Pathogens and Carcinogenesis: A Review
Abstract
Simple Summary
Abstract
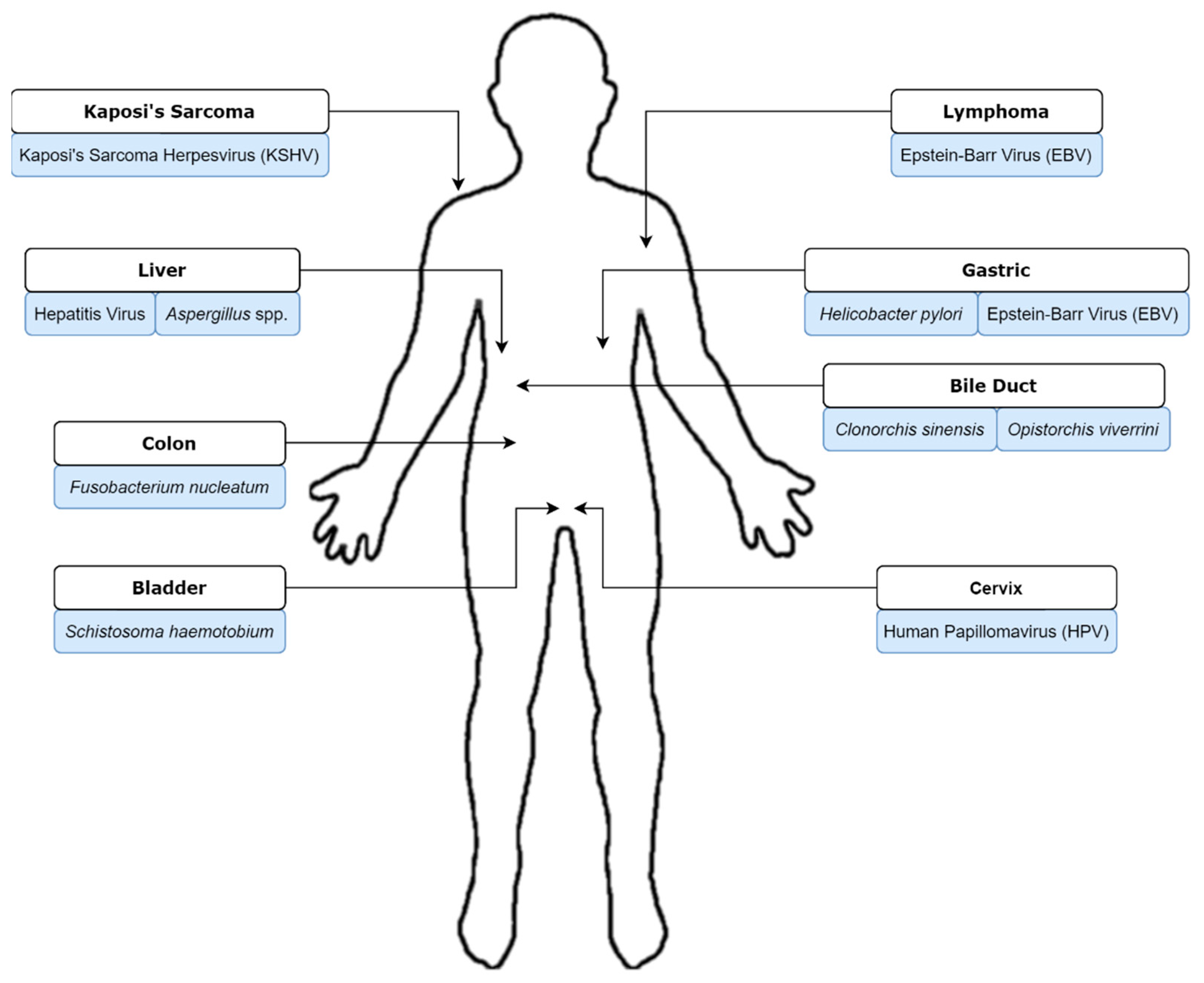
1. Introduction
2. Gastric Cancer
2.1. Helicobacter pylori
2.2. Epstein-Barr Virus (EBV)
3. Liver Cancer
3.1. Hepatitis Virus
3.2. Aspergillus spp.
4. Bile Duct Cancer
4.1. Opisthorchis viverrini
4.2. Clonorchis sinensis
5. Colorectal Cancer
Fusobacterium nucleatum
6. Bladder Cancer
Schistosoma haematobium
7. Cervical Cancer
Human Papillomavirus (HPV)
8. Kaposi’s Sarcoma
Kaposi’s Sarcoma Herpesvirus (KSHV)/Human Herpesvirus 8 (HHV8)
9. Lymphoma
Epstein-Barr Virus (EBV)
10. Implication of Infectious Agents in Carcinogenesis and Future Considerations
11. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Rahman, T. The Difficulties in Cancer Treatment. Ecancermedicalscience 2012, 6, ed16. [Google Scholar] [CrossRef]
- Zeller, G.; Tap, J.; Voigt, A.Y.; Sunagawa, S.; Kultima, J.R.; Costea, P.I.; Amiot, A.; Böhm, J.; Brunetti, F.; Habermann, N.; et al. Potential of fecal microbiota for early-stage detection of colorectal cancer. Mol. Syst. Biol. 2014, 10, 766. [Google Scholar] [CrossRef]
- Khan, N.; Afaq, F.; Mukhtar, H. Lifestyle as risk factor for cancer: Evidence from human studies. Cancer Lett. 2010, 293, 133–143. [Google Scholar] [CrossRef]
- Goldszmid, R.S.; Dzutsev, A.; Trinchieri, G. Host Immune Response to Infection and Cancer: Unexpected Commonalities. Cell Host Microbe 2014, 15, 295–305. [Google Scholar] [CrossRef] [PubMed]
- Blaser, M.J. Understanding Microbe-Induced Cancers. Cancer Prev. Res. 2008, 1, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Tjalsma, H.; Boleij, A.; Marchesi, J.; Dutilh, B.E. A bacterial driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Genet. 2012, 10, 575–582. [Google Scholar] [CrossRef]
- Zhou, Z.; Chen, J.; Yao, H.; Hu, H. Fusobacterium and Colorectal Cancer. Front. Oncol. 2018, 8, 371. [Google Scholar] [CrossRef] [PubMed]
- Chang, A.H.; Parsonnet, J. Role of Bacteria in Oncogenesis. Clin. Microbiol. Rev. 2010, 23, 837–857. [Google Scholar] [CrossRef]
- Howley, P.M. Gordon Wilson Lecture: Infectious Disease Causes of Cancer: Opportunities for Prevention and Treatment. Trans. Am. Clin. Clim. Assoc. 2015, 126, 117–132. [Google Scholar]
- Hausen, H.Z.; Fox, J.G.; Wang, T.C.; Parsonnet, J. Infections Causing Human Cancer; WILEY-VCH Verlag GmbH & Co. KGaA: Weinheim, Germany, 2006; pp. 468–484. ISBN 9783527609314. [Google Scholar] [CrossRef]
- Kumar, P.; Murphy, F.A. Francis Peyton Rous. Emerg. Infect. Dis. 2013, 19, 660–663. [Google Scholar] [CrossRef] [PubMed]
- Pannone, G.; Zamparese, R.; Pace, M.; Pedicillo, M.C.; Cagiano, S.; Somma, P.; Errico, M.E.; Donofrio, V.; Franco, R.; De Chiara, A.; et al. The role of EBV in the pathogenesis of Burkitt’s Lymphoma: An Italian hospital based survey. Infect. Agents Cancer 2014, 9, 1–11. [Google Scholar] [CrossRef]
- Epstein, M.; Achong, B.; Barr, Y. Virus Particles in Cultured Lymphoblasts from Burkitt’s Lymphoma. Lancet 1964, 283, 702–703. [Google Scholar] [CrossRef]
- Dalton-Griffin, L.; Kellam, P. Infectious causes of cancer and their detection. J. Biol. 2009, 8, 67. [Google Scholar] [CrossRef]
- Buckland, G.; Travier, N.; Huerta, J.; Bueno-De-Mesquita, H.; Siersema, P.; Skeie, G.; Weiderpass, E.; Engeset, D.; Ericson, U.; Ohlsson, B.; et al. Healthy lifestyle index and risk of gastric adenocarcinoma in the EPIC cohort study. Int. J. Cancer 2014, 137, 598–606. [Google Scholar] [CrossRef]
- Ladeiras-Lopes, R.; Pereira, A.K.; Nogueira, A.; Pinheiro-Torres, T.; Pinto, I.; Santos-Pereira, R.; Lunet, N. Smoking and gastric cancer: Systematic review and meta-analysis of cohort studies. Cancer Causes Control. 2008, 19, 689–701. [Google Scholar] [CrossRef]
- Nagel, G.; Linseisen, J.; Boshuizen, H.C.; Pera, G.; Giudice, G.D.; Westert, G.P.; Bueno-De-Mesquita, H.B.; Allen, N.E.; Key, T.J.; Numans, M.E.; et al. Socioeconomic position and the risk of gastric and oesophageal cancer in the European Prospective Investigation into Cancer and Nutrition (EPIC-EURGAST). Int. J. Epidemiol. 2007, 36, 66–76. [Google Scholar] [CrossRef]
- Sitarz, R.; Skierucha, M.; Mielko, J.; Offerhaus, J.; Maciejewski, R.; Polkowski, W. Gastric cancer: Epidemiology, prevention, classification, and treatment. Cancer Manag. Res. 2018, 10, 239–248. [Google Scholar] [CrossRef] [PubMed]
- Stewart, O.A.; Wu, F.; Chen, Y. The role of gastric microbiota in gastric cancer. Gut Microbes 2020, 11, 1220–1230. [Google Scholar] [CrossRef] [PubMed]
- Murphy, G.; Pfeiffer, R.; Camargo, M.C.; Rabkin, C.S. Meta-analysis Shows That Prevalence of Epstein–Barr Virus-Positive Gastric Cancer Differs Based on Sex and Anatomic Location. Gastroenterology 2009, 137, 824–833. [Google Scholar] [CrossRef] [PubMed]
- Amieva, M.; Peek, R.M. Pathobiology of Helicobacter pylori–Induced Gastric Cancer. Gastroenterology 2016, 150, 64–78. [Google Scholar] [CrossRef]
- Kusters, J.G.; van Vliet, A.H.M.; Kuipers, E.J. Pathogenesis of Helicobacter pylori Infection. Clin. Microbiol. Rev. 2006, 19, 449–490. [Google Scholar] [CrossRef] [PubMed]
- Amieva, M.R.; El–Omar, E.M. Host-Bacterial Interactions in Helicobacter pylori Infection. Gastroenterology 2008, 134, 306–323. [Google Scholar] [CrossRef] [PubMed]
- Graham, D.Y.; Miftahussurur, M. Helicobacter pylori urease for diagnosis of Helicobacter pylori infection: A mini review. J. Adv. Res. 2018, 13, 51–57. [Google Scholar] [CrossRef]
- Dumrese, C.; Slomianka, L.; Ziegler, U.; Choi, S.S.; Kalia, A.; Fulurija, A.; Lu, W.; Berg, D.E.; Benghezal, M.; Marshall, B.; et al. The secreted Helicobacter cysteine-rich protein A causes adherence of human monocytes and differentiation into a macrophage-like phenotype. FEBS Lett. 2009, 583, 1637–1643. [Google Scholar] [CrossRef] [PubMed]
- Shiotani, A.; Graham, D.Y. Pathogenesis and therapy of gastric and duodenal ulcer disease. Med. Clin. N. Am. 2002, 86, 1447–1466. [Google Scholar] [CrossRef]
- Backert, S.; Selbach, M. Role of type IV secretion in Helicobacter pyloripathogenesis. Cell. Microbiol. 2008, 10, 1573–1581. [Google Scholar] [CrossRef] [PubMed]
- Kao, C.-Y.; Sheu, B.-S.; Wu, J.-J. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed. J. 2016, 39, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Backert, S.; Neddermann, M.; Maubach, G.; Naumann, M. Pathogenesis of Helicobacter pylori infection. Helicobacter 2016, 21, 19–25. [Google Scholar] [CrossRef]
- Viala, J.; Chaput, C.; Boneca, I.G.; Cardona, A.; Girardin, S.E.; Moran, A.P.; Athman, R.; Mémet, S.; Huerre, M.R.; Coyle, A.J.; et al. Nod1 responds to peptidoglycan delivered by the Helicobacter pylori cag pathogenicity island. Nat. Immunol. 2004, 5, 1166–1174. [Google Scholar] [CrossRef]
- Cover, T.L.; Blanke, S.R. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nat. Rev. Genet. 2005, 3, 320–332. [Google Scholar] [CrossRef]
- Zali, H.; Rezaei-Tavirani, M.; Azodi, M.Z. Gastric cancer: Prevention, risk factors and treatment. Gastroenterol. Hepatol. Bed Bench 2011, 4, 175–185. [Google Scholar]
- Marshall, B.J.; Warren, J.R. Unidentified Curved Bacilli in the Stomach Of Patients with Gastritis and Peptic Ulceration. Lancet 1984, 323, 1311–1315. [Google Scholar] [CrossRef]
- Ahn, H.J.; Lee, D.S. Helicobacter pylori in gastric carcinogenesis. World J. Gastrointest. Oncol. 2015, 7, 455–465. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Slack, F.J.; Anastasiadou, E. Epstein-Barr virus: From kisses to cancer, an ingenious immune evader. Oncotarget 2018, 9, 36411–36412. [Google Scholar] [CrossRef] [PubMed]
- Burke, A.P.; Yen, T.S.; Shekitka, K.M.; Sobin, L.H. Lymphoepithelial carcinoma of the stomach with Epstein-Barr virus demonstrated by polymerase chain reaction. Mod. Pathol. 1990, 3, 377–380. [Google Scholar]
- Camargo, M.C.; Murphy, G.; Koriyama, C.; Pfeiffer, R.M.; Kim, W.H.; Herrera-Goepfert, R.; Corvalan, A.H.; Carrascal, E.; Abdirad, A.; Anwar, M.; et al. Determinants of Epstein-Barr virus-positive gastric cancer: An international pooled analysis. Br. J. Cancer 2011, 105, 38–43. [Google Scholar] [CrossRef]
- Iizasa, H.; Nanbo, A.; Nishikawa, J.; Jinushi, M.; Yoshiyama, H. Epstein-Barr Virus (EBV)-associated Gastric Carcinoma. Viruses 2012, 4, 3420–3439. [Google Scholar] [CrossRef]
- Akiba, S.; Koriyama, C.; Herrera-Goepfert, R.; Eizuru, Y. Epstein-Barr virus associated gastric carcinoma: Epidemiological and clinicopathological features. Cancer Sci. 2008, 99, 195–201. [Google Scholar] [CrossRef]
- Shinozaki-Ushiku, A.; Kunita, A.; Aya, S.-U. Update on Epstein-Barr virus and gastric cancer (Review). Int. J. Oncol. 2015, 46, 1421–1434. [Google Scholar] [CrossRef]
- Fukayama, M.; Abe, H.; Kunita, A.; Shinozaki-Ushiku, A.; Matsusaka, K.; Ushiku, T.; Kaneda, A. Thirty years of Epstein-Barr virus-associated gastric carcinoma. Virchows Archiv 2020, 476, 353–365. [Google Scholar] [CrossRef]
- Qu, Y.; Dang, S.; Hou, P. Gene methylation in gastric cancer. Clin. Chim. Acta 2013, 424, 53–65. [Google Scholar] [CrossRef]
- Imai, S.; Nishikawa, J.; Takada, K. Cell-to-Cell Contact as an Efficient Mode of Epstein-Barr Virus Infection of Diverse Human Epithelial Cells. J. Virol. 1998, 72, 4371–4378. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, J.; Cheng, A.S.L.; Yu, J.; To, K.F.; Kang, W. Gastric cancer: Genome damaged by bugs. Oncogene 2020, 39, 3427–3442. [Google Scholar] [CrossRef] [PubMed]
- Iwakiri, D.; Eizuru, Y.; Tokunaga, M.; Takada, K. Autocrine growth of Epstein-Barr virus-positive gastric carcinoma cells mediated by an Epstein-Barr virus-encoded small RNA. Cancer Res. 2003, 63, 7062–7067. [Google Scholar] [PubMed]
- Banerjee, A.S.; Pal, A.D.; Banerjee, S. Epstein–Barr virus-encoded small non-coding RNAs induce cancer cell chemoresistance and migration. Virology 2013, 443, 294–305. [Google Scholar] [CrossRef] [PubMed]
- Sivachandran, N.; Dawson, C.W.; Young, L.S.; Liu, F.-F.; Middeldorp, J.; Frappier, L. Contributions of the Epstein-Barr Virus EBNA1 Protein to Gastric Carcinoma. J. Virol. 2011, 86, 60–68. [Google Scholar] [CrossRef]
- Kim, S.-M.; Hur, D.Y.; Hong, S.-W.; Kim, J.H. EBV-encoded EBNA1 regulates cell viability by modulating miR34a-NOX2-ROS signaling in gastric cancer cells. Biochem. Biophys. Res. Commun. 2017, 494, 550–555. [Google Scholar] [CrossRef]
- Hino, R.; Uozaki, H.; Murakami, N.; Ushiku, T.; Shinozaki, A.; Ishikawa, S.; Morikawa, T.; Nakaya, T.; Sakatani, T.; Takada, K.; et al. Activation of DNA Methyltransferase 1 by EBV Latent Membrane Protein 2A Leads to Promoter Hypermethylation of PTEN Gene in Gastric Carcinoma. Cancer Res. 2009, 69, 2766–2774. [Google Scholar] [CrossRef]
- Tsai, C.-Y.; Liu, Y.Y.; Liu, K.-H.; Hsu, J.-T.; Cheng-Tang, C.; Chiu, C.-T.; Yeh, T.-S. Comprehensive profiling of virus microRNAs of Epstein-Barr virus-associated gastric carcinoma: Highlighting the interactions of ebv-Bart9 and host tumor cells. J. Gastroenterol. Hepatol. 2017, 32, 82–91. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, J.; Wei, L.; Peng, Q.; Gao, Y.; Fu, Y.; Lu, Y.; Qin, Z.; Zhang, X.; Lu, J.; et al. Epstein-Barr Virus MicroRNA miR-BART5-3p Inhibits p53 Expression. J. Virol. 2018, 92, 92. [Google Scholar] [CrossRef]
- Hooykaas, M.J.G.; van Gent, M.; Soppe, J.A.; Kruse, E.; Boer, I.G.J.; Van Leenen, D.; Koerkamp, M.J.A.G.; Holstege, F.C.P.; Ressing, M.E.; Wiertz, E.J.H.J.; et al. EBV MicroRNA BART16 Suppresses Type I IFN Signaling. J. Immunol. 2017, 198, 4062–4073. [Google Scholar] [CrossRef] [PubMed]
- Byam, J.; Renz, J.; Millis, J.M. Liver transplantation for hepatocellular carcinoma. HepatoBiliary Surg. Nutr. 2013, 2, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Bray, F.; Ferlay, J.; Soerjomataram, I.; Siegel, R.L.; Torre, L.A.; Jemal, A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018, 68, 394–424. [Google Scholar] [CrossRef]
- Chuang, S.-C.; Vecchia, C.L.; Boffetta, P. Liver cancer: Descriptive epidemiology and risk factors other than HBV and HCV infection. Cancer Lett. 2009, 286, 9–14. [Google Scholar] [CrossRef]
- GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet 2018, 392, 1736–1788. [Google Scholar] [CrossRef]
- Fattovich, G.; Stroffolini, T.; Zagni, I.; Donato, F. Hepatocellular Carcinoma in Albania: Incidence and Risk Factors. Int. J. Sci. Res. 2016, 5, 1178–1183. [Google Scholar] [CrossRef]
- Tu, T.; Bühler, S.; Bartenschlager, R. Chronic viral hepatitis and its association with liver cancer. Biol. Chem. 2017, 398, 817–837. [Google Scholar] [CrossRef]
- Handsfield, H.H. Hepatitis A and B immunization in persons being evaluated for sexually transmitted diseases. Am. J. Med. 2005, 118, 69–74. [Google Scholar] [CrossRef]
- El-Serag, H.B. Epidemiology of Viral Hepatitis and Hepatocellular Carcinoma. Gastroenterology 2012, 142, 1264–1273. [Google Scholar] [CrossRef]
- El–Serag, H.B.; Rudolph, K.L. Hepatocellular Carcinoma: Epidemiology and Molecular Carcinogenesis. Gastroenterology 2007, 132, 2557–2576. [Google Scholar] [CrossRef]
- Lamontagne, R.J.; Bagga, S.; Bouchard, M.J. Hepatitis B virus molecular biology and pathogenesis. Hepatoma Res. 2016, 2, 163–186. [Google Scholar] [CrossRef]
- Ringelhan, M.; McKeating, J.A.; Protzer, U. Correction to ‘Viral hepatitis and liver cancer’. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20170339. [Google Scholar] [CrossRef]
- Watashi, K.; Urban, S.; Li, W.; Wakita, T. NTCP and Beyond: Opening the Door to Unveil Hepatitis B Virus Entry. Int. J. Mol. Sci. 2014, 15, 2892–2905. [Google Scholar] [CrossRef]
- Kgatle, M.M.; Spearman, C.W.; Kalla, A.A.; Hairwadzi, H.N. DNA Oncogenic Virus-Induced Oxidative Stress, Genomic Damage, and Aberrant Epigenetic Alterations. Oxidative Med. Cell. Longev. 2017, 2017, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Rivière, L.; Gerossier, L.; Ducroux, A.; Dion, S.; Deng, Q.; Michel, M.-L.; Buendia, M.-A.; Hantz, O.; Neuveut, C. HBx relieves chromatin-mediated transcriptional repression of hepatitis B viral cccDNA involving SETDB1 histone methyltransferase. J. Hepatol. 2015, 63, 1093–1102. [Google Scholar] [CrossRef]
- Fattovich, G.; Bortolotti, F.; Donato, F. Natural history of chronic hepatitis B: Special emphasis on disease progression and prognostic factors. J. Hepatol. 2008, 48, 335–352. [Google Scholar] [CrossRef] [PubMed]
- Verme, G.; Amoroso, P.; Lettieri, G.; Pierri, P.; David, E.; Sessa, F.; Rizzi, R.; Bonino, F.; Recchia, S.; Rizzetto, M. A histological study of hepatitis delta virus liver disease. Hepatology 1986, 6, 1303–1307. [Google Scholar] [CrossRef]
- European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of chronic hepatitis B virus infection. J. Hepatol. 2012, 57, 167–185. [Google Scholar] [CrossRef] [PubMed]
- Mason, W.S.; Gill, U.S.; Litwin, S.; Zhou, Y.; Peri, S.; Pop, O.; Hong, M.; Naik, S.; Quaglia, A.; Bertoletti, A.; et al. HBV DNA Integration and Clonal Hepatocyte Expansion in Chronic Hepatitis B Patients Considered Immune Tolerant. Gastroenterology 2016, 151, 986–998.e4. [Google Scholar] [CrossRef] [PubMed]
- Lin, M.V.; King, L.Y.; Chung, R.T. Hepatitis C Virus–Associated Cancer. Annu. Rev. Pathol. Mech. Dis. 2015, 10, 345–370. [Google Scholar] [CrossRef] [PubMed]
- Shi, G.; Suzuki, T. Molecular Basis of Encapsidation of Hepatitis C Virus Genome. Front. Microbiol. 2018, 9, 396. [Google Scholar] [CrossRef] [PubMed]
- Ashfaq, U.A.; Javed, T.; Rehman, S.; Nawaz, Z.; Riazuddin, S. An overview of HCV molecular biology, replication and immune responses. Virol. J. 2011, 8, 161. [Google Scholar] [CrossRef]
- Bartosch, B.; Thimme, R.; Blum, H.E.; Zoulim, F. Hepatitis C virus-induced hepatocarcinogenesis. J. Hepatol. 2009, 51, 810–820. [Google Scholar] [CrossRef] [PubMed]
- Pekow, J.R.; Bhan, A.K.; Zheng, H.; Chung, R.T. Hepatic steatosis is associated with increased frequency of hepatocellular carcinoma in patients with hepatitis C-related cirrhosis. Cancer 2007, 109, 2490–2496. [Google Scholar] [CrossRef]
- De Mochel, N.S.R.; Seronello, S.; Wang, S.H.; Ito, C.; Zheng, J.X.; Liang, T.J.; Lambeth, J.D.; Choi, J. Hepatocyte NAD(P)H oxidases as an endogenous source of reactive oxygen species during hepatitis C virus infection. Hepatology 2010, 52, 47–59. [Google Scholar] [CrossRef]
- Irshad, M.; Gupta, P.; Irshad, K. Molecular basis of hepatocellular carcinoma induced by hepatitis C virus infection. World J. Hepatol. 2017, 9, 1305–1314. [Google Scholar] [CrossRef]
- McGivern, D.R.; Lemon, S.M. Virus-specific mechanisms of carcinogenesis in hepatitis C virus associated liver cancer. Oncogene 2011, 30, 1969–1983. [Google Scholar] [CrossRef]
- Okuda, M.; Li, K.; Beard, M.R.; Showalter, L.A.; Scholle, F.; Lemon, S.M.; Weinman, S.A. Mitochondrial injury, oxidative stress, and antioxidant gene expression are induced by hepatitis C virus core protein. Gastroenterology 2002, 122, 366–375. [Google Scholar] [CrossRef] [PubMed]
- Hoshida, Y.; Fuchs, B.C.; Bardeesy, N.; Baumert, T.F.; Chung, R.T. Pathogenesis and prevention of hepatitis C virus-induced hepatocellular carcinoma. J. Hepatol. 2014, 61, S79–S90. [Google Scholar] [CrossRef] [PubMed]
- Kew, M.C. Aflatoxins as a cause of hepatocellular carcinoma. J. Gastrointest. Liver Dis. 2013, 22, 305–310. [Google Scholar]
- Jalili, M. A Review on Aflatoxins Reduction in Food. Iran. J. Health Saf. Environ. 2016, 3, 445–459. [Google Scholar]
- Waliyar, F.; Osiru, M.; Ntare, B.; Kumar, K.V.K.; Sudini, H.; Traore, A.; Diarra, B. Post-harvest management of aflatoxin contamination in groundnut. World Mycotoxin J. 2015, 8, 245–252. [Google Scholar] [CrossRef]
- Kensler, T.W.; Roebuck, B.D.; Wogan, G.N.; Groopman, J.D. Aflatoxin: A 50-Year Odyssey of Mechanistic and Translational Toxicology. Toxicol. Sci. 2010, 120, S28–S48. [Google Scholar] [CrossRef] [PubMed]
- Strosnider, H.; Azziz-Baumgartner, E.; Banziger, M.; Bhat, R.V.; Breiman, R.; Brune, M.-N.; Decock, K.; Dilley, A.; Groopman, J.; Hell, K.; et al. Workgroup Report: Public Health Strategies for Reducing Aflatoxin Exposure in Developing Countries. Environ. Health Perspect. 2006, 114, 1898–1903. [Google Scholar] [CrossRef]
- Phillips, T.D.; Afriyie-Gyawu, E.; Williams, J.; Huebner, H.; Ankrah, N.-A.; Ofori-Adjei, D.; Jolly, P.; Johnson, N.; Taylor, J.; Marroquin-Cardona, A.; et al. Reducing human exposure to aflatoxin through the use of clay: A review. Food Addit. Contam. Part. A 2008, 25, 134–145. [Google Scholar] [CrossRef]
- Yang, J.D.; Hainaut, P.; Gores, G.J.; Amadou, A.; Plymoth, A.; Roberts, L.R. A global view of hepatocellular carcinoma: Trends, risk, prevention and management. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 589–604. [Google Scholar] [CrossRef]
- McCullough, A.K.; Lloyd, R.S. Mechanisms underlying aflatoxin-associated mutagenesis—Implications in carcinogenesis. DNA Repair 2019, 77, 76–86. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Li, L.; Makarova, A.V.; Burgers, P.M.; Stone, M.; Lloyd, R.S. Error-prone Replication Bypass of the Primary Aflatoxin B1 DNA Adduct, AFB1-N7-Gua. J. Biol. Chem. 2014, 289, 18497–18506. [Google Scholar] [CrossRef] [PubMed]
- Lin, Y.-C.; Li, L.; Makarova, A.V.; Burgers, P.M.; Stone, M.; Lloyd, R.S. Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct. Carcinogenesis 2014, 35, 1461–1468. [Google Scholar] [CrossRef]
- Hamid, A.S.; Tesfamariam, I.G.; Zhang, Y.; Zhang, Z.G. Aflatoxin B1-induced hepatocellular carcinoma in developing countries: Geographical distribution, mechanism of action and prevention. Oncol. Lett. 2013, 5, 1087–1092. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.-H.; Sun, L.-H.; Lu, D.-D.; Sun, Y.; Ma, L.-J.; Zhang, X.-R.; Huang, J.; Yu, L. Codon 249 mutation in exon 7 ofp53gene in plasma DNA: Maybe a new early diagnostic marker of hepatocellular carcinoma in Qidong risk area, China. World J. Gastroenterol. 2003, 9, 692–695. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Tavolari, S.; Brandi, G. Cholangiocarcinoma: Epidemiology and risk factors. Liver Int. 2019, 39, 19–31. [Google Scholar] [CrossRef]
- Prueksapanich, P.; Piyachaturawat, P.; Aumpansub, P.; Ridtitid, W.; Chaiteerakij, R.; Rerknimitr, R. Liver Fluke-Associated Biliary Tract Cancer. Gut Liver 2018, 12, 236–245. [Google Scholar] [CrossRef]
- Suk, W.A.; Bhudhisawasdi, V.; Ruchirawat, M. The Curious Case of Cholangiocarcinoma: Opportunities for Environmental Health Scientists to Learn about a Complex Disease. J. Environ. Public Health 2018, 2018, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Saijuntha, W.; Sithithaworn, P.; Kaitsopit, N.; Andrews, R.H.; Petney, T.N. Liver Flukes: Clonorchis and Opisthorchis. In Advances in Experimental Medicine and Biology; Springer Science and Business Media LLC: New York, NY, USA, 2014; Volume 766, pp. 153–199. [Google Scholar]
- Jex, A.R.; Young, N.; Sripa, J.; Hall, R.S.; Scheerlinck, J.-P.; Laha, T.; Sripa, B.; Gasser, R.B. Molecular Changes in Opisthorchis viverrini (Southeast Asian Liver Fluke) during the Transition from the Juvenile to the Adult Stage. PLoS Negl. Trop. Dis. 2012, 6, e1916. [Google Scholar] [CrossRef]
- Kaewpitoon, N. Opisthorchis viverrini: The carcinogenic human liver fluke. World J. Gastroenterol. 2008, 14, 666–674. [Google Scholar] [CrossRef]
- Young, N.D.; Campbell, B.E.; Hall, R.S.; Jex, A.R.; Cantacessi, C.; Laha, T.; Sohn, W.-M.; Sripa, B.; Loukas, A.; Brindley, P.J.; et al. Unlocking the Transcriptomes of Two Carcinogenic Parasites, Clonorchis sinensis and Opisthorchis viverrini. PLoS Negl. Trop. Dis. 2010, 4, e719. [Google Scholar] [CrossRef]
- Sripa, B.; Brindley, P.J.; Mulvenna, J.; Laha, T.; Smout, M.; Mairiang, E.; Bethony, J.M.; Loukas, A. The tumorigenic liver fluke Opisthorchis viverrini—Multiple pathways to cancer. Trends Parasitol. 2012, 28, 395–407. [Google Scholar] [CrossRef]
- Smout, M.; Laha, T.; Mulvenna, J.; Sripa, B.; Suttiprapa, S.; Jones, A.; Brindley, P.J.; Loukas, A. A Granulin-Like Growth Factor Secreted by the Carcinogenic Liver Fluke, Opisthorchis viverrini, Promotes Proliferation of Host Cells. PLoS Pathog. 2009, 5, e1000611. [Google Scholar] [CrossRef]
- van Tong, H.; Brindley, P.J.; Meyer, C.G.; Velavan, T.P. Parasite Infection, Carcinogenesis and Human Malignancy. EBioMedicine 2017, 15, 12–23. [Google Scholar] [CrossRef]
- Chan-On, W.; Nairismägi, M.-L.; Ong, C.K.; Lim, W.K.; Dima, S.; Pairojkul, C.; Lim, K.H.; McPherson, J.R.; Cutcutache, I.; Heng, H.L.; et al. Exome sequencing identifies distinct mutational patterns in liver fluke–related and non-infection-related bile duct cancers. Nat. Genet. 2013, 45, 1474–1478. [Google Scholar] [CrossRef]
- Jusakul, A.; Kongpetch, S.; Teh, B.T. Genetics of Opisthorchis viverrini-related cholangiocarcinoma. Curr. Opin. Gastroenterol. 2015, 31, 258–263. [Google Scholar] [CrossRef]
- Flavell, D.J.; Lucas, S.B. Potentiation by the human liver fluke, Opisthorchis viverrini, of the carcinogenic action of N-nitrosodimethylamine upon the biliary epithelium of the hamster. Br. J. Cancer 1982, 46, 985–989. [Google Scholar] [CrossRef][Green Version]
- Deenonpoe, R.; Chomvarin, C.; Pairojkul, C.; Chamgramol, Y.; Loukas, A.; Brindley, P.J.; Sripa, B. The Carcinogenic Liver Fluke Opisthorchis viverrini is a Reservoir for Species of Helicobacter. Asian Pac. J. Cancer Prev. 2015, 16, 1751–1758. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.-Y.; Murrell, K.D.; Lymbery, A.J. Fish-borne parasitic zoonoses: Status and issues. Int. J. Parasitol. 2005, 35, 1233–1254. [Google Scholar] [CrossRef]
- Kim, T.-S.; Pak, J.H.; Kim, J.-B.; Bahk, Y.Y. Clonorchis sinensis, an oriental liver fluke, as a human biological agent of cholangiocarcinoma: A brief review. BMB Rep. 2016, 49, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Yoo, K.-S.; Lim, W.T.; Choi, H.S. Biology of Cholangiocytes: From Bench to Bedside. Gut Liver 2016, 10, 687–698. [Google Scholar] [CrossRef] [PubMed]
- Nam, J.-H.; Moon, J.H.; Kim, I.K.; Lee, M.-R.; Hong, S.-J.; Ahn, J.H.; Chung, J.W.; Pak, J.H. Free radicals enzymatically triggered by Clonorchis sinensis excretory–secretory products cause NF-κB-mediated inflammation in human cholangiocarcinoma cells. Int. J. Parasitol. 2012, 42, 103–113. [Google Scholar] [CrossRef]
- Won, J.; Ju, J.-W.; Kim, S.M.; Shin, Y.; Chung, S.; Pak, J.H. Clonorchis sinensis Infestation Promotes Three-Dimensional Aggregation and Invasion of Cholangiocarcinoma Cells. PLoS ONE 2014, 9, e110705. [Google Scholar] [CrossRef]
- Han, C.; Leng, J.; Demetris, A.J.; Wu, T. Cyclooxygenase-2 Promotes Human Cholangiocarcinoma Growth. Cancer Res. 2004, 64, 1369–1376. [Google Scholar] [CrossRef]
- Jaiswal, M.; LaRusso, N.F.; Shapiro, R.A.; Billiar, T.R.; Gores, G.J. Nitric oxide–mediated inhibition of DNA repair potentiates oxidative DNA damage in cholangiocytes. Gastroenterology 2001, 120, 190–199. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.-W.; Kim, J.-Y.; Moon, J.H.; Kim, K.-B.; Kim, T.-S.; Hong, S.-J.; Cheon, Y.P.; Pak, J.H.; Seo, S.-B. Transcriptional induction of minichromosome maintenance protein 7 (Mcm7) in human cholangiocarcinoma cells treated with Clonorchis sinensis excretory–secretory products. Mol. Biochem. Parasitol. 2010, 173, 10–16. [Google Scholar] [CrossRef]
- Shang, F.-M.; Liu, H.-L. Fusobacterium nucleatum and colorectal cancer: A review. World J. Gastrointest. Oncol. 2018, 10, 71–81. [Google Scholar] [CrossRef]
- Kostic, A.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.; et al. Fusobacterium nucleatum Potentiates Intestinal Tumorigenesis and Modulates the Tumor-Immune Microenvironment. Cell Host Microbe 2013, 14, 207–215. [Google Scholar] [CrossRef]
- Rubinstein, M.R.; Wang, X.; Liu, W.; Hao, Y.; Cai, G.; Han, Y.W. Fusobacterium nucleatum Promotes Colorectal Carcinogenesis by Modulating E-Cadherin/β-Catenin Signaling via its FadA Adhesin. Cell Host Microbe 2013, 14, 195–206. [Google Scholar] [CrossRef] [PubMed]
- Castellarin, M.; Warren, R.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A.; et al. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2011, 22, 299–306. [Google Scholar] [CrossRef]
- Ma, C.; Luo, H.; Gao, F.; Tang, Q.; Chen, W. Fusobacterium nucleatum promotes the progression of colorectal cancer by interacting with E-cadherin. Oncol. Lett. 2018, 16, 2606–2612. [Google Scholar] [CrossRef]
- Dharmani, P.; Strauss, J.; Ambrose, C.; Allen-Vercoe, E.; Chadee, K. Fusobacterium nucleatum Infection of Colonic Cells Stimulates MUC2 Mucin and Tumor Necrosis Factor Alpha. Infect. Immun. 2011, 79, 2597–2607. [Google Scholar] [CrossRef]
- Park, H.E.; Kim, J.H.; Cho, N.-Y.; Lee, H.S.; Kang, G.H. Intratumoral Fusobacterium nucleatum abundance correlates with macrophage infiltration and CDKN2A methylation in microsatellite-unstable colorectal carcinoma. Virchows Archiv 2017, 471, 329–336. [Google Scholar] [CrossRef]
- Koul, H.K.; Pal, M.; Koul, S. Role of p38 MAP Kinase Signal Transduction in Solid Tumors. Genes Cancer 2013, 4, 342–359. [Google Scholar] [CrossRef] [PubMed]
- Huynh, T.; Kapur, R.V.; Kaplan, C.W.; Cacalano, N.; Haake, S.K.; Shi, W.; Sieling, P.; Jewett, A. The Role of Aggregation in Fusobacterium nucleatum–Induced Immune Cell Death. J. Endod. 2011, 37, 1531–1535. [Google Scholar] [CrossRef] [PubMed]
- Shenker, B.J.; Datar, S. Fusobacterium nucleatum inhibits human T-cell activation by arresting cells in the mid-G1 phase of the cell cycle. Infect. Immun. 1995, 63, 4830–4836. [Google Scholar] [CrossRef] [PubMed]
- Nistal, E.; Fernández-Fernández, N.; Vivas, S.; Olcoz, J.L. Factors Determining Colorectal Cancer: The Role of the Intestinal Microbiota. Front. Oncol. 2015, 5, 220. [Google Scholar] [CrossRef]
- Osman, M.A.; Neoh, H.-M.; Mutalib, N.-S.A.; Chin, S.-F.; Mazlan, L.; Ali, R.A.R.; Zakaria, A.D.; Ngiu, C.S.; Ang, M.Y.; Jamal, R. Parvimonas micra, Peptostreptococcus stomatis, Fusobacterium nucleatum and Akkermansia muciniphila as a four-bacteria biomarker panel of colorectal cancer. Sci. Rep. 2021, 11, 1–12. [Google Scholar] [CrossRef]
- Drewes, J.L.; White, J.R.; Dejea, C.M.; Fathi, P.; Iyadorai, T.; Vadivelu, J.; Roslani, A.C.; Wick, E.C.; Mongodin, E.F.; Loke, M.F.; et al. High-resolution bacterial 16S rRNA gene profile meta-analysis and biofilm status reveal common colorectal cancer consortia. NPJ Biofilms Microbiomes 2017, 3, 1–12. [Google Scholar] [CrossRef]
- Baxter, N.T.; Iv, M.T.R.; Rogers, M.A.M.; Schloss, P.D. Microbiota-based model improves the sensitivity of fecal immunochemical test for detecting colonic lesions. Genome Med. 2016, 8, 1–10. [Google Scholar] [CrossRef]
- Mostafa, M.H.; Sheweita, S.; O’Connor, P.J. Relationship between Schistosomiasis and Bladder Cancer. Clin. Microbiol. Rev. 1999, 12, 97–111. [Google Scholar] [CrossRef]
- Gray, D.; Ross, A.G.; Li, Y.-S.; McManus, D.P. Diagnosis and management of schistosomiasis. BMJ 2011, 342, d2651. [Google Scholar] [CrossRef]
- Bella, S.D.; Riccardi, N.; Giacobbe, D.R.; Luzzati, R. History of schistosomiasis (bilharziasis) in humans: From Egyptian medical papyri to molecular biology on mummies. Pathog. Glob. Health 2018, 112, 268–273. [Google Scholar] [CrossRef]
- Barakat, R.M. Epidemiology of Schistosomiasis in Egypt: Travel through Time: Review. J. Adv. Res. 2013, 4, 425–432. [Google Scholar] [CrossRef]
- Ross, A.G.; Inobaya, M.T.; Olveda, R.M.; Chau, T.N.; Olveda, D.U. Prevention and control of schistosomiasis: A current perspective. Res. Rep. Trop. Med. 2014, 5, 65–75. [Google Scholar] [CrossRef]
- Ishida, K.; Hsieh, M.H. Understanding Urogenital Schistosomiasis-Related Bladder Cancer: An Update. Front. Med. 2018, 5, 223. [Google Scholar] [CrossRef]
- Rosin, M.P.; Zaki, S.S.E.D.; Ward, A.J.; Anwar, W.A. Involvement of inflammatory reactions and elevated cell proliferation in the development of bladder cancer in schistosomiasis patients. Mutat. Res. Mol. Mech. Mutagen. 1994, 305, 283–292. [Google Scholar] [CrossRef]
- Pearce, E.J.; Macdonald, A.S. The immunobiology of schistosomiasis. Nat. Rev. Immunol. 2002, 2, 499–511. [Google Scholar] [CrossRef] [PubMed]
- Adebayo, A.S.; Survayanshi, M.; Bhute, S.; Agunloye, A.M.; Isokpehi, R.D.; Anumudu, C.I.; Shouche, Y.S. The microbiome in urogenital schistosomiasis and induced bladder pathologies. PLoS Neglected Trop. Dis. 2017, 11, e0005826. [Google Scholar] [CrossRef]
- Botelho, M.C.; Alves, H.; Richter, J. Halting Schistosoma haematobium—Associated Bladder Cancer. Int. J. Cancer Manag. 2017, 10, 9430. [Google Scholar] [CrossRef] [PubMed]
- Gouveia, M.J.; Santos, J.; Brindley, P.J.; Rinaldi, G.; Lopes, C.; Santos, L.L.; Costa, J.M.C.D.; Vale, N. Estrogen-like metabolites and DNA-adducts in urogenital schistosomiasis-associated bladder cancer. Cancer Lett. 2015, 359, 226–232. [Google Scholar] [CrossRef]
- Panani, A.D.; Ferti, A.D.; Raptis, S.A.; Roussos, C. Novel recurrent structural chromosomal aberrations in primary bladder cancer. Anticancer. Res. 2004, 24, 2967–2974. [Google Scholar] [PubMed]
- Fadl-Elmula, I. Chromosomal changes in uroepithelial carcinomas. Cell Chromosom. 2005, 4, 1. [Google Scholar] [CrossRef]
- Aly, M.S.; Khaled, H.M.; Emara, M.; Hussein, T.D. Cytogenetic profile of locally advanced and metastatic schistosoma-related bladder cancer and response to chemotherapy. Cancer Genet. 2012, 205, 156–162. [Google Scholar] [CrossRef]
- McConkey, D.J.; Lee, S.; Choi, W.; Tran, M.; Majewski, T.; Lee, S.; Siefker-Radtke, A.; Dinney, C.; Czerniak, B. Molecular genetics of bladder cancer: Emerging mechanisms of tumor initiation and progression. Urol. Oncol. Semin. Orig. Investig. 2010, 28, 429–440. [Google Scholar] [CrossRef] [PubMed]
- Botelho, M.C.; Veiga, I.; Oliveira, P.A.; Lopes, C.; Teixeira, M.; Costa, J.M.C.D.; Machado, J.C. Carcinogenic ability of Schistosoma haematobium possibly through oncogenic mutation of KRAS gene. Adv. Cancer Res. Treat. 2013, 2013, 876585. [Google Scholar] [PubMed]
- Zaghloul, M.S. Bladder cancer and schistosomiasis. J. Egypt. Natl. Cancer Inst. 2012, 24, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Chala, B.; Choi, M.-H.; Moon, K.C.; Kim, H.S.; Kwak, C.; Hong, S.-T. Development of Urinary Bladder Pre-Neoplasia by Schistosoma haematobium Eggs and Chemical Carcinogen in Mice. Korean J. Parasitol. 2017, 55, 21–29. [Google Scholar] [CrossRef] [PubMed]
- Honeycutt, J.; Hammam, O.; Fu, C.-L.; Hsieh, M.H. Controversies and challenges in research on urogenital schistosomiasis-associated bladder cancer. Trends Parasitol. 2014, 30, 324–332. [Google Scholar] [CrossRef]
- Vu, M.; Yu, J.; Awolude, O.A.; Chuang, L. Cervical cancer worldwide. Curr. Probl. Cancer 2018, 42, 457–465. [Google Scholar] [CrossRef]
- Madeleine, M.M.; Anttila, T.; Schwartz, S.M.; Saikku, P.; Leinonen, M.; Carter, J.J.; Wurscher, M.; Johnson, L.G.; Galloway, D.A.; Daling, J.R. Risk of cervical cancer associated withChlamydia trachomatis antibodies by histology, HPV type and HPV cofactors. Int. J. Cancer 2007, 120, 650–655. [Google Scholar] [CrossRef]
- Schiffman, M.; Wentzensen, N.; Wacholder, S.; Kinney, W.; Gage, J.C.; Castle, P.E. Human Papillomavirus Testing in the Prevention of Cervical Cancer. J. Natl. Cancer Inst. 2011, 103, 368–383. [Google Scholar] [CrossRef]
- zur Hausen, H.; Francoise, B.-S. The Discoveries of Human Papilloma Viruses That Cause Cervical Cancer and of Human Immunodeficiency Virus. Nobel Prize 2008, 8, 1–26. Available online: https://www.nobelprize.org/uploads/2018/06/advanced-medicineprize2008.pdf (accessed on 24 January 2021).
- Gravitt, P.E. The known unknowns of HPV natural history. J. Clin. Investig. 2011, 121, 4593–4599. [Google Scholar] [CrossRef]
- Okunade, K.S. Human papillomavirus and cervical cancer. J. Obstet. Gynaecol. 2020, 40, 602–608. [Google Scholar] [CrossRef]
- Schiller, J.T.; Day, P.M.; Kines, R.C. Current understanding of the mechanism of HPV infection. Gynecol. Oncol. 2010, 118, S12–S17. [Google Scholar] [CrossRef] [PubMed]
- Kines, R.C.; Cerio, R.J.; Roberts, J.N.; Thompson, C.D.; Pinos, E.D.L.; Lowy, D.R.; Schiller, J.T. Human papillomavirus capsids preferentially bind and infect tumor cells. Int. J. Cancer 2016, 138, 901–911. [Google Scholar] [CrossRef] [PubMed]
- Doorbar, J.; Egawa, N.; Griffin, H.; Kranjec, C.; Murakami, I. Human papillomavirus molecular biology and disease association. Rev. Med. Virol. 2015, 25, 2–23. [Google Scholar] [CrossRef]
- McMurray, H.R.; Nguyen, D.; Westbrook, T.F.; Mcance, D.J. Biology of human papillomaviruses. Int. J. Exp. Pathol. 2001, 82, 15–33. [Google Scholar] [CrossRef]
- Doorbar, J.; Quint, W.; Banks, L.; Bravo, I.G.; Stoler, M.; Broker, T.R.; Stanley, M.A. The Biology and Life-Cycle of Human Papillomaviruses. Vaccine 2012, 30, F55–F70. [Google Scholar] [CrossRef] [PubMed]
- Horvath, C.A.J.; Boulet, G.A.V.; Renoux, V.M.; Delvenne, P.O.; Bogers, J.-P.J. Mechanisms of cell entry by human papillomaviruses: An overview. Virol. J. 2010, 7, 11. [Google Scholar] [CrossRef] [PubMed]
- Yeo-Teh, N.S.L.; Ito, Y.; Jha, S. High-Risk Human Papillomaviral Oncogenes E6 and E7 Target Key Cellular Pathways to Achieve Oncogenesis. Int. J. Mol. Sci. 2018, 19, 1706. [Google Scholar] [CrossRef]
- Longworth, M.S.; Laimins, L.A. Pathogenesis of Human Papillomaviruses in Differentiating Epithelia. Microbiol. Mol. Biol. Rev. 2004, 68, 362–372. [Google Scholar] [CrossRef]
- Balasubramaniam, S.D.; Balakrishnan, V.; Oon, C.E.; Kaur, G. Key Molecular Events in Cervical Cancer Development. Medicine 2019, 55, 384. [Google Scholar] [CrossRef]
- Boshoff, C. Kaposi’s Sarcoma Biology. IUBMB Life 2002, 53, 259–261. [Google Scholar] [CrossRef] [PubMed]
- Torrence, G.M.; Wrobel, J.S. A case of mistaken identity: Classic Kaposi sarcoma misdiagnosed as a diabetic foot ulcer in an atypical patient. Clin. Diabetes Endocrinol. 2019, 5, 1–6. [Google Scholar] [CrossRef]
- Cesarman, E.; Damania, B.; Krown, S.E.; Martin, J.; Bower, M.; Whitby, D. Kaposi sarcoma. Nat. Rev. Dis. Prim. 2019, 5, 1–21. [Google Scholar] [CrossRef]
- Chang, Y.; Cesarman, E.; Pessin, M.; Lee, F.; Culpepper, J.; Knowles, D.; Moore, P. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi’s sarcoma. Science 1994, 266, 1865–1869. [Google Scholar] [CrossRef]
- Schulz, T.F. The pleiotropic effects of Kaposi’s sarcoma herpesvirus. J. Pathol. 2005, 208, 187–198. [Google Scholar] [CrossRef] [PubMed]
- Henke-Gendo, C.; Schulz, T. Transmission and disease association of Kaposi’s sarcoma-associated herpesvirus: Recent developments. Curr. Opin. Infect. Dis. 2004, 17, 53–57. [Google Scholar] [CrossRef] [PubMed]
- You, H.; Asiimwe, S.; Byakwaga, H.; Glidden, D.; Shiboski, C.; Dollard, S.; Martin, J. Low Prevalence of KSHV DNA in Female Genital Fluid: Findings from Zimbabwean Women and Systematic Review of the Literature of KSHV Shedding in Body Fluids and Mucous Membranse Surfaces. In Proceedings of the 16th International Conference on Malignancies in HIV/AIDS, Bethesda, Maryland, 23–24 October 2017; p. 115. [Google Scholar]
- Bechtel, J.T.; Liang, Y.; Hvidding, J.; Ganem, D. Host Range of Kaposi’s Sarcoma-Associated Herpesvirus in Cultured Cells. J. Virol. 2003, 77, 6474–6481. [Google Scholar] [CrossRef]
- Dittmer, D.P.; Damania, B. Kaposi sarcoma–associated herpesvirus: Immunobiology, oncogenesis, and therapy. J. Clin. Investig. 2016, 126, 3165–3175. [Google Scholar] [CrossRef]
- Ballon, G.; Akar, G.; Cesarman, E. Systemic Expression of Kaposi Sarcoma Herpesvirus (KSHV) Vflip in Endothelial Cells Leads to a Profound Proinflammatory Phenotype and Myeloid Lineage Remodeling In Vivo. PLoS Pathog. 2015, 11, e1004581. [Google Scholar] [CrossRef]
- Aoki, Y.; Yarchoan, R.; Wyvill, K.; Okamoto, S.-I.; Little, R.F.; Tosato, G. Detection of viral interleukin-6 in Kaposi sarcoma–associated herpesvirus–linked disorders. Blood 2001, 97, 2173–2176. [Google Scholar] [CrossRef]
- Arvanitakis, L.; Geras-Raaka, E.; Varma, A.; Gershengorn, M.C.; Cesarman, E. Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nat. Cell Biol. 1997, 385, 347–350. [Google Scholar] [CrossRef] [PubMed]
- Cavallin, L.E.; Ma, Q.; Naipauer, J.; Gupta, S.; Kurian, M.; Locatelli, P.; Romanelli, P.; Nadji, M.; Goldschmidt-Clermont, P.J.; Mesri, E.A. KSHV-induced ligand mediated activation of PDGF receptor-alpha drives Kaposi’s sarcomagenesis. PLoS Pathog. 2018, 14, e1007175. [Google Scholar] [CrossRef] [PubMed]
- Thakker, S.; Verma, S.C. Co-infections and Pathogenesis of KSHV-Associated Malignancies. Front. Microbiol. 2016, 7, 151. [Google Scholar] [CrossRef] [PubMed]
- Dabrowska, A.; Kim, N.; Aldovini, A. Tat-Induced FOXO3a Is a Key Mediator of Apoptosis in HIV-1-Infected Human CD4+T Lymphocytes. J. Immunol. 2008, 181, 8460–8477. [Google Scholar] [CrossRef]
- Zhu, X.; Guo, Y.; Yao, S.; Yan, Q.; Xue, M.; Hao, T.; Zhou, F.; Zhu, J.; Qin, D.; Lu, C. Synergy between Kaposi’s sarcoma-associated herpesvirus (KSHV) vIL-6 and HIV-1 Nef protein in promotion of angiogenesis and oncogenesis: Role of the AKT signaling pathway. Oncogene 2013, 33, 1986–1996. [Google Scholar] [CrossRef] [PubMed]
- Qin, J.; Lu, C. Infection of KSHV and Interaction with HIV: The Bad Romance. In Infectious Agents Associated Cancers: Epidemi-ology and Molecular Biology; Cai, Q., Yuan, Z., Lan, K., Eds.; Springer Nature Singapore Pte. Ltd.: Singapore, 2017; p. 237. ISBN 9789811057656. [Google Scholar]
- Zeng, Y.; Zhang, X.; Huang, Z.; Cheng, L.; Yao, S.; Qin, D.; Chen, X.; Tang, Q.; Lv, Z.; Zhang, L.; et al. Intracellular Tat of Human Immunodeficiency Virus Type 1 Activates Lytic Cycle Replication of Kaposi’s Sarcoma-Associated Herpesvirus: Role of JAK/STAT Signaling. J. Virol. 2006, 81, 2401–2417. [Google Scholar] [CrossRef]
- Shannon-Lowe, C.; Rickinson, A.B.; Bell, A.I. Epstein–Barr virus-associated lymphomas. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160271. [Google Scholar] [CrossRef] [PubMed]
- Malcolm, T.I.M.; Hodson, D.J.; Macintyre, E.A.; Turner, S.D. Challenging perspectives on the cellular origins of lymphoma. Open Biol. 2016, 6, 160232. [Google Scholar] [CrossRef] [PubMed]
- Chihara, D.; Nastoupil, L.J.; Williams, J.N.; Lee, P.; Koff, J.L.; Flowers, C.R. New insights into the epidemiology of non-Hodgkin lymphoma and implications for therapy. Expert Rev. Anticancer. Ther. 2015, 15, 531–544. [Google Scholar] [CrossRef]
- Zhang, Y.; Dai, Y.; Zheng, T.; Ma, S. Risk factors of non-Hodgkin’s lymphoma. Expert Opin. Med. Diagn. 2011, 5, 539–550. [Google Scholar] [CrossRef] [PubMed]
- Farrell, P.J. Epstein–Barr Virus and Cancer. Annu. Rev. Pathol. Mech. Dis. 2019, 14, 29–53. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Hawkins, J.B.; Tracy, S.I.; Shapiro, M. The pathogenesis of Epstein–Barr virus persistent infection. Curr. Opin. Virol. 2013, 3, 227–232. [Google Scholar] [CrossRef] [PubMed]
- Pattle, S.B.; Farrell, P. The role of Epstein–Barr virus in cancer. Expert Opin. Biol. Ther. 2006, 6, 1193–1205. [Google Scholar] [CrossRef]
- Klein, G.; Klein, E.; Kashuba, E. Interaction of Epstein-Barr virus (EBV) with human B-lymphocytes. Biochem. Biophys. Res. Commun. 2010, 396, 67–73. [Google Scholar] [CrossRef]
- Thorley-Lawson, D.A.; Allday, M.J. The curious case of the tumour virus: 50 years of Burkitt’s lymphoma. Nat. Rev. Genet. 2008, 6, 913–924. [Google Scholar] [CrossRef]
- McMahon, S.B. MYC and the Control of Apoptosis. Cold Spring Harb. Perspect. Med. 2014, 4, a014407. [Google Scholar] [CrossRef]
- Gruhne, B.; Sompallae, R.; Marescotti, D.; Kamranvar, S.A.; Gastaldello, S.; Masucci, M.G. The Epstein-Barr virus nuclear antigen-1 promotes genomic instability via induction of reactive oxygen species. Proc. Natl. Acad. Sci. USA 2009, 106, 2313–2318. [Google Scholar] [CrossRef]
- Larcher, C.; Kempkes, B.; Kremmer, E.; Prodinger, W.M.; Pawlita, M.; Bornkamm, G.W.; Dierich, M.P. Expression of Epstein-Barr virus nuclear antigen-2 (EBNA2) induces CD21/CR2 on B and T cell lines and shedding of soluble CD21. Eur. J. Immunol. 1995, 25, 1713–1719. [Google Scholar] [CrossRef]
- Hatton, O.L.; Harris-Arnold, A.; Schaffert, S.; Krams, S.M.; Martinez, O.M. The interplay between Epstein–Barr virus and B lymphocytes: Implications for infection, immunity, and disease. Immunol. Res. 2014, 58, 268–276. [Google Scholar] [CrossRef]
- Lamb, R.; Parks, G. Paramyxoviridae: The viruses and their replication. In Fields Virology, 5th ed.; Knipe, D.M., Howley, P.M., Griffin, D.E., Lamb, R.A., Martin, M.A., Roizman, B., Straus, S.E., Eds.; Lippincott Williams & Wilkins: Philadelphia, PA, USA, 2007; Volume 1, pp. 1449–1496. ISBN 9780781760607. [Google Scholar]
- Küppers, R. Molecular biology of Hodgkin lymphoma. Hematol. Am. Soc. Hematol. Educ. Progr. 2009, 2009, 491–496. [Google Scholar] [CrossRef] [PubMed]
- de la Cruz-Merino, L.; Lejeune, M.; Fernández, E.N.; Carrasco, F.H.; López, A.G.; Vacas, A.I.; Pulla, M.P.; Callau, C.; Álvaro, T. Role of Immune Escape Mechanisms in Hodgkin’s Lymphoma Development and Progression: A Whole New World with Therapeutic Implications. Clin. Dev. Immunol. 2012, 2012, 1–24. [Google Scholar] [CrossRef]
- White, M.K.; Pagano, J.S.; Khalili, K. Viruses and Human Cancers: A Long Road of Discovery of Molecular Paradigms. Clin. Microbiol. Rev. 2014, 27, 463–481. [Google Scholar] [CrossRef]
- Steidl, C.; Shah, S.P.; Woolcock, B.W.; Rui, L.; Kawahara, M.; Farinha, P.; Johnson, N.A.; Zhao, Y.; Telenius, A.; Neriah, S.B.; et al. MHC class II transactivator CIITA is a recurrent gene fusion partner in lymphoid cancers. Nat. Cell Biol. 2011, 471, 377–381. [Google Scholar] [CrossRef]
- Lin, C.-L.; Kao, J.-H. Hepatitis B: Immunization and Impact on Natural History and Cancer Incidence. Gastroenterol. Clin. N. Am. 2020, 49, 201–214. [Google Scholar] [CrossRef] [PubMed]
- Bigaard, J.; Franceschi, S. Vaccination against HPV: Boosting coverage and tackling misinformation. Mol. Oncol. 2020, 15, 770–778. [Google Scholar] [CrossRef]
- Zhou, Y.-B.; Chen, Y.; Liang, S.; Song, X.-X.; Chen, G.-X.; He, Z.; Cai, B.; Yihuo, W.-L.; He, Z.-G.; Jiang, Q.-W. Multi-host model and threshold of intermediate host Oncomelania snail density for eliminating schistosomiasis transmission in China. Sci. Rep. 2016, 6, 31089. [Google Scholar] [CrossRef]
- Lo, N.C.; Gurarie, D.; Yoon, N.; Coulibaly, J.T.; Bendavid, E.; Andrews, J.R.; King, C. Impact and cost-effectiveness of snail control to achieve disease control targets for schistosomiasis. Proc. Natl. Acad. Sci. USA 2018, 115, E584–E591. [Google Scholar] [CrossRef] [PubMed]
- Suwannahitatorn, P.; Webster, J.; Riley, S.; Mungthin, M.; Donnelly, C.A. Uncooked fish consumption among those at risk of Opisthorchis viverrini infection in central Thailand. PLoS ONE 2019, 14, e0211540. [Google Scholar] [CrossRef]
- MacLaughlin, K.L.; Jacobson, R.M.; Breitkopf, C.R.; Wilson, P.M.; Jacobson, D.J.; Fan, C.; Sauver, J.L.S.; Rutten, L.J.F. Trends Over Time in Pap and Pap-HPV Cotesting for Cervical Cancer Screening. J. Women’s Health 2019, 28, 244–249. [Google Scholar] [CrossRef]
- Wroblewski, L.E.; Peek, R.M.; Wilson, K.T. Helicobacter pylori and Gastric Cancer: Factors That Modulate Disease Risk. Clin. Microbiol. Rev. 2010, 23, 713–739. [Google Scholar] [CrossRef]
- Pawlotsky, J.-M.; Feld, J.J.; Zeuzem, S.; Hoofnagle, J.H. From non-A, non-B hepatitis to hepatitis C virus cure. J. Hepatol. 2015, 62, S87–S99. [Google Scholar] [CrossRef] [PubMed]
- Krown, S.E.; Dittmer, D.P.; Cesarman, E. Pilot Study of Oral Valganciclovir Therapy in Patients with Classic Kaposi Sarcoma. J. Infect. Dis. 2011, 203, 1082–1086. [Google Scholar] [CrossRef]
- Hong, S.-T. Albendazole and Praziquantel: Review and Safety Monitoring in Korea. Infect. Chemother. 2018, 50, 1–10. [Google Scholar] [CrossRef] [PubMed]
| Pathogen | Type of Cancer | Carcinogenesis Mechanism | Prevention/Treatment |
|---|---|---|---|
| Helicobacter pylori | Gastric |
|
|
| Epstein-Barr Virus (EBV) | Gastric |
|
|
| Hepatitis B Virus (HBV) | Liver |
|
|
| Hepatitis C Virus (HCV) | Liver |
|
|
| Aspergillus spp. | Liver |
|
|
| Opisthorchis viverrini | Bile Duct |
|
|
| Clonorchis sinensis | Bile Duct |
|
|
| Fusobacterium nucleatum | Colorectal |
|
|
| Schitosoma haematobium | Bladder |
|
|
| Human Papillomavirus (HPV) | Cervical |
|
|
| Kaposi’s Sarcoma Herpesvirus (KSHV) | Kaposi’s Sarcoma |
|
|
| Epstein-Barr Virus (EBV) | Lymphoma | Burkitt’s Lymphoma
|
|
Hodgkin’s Lymphoma
|
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hatta, M.N.A.; Mohamad Hanif, E.A.; Chin, S.-F.; Neoh, H.-m. Pathogens and Carcinogenesis: A Review. Biology 2021, 10, 533. https://doi.org/10.3390/biology10060533
Hatta MNA, Mohamad Hanif EA, Chin S-F, Neoh H-m. Pathogens and Carcinogenesis: A Review. Biology. 2021; 10(6):533. https://doi.org/10.3390/biology10060533
Chicago/Turabian StyleHatta, Muhammad Nur Adam, Ezanee Azlina Mohamad Hanif, Siok-Fong Chin, and Hui-min Neoh. 2021. "Pathogens and Carcinogenesis: A Review" Biology 10, no. 6: 533. https://doi.org/10.3390/biology10060533
APA StyleHatta, M. N. A., Mohamad Hanif, E. A., Chin, S.-F., & Neoh, H.-m. (2021). Pathogens and Carcinogenesis: A Review. Biology, 10(6), 533. https://doi.org/10.3390/biology10060533






