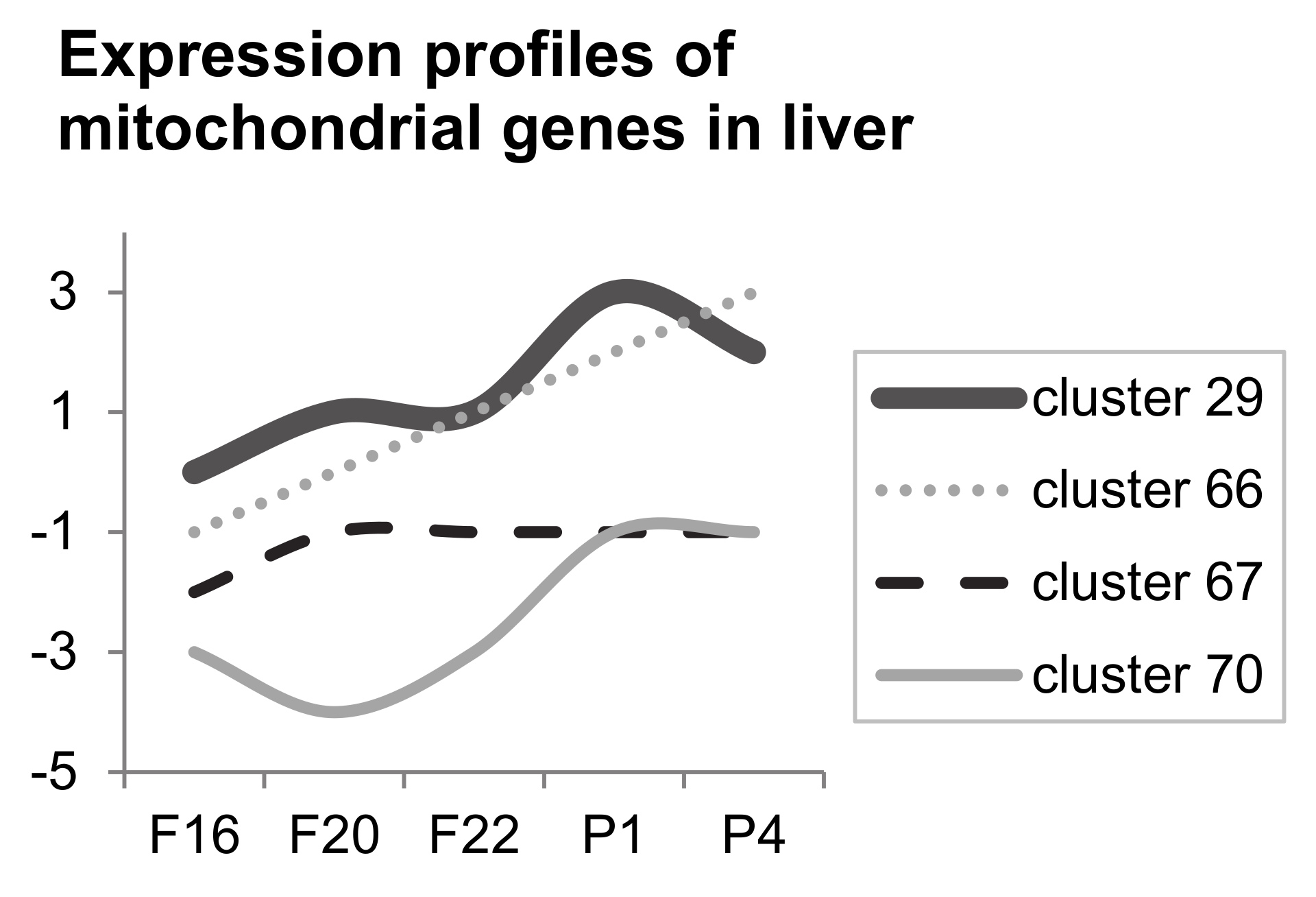
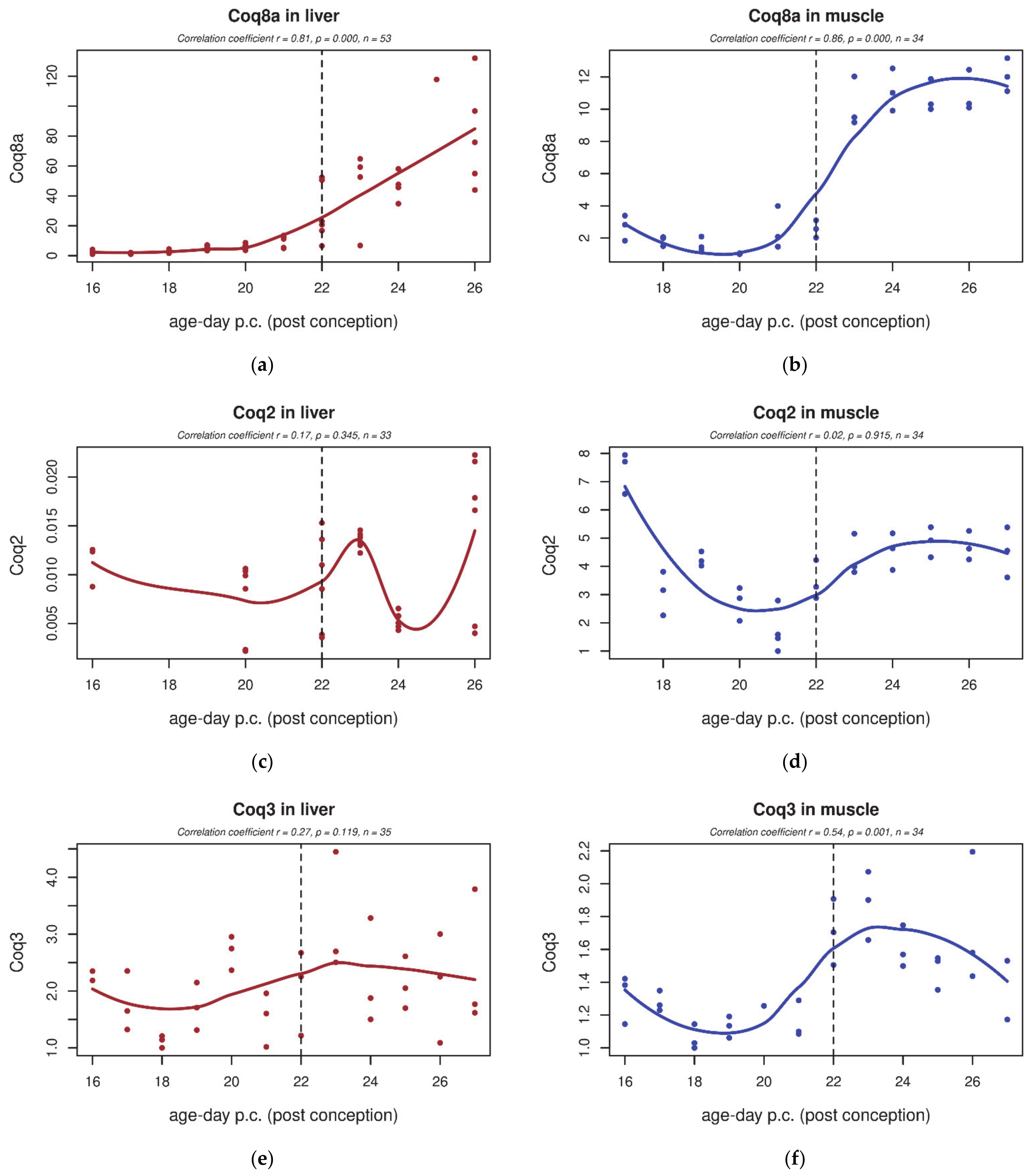
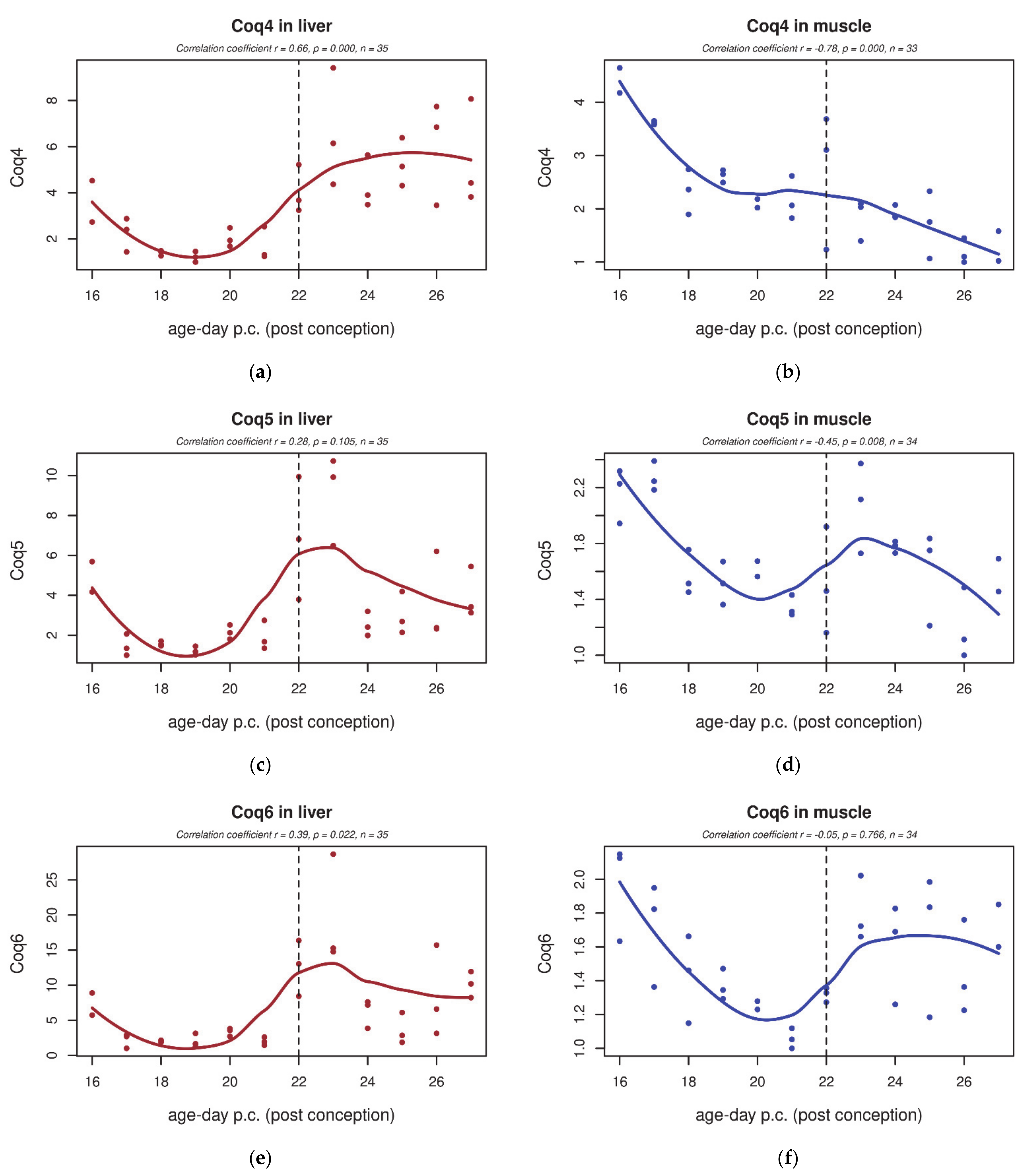
| GO Term | Trend L/M | Genes | Total Count |
|---|---|---|---|
| Mitochondrial genome maintenance | −0.003 (L) −0.017 (M) | Peo1, Polg, Mrpl17, Tp53, Lig3, Dnaja3, Flcn, Opa1, Parp1, Akt3, Ppargc1a, Primpol, Tk2, Tfam, Slc25a16, Dna2, Mgme1, Stoml2, Slc25a33, Rnaseh1, Rrm2b, Pif1, Pid1, Pif1, Mef2a | 25 |
| Amino-acid betaine metabolic process | 0.208 (L) 0.03 (M) | Cpt1a, Cpt1c, Por, Chdh, Aldh7a1, Dmgdh, Acadm, Tmlhe, Crat, Bbox1, Crot, Acadl | 12 |
| Endocytosis | 0.022 (L) 0.021 (M) | Mex3b, Unc119, Cd9, Sfrp4, Cd36, Mapkapk3, Tinagl1, Wnt5a, Gsn, Cav2, Cav2, Eef2k, Ubqln2, Pycard, Cav1, Cdc7, Gas6, Mrc1, Ap2b1, Cd163, Tgfbr2, Dnm2, Tub, Nlgn3, Enpp3 and others | 335 |
| Regulation of peptidyl–serine phosphorylation | 0.009 (L) −0.001 (M) | Dmd, Wnt5a, Cav1, Gas6, Pde4d, Nos1, Sfrp2, Akt2, Fnip1, Prkd1, Cd44, Gpd1l, Hrc, Rassf2, Gsk3b, Mif, Ntf3, Tgfb1, Txn1, Bcl2, Gsk3a, Camk1, Arrb2, Ogt, Gfra2 and others | 86 |
| GO Term | Genes | Enrichment | Count |
|---|---|---|---|
| Fatty acid β-oxidation using acyl-CoA dehydrogenase | Acox2, Gcdh, Acadsb, Acadm, Acads, Acadl, Acox3, Acadvl, Ivd, Etfdh, Acad10, Etfb, Etfa | 9.68 | 13 |
| Protein import into mitochondrial matrix | Grpel1, Tomm7, Pam16, Timm17a, Tomm40l, Tomm20, Tomm40, Dnlz, Tomm22, Timm50, Timm44, Timm21 | 6.54 | 12 |
| Mitochondrial fission | Mff, Fis1, Dnm1l, Opa1, Mief1, Mtfp1, Mul1, Park2, Ppp2r2b, Mtfr1l, Mtfr1 | 6.17 | 11 |
| Long-chain fatty acid metabolic process | Slc27a1, Acsl1, Cd36, Acot2, Slc27a3, Acsl4, Acsl3, Slc27a2, Cpt1a, Acsl5 | 3.9 | 10 |
| ATP metabolic process | Atp5d, Atp5e, Ndufaf7, Atp5b, Ak3, Ak2, Atp5g1, Ak4, Bad, Atp6v1a, Slc25a25, Atp5l, Atp5o, Atp5a1, Atp5i, Atp5h, Ndufs1 | 3.88 | 17 |
| NADH metabolic process | Gpd2, Gpd1, Dlst, Idh3g, Idh3b, Ogdh, Idh3a, Mdh2, Mdh1 | 3.53 | 9 |
| Release of cytochrome c from mitochondria | Mff, Bak1, Fis1, Dnm1l, Bcl2, Bax, Bcl2a1, Tp53, Mapk9, Timm50, Bad, Bcl2l1 | 3.17 | 12 |
| Tetrahydrofolate metabolic process | Mthfd1, Shmt1, Mthfd2, Tyms, Mthfs, Shmt2, Mthfd1l | 3.08 | 7 |
| Positive regulation of mitochondrial Ca2+ concentration | Micu1, Fis1, Mcur1, Micu2, Rap1gds1, Tgm2, Mcu, Bcap31 | 2.98 | 8 |
| Glutathione metabolic process | Gsta4, Aldh5a1, Ethe1, Clic1, Sod1, Hagh, Gsr, Gpx1, Clic4, Gstk1, Gpx4, Idh1, Txnrd1, Gstp1, Mgst1 | 2.86 | 15 |
| Human gene | Rat gene | Function |
|---|---|---|
| PDSS1 * | Pdss1 | polyisoprenoid chain synthesis |
| PDSS2 * | Pdss2 | polyisoprenoid chain synthesis |
| COQ2 * | Coq2† | p-HB prenylation |
| COQ3 | Coq3† | modification step—O-methylation |
| COQ4 * | Coq4† | scaffold protein |
| COQ5 * | Coq5† | modification step—C-methylation |
| COQ6 * | Coq6† | modification step—C5-hydroxylation |
| COQ7 * | Coq7† | modification step—hydroxylation |
| COQ8A * | Coq8a† | ATPase/kinase |
| COQ8B * | Coq8b | ATPase/kinase |
| COQ9 * | Coq9† | lipid binding/scaffold protein/C4-hydroxylation |
| COQ10A | Coq10a | lipid or CoQ-intermediate binding |
| COQ10B | Coq10b | lipid or CoQ-intermediate binding |
| NDUFA9 | Ndufa9† | subunit A9 in NADH:ubiquinone oxidoreductase (complex I) |
| UBIAD1 | Ubiad1† | cholesterol and phospholipid metabolism |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Krizova, J.; Hulkova, M.; Capek, V.; Mlejnek, P.; Silhavy, J.; Tesarova, M.; Zeman, J.; Hansikova, H. Microarray and qPCR Analysis of Mitochondrial Metabolism Activation during Prenatal and Early Postnatal Development in Rats and Humans with Emphasis on CoQ10 Biosynthesis. Biology 2021, 10, 418. https://doi.org/10.3390/biology10050418
Krizova J, Hulkova M, Capek V, Mlejnek P, Silhavy J, Tesarova M, Zeman J, Hansikova H. Microarray and qPCR Analysis of Mitochondrial Metabolism Activation during Prenatal and Early Postnatal Development in Rats and Humans with Emphasis on CoQ10 Biosynthesis. Biology. 2021; 10(5):418. https://doi.org/10.3390/biology10050418
Chicago/Turabian StyleKrizova, Jana, Martina Hulkova, Vaclav Capek, Petr Mlejnek, Jan Silhavy, Marketa Tesarova, Jiri Zeman, and Hana Hansikova. 2021. "Microarray and qPCR Analysis of Mitochondrial Metabolism Activation during Prenatal and Early Postnatal Development in Rats and Humans with Emphasis on CoQ10 Biosynthesis" Biology 10, no. 5: 418. https://doi.org/10.3390/biology10050418
APA StyleKrizova, J., Hulkova, M., Capek, V., Mlejnek, P., Silhavy, J., Tesarova, M., Zeman, J., & Hansikova, H. (2021). Microarray and qPCR Analysis of Mitochondrial Metabolism Activation during Prenatal and Early Postnatal Development in Rats and Humans with Emphasis on CoQ10 Biosynthesis. Biology, 10(5), 418. https://doi.org/10.3390/biology10050418






