Construction of Infectious Clones of Begomoviruses: Strategies, Techniques and Applications
Abstract
:Simple Summary
Abstract
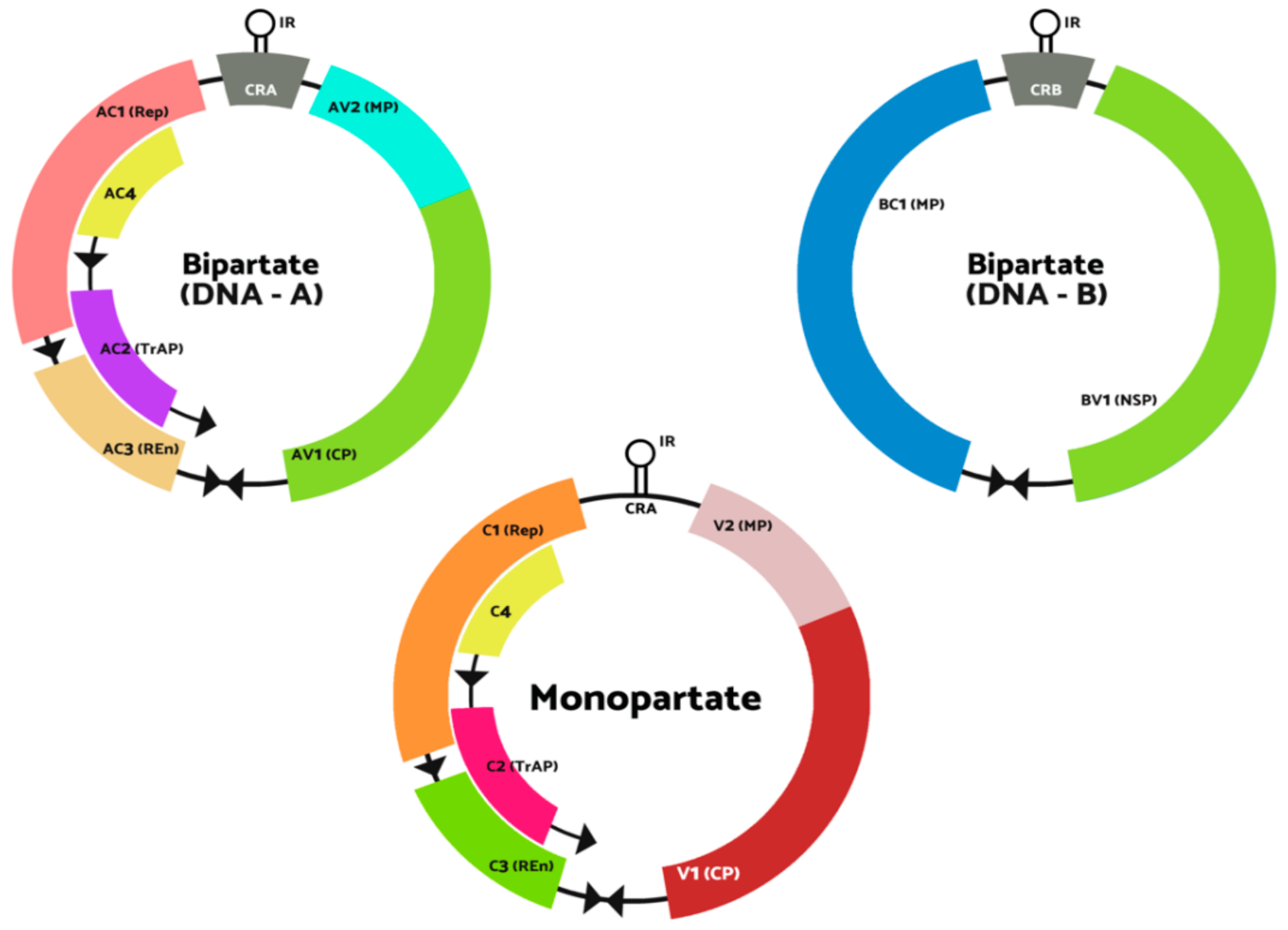
1. Introduction
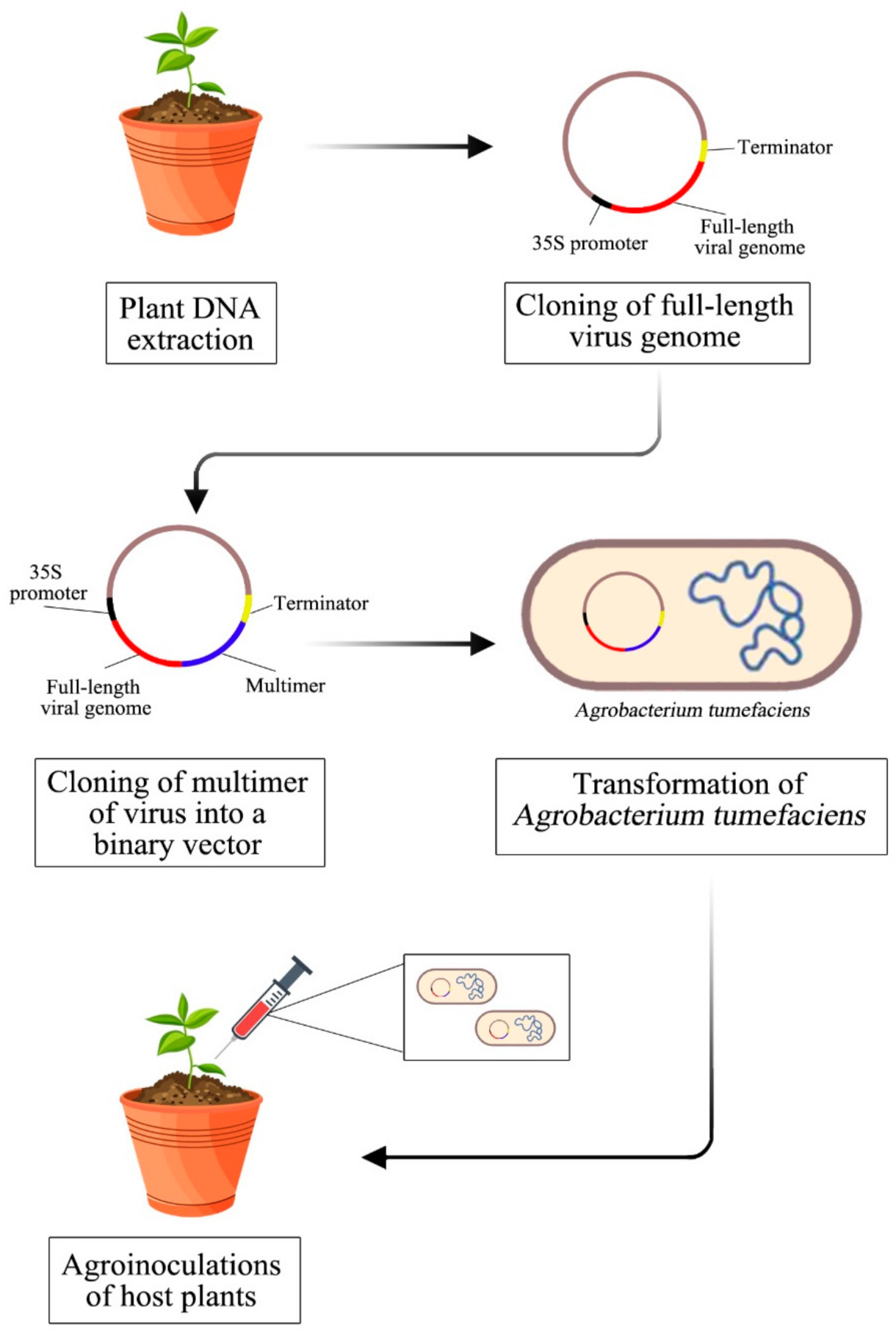
2. Full-Length Infectious Clones Construction Strategy
2.1. Isolation of Total Plant DNA
2.2. Amplification of Viral Genomes
2.3. Cloning of Multimer and Full-Length Genome
2.4. Transformation into Agrobacterium Tumafaciens
2.5. Agroinoculation of Host Plant
3. The Significance of Established Infectious Clones in the Begomovirus Study
3.1. Functional Study on the Begomovirus Genomes Using Infectious Clones
3.1.1. V2 Protein Functional Analysis
3.1.2. Functional Studies of C1 Protein
3.1.3. Functional Studies of C2 Protein
3.1.4. C4 Protein Functional Analysis
3.1.5. Functional Studies on Betasatellite DNAβ of Monopartite Begomovirus
3.2. Infectious Clones for Screening Resistant Varieties
4. Future Perspectives
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Taniguchi, T.; Palmieri, M.; Weissmann, C. QB DNA-containing hybrid plasmids giving rise to QB phage formation in the bacterial host. Nature 1978, 274, 223–228. [Google Scholar] [CrossRef] [PubMed]
- Lowy, D.R.; Rands, E.; Chattopadhyay, S.K.; Garon, C.F.; Hager, G.L. Molecular cloning of infectious integrated murine leukemia virus DNA from infected mouse cells. Proc. Natl. Acad. Sci. USA 1980, 77, 614–618. [Google Scholar] [CrossRef] [Green Version]
- Racaniello, V.R.; Baltimore, D. Cloned poliovirus complementary DNA is infectious in mammalian cells. Science 1981, 214, 916–919. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.; Fang, X.-D.; Qiao, J.-H.; Gao, Q.; Wang, X.-B. Reverse genetics system of plant negative-strand RNA viruses are difficult to developed but powerful for virus-host interaction studies and virus-based vector applications. Phytopathol. Res. 2020, 2. [Google Scholar] [CrossRef]
- Kannan, M.; Zainal, Z.; Ismail, I.; Baharum, S.N.; Bunawan, H. Application of reverse genetics in functional genomics of Potyvirus. Viruses 2020, 12, 803. [Google Scholar] [CrossRef]
- Aubry, F.; Nougairede, A.; Gould, E.A.; Lamballerie, X. Flavivirus reverse genetic systems, construction techniques and applications: A historical perspective. Antivir. Res. 2015, 114, 67–85. [Google Scholar] [CrossRef] [Green Version]
- Kannan, M.; Ismail, I.; Bunawan, H. Maize Dwarf Mosaic Virus: From Genome to Disease Management. Viruses 2018, 10, 492. [Google Scholar] [CrossRef] [Green Version]
- Brown, J.K.; Zerbini, F.M.; Navas-Castillo, J.; Moriones, E.; Ramos-Sobrinho, R.; Silva, J.C.; Fiallo-Olivé, E.; Briddon, R.W.; Hernández-Zepeda, C.; Idris, A.; et al. Revision of Begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 2015, 160, 1593–1619. [Google Scholar] [CrossRef]
- Sau, A.R.; Nazmie, N.M.F.; Yusop, M.S.M.; Akbar, M.A.; Saad, M.F.M.; Baharum, S.N.; Talip, N.; Goh, H.H.; Kassim, H.; Bunawan, H. First report of pepper vein yellows virus (PeVYV) and pepper yellow leaf curl virus (PepYLCV) infecting Chili Pepper (Capsicum annuum L.) in Malaysia. Plant Dis. 2020, 104. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X. Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 2013, 51, 357–381. [Google Scholar] [CrossRef]
- Lozano, G.; Trenado, H.P.; Fiallo-Olivé, E.; Chirinos, D.; Geraud-Pouey, F.; Briddon, R.W.; Navas-Castillo, J. Characterization of Non-coding DNA Satellites Associated with Sweepoviruses (Genus Begomovirus, Geminiviridae)—Definition of a Distinct Class of Begomovirus-Associated Satellites. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef] [Green Version]
- Fiallo-Olivé, E.; Navas-Castillo, J. Molecular and Biological Characterization of a New World Mono-/Bipartite Begomovirus/Deltasatellite Complex Infecting Corchorus siliquosus. Front. Microbiol. 2020, 11, 1755. [Google Scholar] [CrossRef]
- Pandey, V.; Srivastava, A.; Gaur, R.K. Begomovirus: A curse for the agricultural crops. Arch. Phytopathol. Plant Prot. 2021. [Google Scholar] [CrossRef]
- Hanley-Bowdoin, L.; Settlage, S.B.; Orozco, B.M.; Nagar, S.; Robertson, D. Geminiviruses: Models for plant DNA replication, transcription, and cell cycle regulation. Crit. Rev. Biochem. Mol. Biol. 2000, 35, 105–140. [Google Scholar] [CrossRef]
- Laufs, J.; Traut, W.; Heyraud, F.; Matzeit, V.; Rogers, S.G.; Schell, J.; Gronenborn, B. In vitro cleavage and joining at the viral origin of replication by the replication initiator protein of tomato yellow leaf curl virus. Proc. Natl. Acad. Sci. USA 1995, 92, 3879–3883. [Google Scholar] [CrossRef] [Green Version]
- Fontes, E.P.; Eagle, P.A.; Sipe, P.S.; Luckow, V.A.; Hanley-Bowdoin, L. Interaction between a geminivirus replication protein and origin DNA is essential for viral replication. J. Biol. Chem. 1994, 269, 8459–8465. [Google Scholar] [CrossRef]
- Arguello-Astorga, G.; Lopez-Ochoa, L.; Kong, L.J.; Orozco, B.M.; Settlage, S.B.; Hanley-Bowdoin, L. A novel motif in geminivirus replication proteins interacts with the plant retinoblastoma-related protein. J. Virol. 2004, 78, 4817–4826. [Google Scholar] [CrossRef] [Green Version]
- Sunter, G.; Bisaro, D.M. Transactivation of geminivirus AR1 and BR1 gene expression by the viral AL2 gene product occurs at the level of transcription. Plant Cell 1992, 4, 1321–1331. [Google Scholar] [CrossRef] [Green Version]
- Bisaro, D.M. Silencing suppression by geminivirus proteins. Virology 2006, 344, 158–168. [Google Scholar] [CrossRef] [Green Version]
- Hanley-Bowdoin, L.; Bejarano, E.R.; Robertson, D.; Mansoor, S. Geminiviruses: Masters at redirecting and reprogramming plant processes. Nat. Reviews. Microbiol. 2013, 11, 777–788. [Google Scholar] [CrossRef]
- Noueiry, A.O.; Lucas, W.J.; Gilbertson, R.L. Two proteins of a plant DNA virus coordinate nuclear and plasmodesmal transport. Cell 1994, 76, 925–932. [Google Scholar] [CrossRef]
- Sanderfoot, A.A.; Lazarowitz, S.G. Getting it together in plant virus movement: Cooperative interactions between bipartite geminivirus movement proteins. Trends Cell Biol. 1996, 6, 353–358. [Google Scholar] [CrossRef]
- Kushawaha, A.K.; Dasgupta, I. Infectivity of cloned begomoviral DNAs: An appraisal. Virus Dis. 2018, 30, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Dellaporta, S.L.; Wood, J.; Hicks, J.B. A plant DNA minipreparation: Version II. Plant Molacular Biol. Rep. 1983, 1, 19–21. [Google Scholar] [CrossRef]
- Doyle, J.J.; Doyle, J.L. A rapid DNA isolation from small amount of fresh leaf tissue. Phytochemistry 1987, 19, 11–15. [Google Scholar]
- Palmer, K.E.; Schnippenkoetter, W.H.; Rybicki, E.P. Geminivirus isolation and DNA extraction. Methods Mol. Biol. 1998, 81, 41–52. [Google Scholar] [CrossRef]
- Inoue-Nagata, A.K.; Albuquerque, L.C.; Rocha, W.B.; Nagata, T. A simple method for cloning the complete begomovirus genome using the bacteriophage phi29 DNA polymerase. J. Virol. Methods 2004, 116, 209–211. [Google Scholar] [CrossRef]
- Kiss, M.M.; Ortoleva-Donnelly, L.; Beer, N.R.; Warner, J.; Bailey, C.G.; Colston, B.W.; Rothberg, J.M.; Link, D.R.; Leamon, J.H. High-throughput quantitative polymerase chain reaction in picoliter droplets. Anal. Chem. 2008, 80, 8975–8981. [Google Scholar] [CrossRef]
- Nagata, T.; Inoue-Nagata, A.K. Simplified methods for the construction of RNA and DNA virus infectious clones. Methods Mol. Biol. (Cliftonn. J.) 2015, 1236, 241–254. [Google Scholar] [CrossRef]
- Wu, C.Y.; Lai, Y.C.; Lin, N.S.; Hsu, Y.H.; Tsai, H.T.; Liao, J.Y.; Hu, C.C. A simplified method of constructing infectious clones of begomovirus employing limited restriction enzyme digestion of products of rolling circle amplification. J. Virol. Methods 2008, 147, 355–359. [Google Scholar] [CrossRef]
- Bang, B.; Lee, J.; Kim, S.; Park, J.; Nguyen, T.T.; Seo, Y.S. A Rapid and Efficient Method for Construction of an Infectious Clone of Tomato yellow leaf curl virus. Plant Pathol. J. 2014, 30, 310–315. [Google Scholar] [CrossRef] [Green Version]
- Trenado, H.P.; Orílio, A.F.; Márquez-Martín, B.; Moriones, E.; Navas-Castillo, J. Sweepoviruses cause disease in sweet potato and related Ipomoea spp.: Fulfilling Koch’s postulates for a divergent group in the genus begomovirus. PLoS ONE 2011, 6, e27329. [Google Scholar] [CrossRef] [Green Version]
- Wise, A.A.; Liu, Z.; Binns, A.N. Three methods for the introduction of foreign DNA into Agrobacterium. Methods Mol. Biol. (Cliftonn. J.) 2006, 343, 43–53. [Google Scholar] [CrossRef]
- Dower, W.J.; Miller, J.F.; Ragsdale, C.W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988, 16, 6127–6145. [Google Scholar] [CrossRef] [Green Version]
- Mersereau, M.; Pazour, G.J.; Das, A. Efficient transformation of Agrobacterium tumefaciens by electroporation. Gene 1990, 90, 149–151. [Google Scholar] [CrossRef]
- Nair, G.R.; Liu, Z.; Binns, A.N. Reexamining the role of the accessory plasmid pAtC58 in the virulence of Agrobacterium tumefaciens strain C58. Plant Physiol. 2003, 133, 989–999. [Google Scholar] [CrossRef] [Green Version]
- An, G. Binary TI vectors for plant transformation and promoter analysis. Methods Enzymol. 1987, 153, 292–293. [Google Scholar] [CrossRef]
- Chen, H.; Nelson, R.S.; Sherwood, J.L. Enhanced recovery of transformants of Agrobacterium tumefaciens after freeze-thaw transformation and drug selection. Biotechniques 1994, 16, 664–670. [Google Scholar]
- Yusop, M.; Saad, M.; Talip, N.; Baharum, S.N.; Bunawan, H. A Review on Viruses Infecting Taro (Colocasia esculenta (L.) Schott). Pathogens 2019, 8, 56. [Google Scholar] [CrossRef] [Green Version]
- Waters, V.L. Conjugative transfer in the dissemination of beta-lactam and aminoglycoside resistance. Front. Biosci. 1999, 4, D433–D456. [Google Scholar] [CrossRef] [Green Version]
- Srivastava, A.; Raj, S.K.; Kumar, S.; Snehi, S.K.; Kulshreshtha, A.; Hallan, V.; Pande, S.S. Molecular identification of Ageratum enation virus, betasatellite and alphasatellite molecules isolated from yellow vein diseased Amaranthus cruentus in India. Virus Genes 2013, 47, 584–590. [Google Scholar] [CrossRef]
- Xiong, Q.; Fan, S.; Wu, J.; Zhou, X. Ageratum yellow vein China virus Is a Distinct Begomovirus Species Associated with a DNAbeta Molecule. Phytopathology 2007, 97, 405–411. [Google Scholar] [CrossRef] [Green Version]
- Briddon, R.W.; Mansoor, S.; Bedford, I.D.; Pinner, M.S.; Markham, P.G. Clones of cotton leaf curl geminivirus induce symptoms atypical of cotton leaf curl disease. Virus Genes 2000, 20, 19–26. [Google Scholar] [CrossRef]
- Wu, J.; Zulfiqar, A.; Huang, C. Infectivity of Euphorbia leaf curl virus and interaction with Tomato yellow leaf curl China betasatellite. Arch. Virol. 2011, 156, 517–521. [Google Scholar] [CrossRef]
- Marwal, A.; Kumar, R.; Paul Khurana, S.M.; Gaur, R.K. Complete nucleotide sequence of a new geminivirus isolated from Vitis vinifera in India: A symptomless host of Grapevine red blotch virus. Virus Dis. 2019, 30, 106–111. [Google Scholar] [CrossRef]
- Wu, J.; Mugiira, R.B.; Zhou, X. Malvastrum leaf curl Guangdong virus is a distinct monopartite begomovirus. Plant Pathol. 2007, 56, 771–776. [Google Scholar] [CrossRef]
- Guo, W.; Jiang, T.; Zhang, X.; Li, G.; Zhou, X. Molecular variation of satellite DNA beta molecules associated with Malvastrum yellow vein virus and their role in pathogenicity. Appl. Environ. Microbiol. 2008, 74, 1909–1913. [Google Scholar] [CrossRef] [Green Version]
- Jiang, T.; Liu, P.; Liao, B.L.; Wu, J.X.; Huang, C.J. Malvastrum yellow vein Yunnan virus is amonopartite begomovirus. Acta Virol. 2010, 54, 21–26. [Google Scholar] [CrossRef]
- Zhang, H.; Ma, X.Y.; Qian, Y.J.; Zhou, X.P. Molecular characterization and infectivity of Papaya leaf curl China virus infecting tomato in China. J. Zhejiang Univ. Sci. B 2010, 11, 109–114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontenele, R.S.; Abreu, R.A.; Lamas, N.S.; Alves-Freitas, D.; Vidal, A.H.; Poppiel, R.R.; Melo, F.L.; Lacorte, C.; Martin, D.P.; Campos, M.A.; et al. Passion Fruit Chlorotic Mottle Virus: Molecular Characterization of a New Divergent Geminivirus in Brazil. Viruses 2018, 10, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, Q.; Fan, S.; Guo, X.; Zhou, X. Stachytarpheta leaf curl virus is a novel monopartite begomovirus species. Arch. Virol. 2005, 150, 2257–2270. [Google Scholar] [CrossRef] [PubMed]
- Choi, E.; Lee, G.; Park, J.; Lee, T.K.; Choi, H.S.; Lee, S. Molecular characterization and an infectious clone construction of sweet potato leaf curl virus (SPLCV) isolated from Korea. Acta Virol. 2012, 56, 187–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bi, H.; Zhang, P. Molecular characterization of two sweepoviruses from China and evaluation of the infectivity of cloned SPLCV-JS in Nicotiana benthamiana. Arch. Virol. 2012, 157, 441–454. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Xie, Y.; Zhou, X. Tobacco curly shoot virus DNAbeta Is Not Necessary for Infection but Intensifies Symptoms in a Host-Dependent Manner. Phytopathology 2005, 95, 902–908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, Y.; Jiang, T.; Zhou, X. Agroinoculation Shows Tobacco leaf curl Yunnan virus is a Monopartite Begomovirus. Eur. J. Plant Pathol. 2006, 115, 369–375. [Google Scholar] [CrossRef]
- Khan, A.J.; Akhtar, S.; Singh, A.K.; Al-Shehi, A.A.; Al-Matrushi, A.M.; Ammara, U.; Briddon, R.W. Recent evolution of a novel begomovirus causing tomato leaf curl disease in the Al-Batinah region of Oman. Arch. Virol. 2014, 159, 445–455. [Google Scholar] [CrossRef]
- Khan, A.J.; Akhtar, S.; Singh, A.K.; Briddon, R.W. A Distinct Strain of Tomato leaf curl Sudan virus Causes Tomato Leaf Curl Disease in Oman. Plant Dis. 2013, 97, 1396–1402. [Google Scholar] [CrossRef] [Green Version]
- Al-Saleh, M.A.; Al-Shahwan, I.M.; Brown, J.K.; Idris, A.M. Molecular characterization of a naturally occurring intraspecific recombinant begomovirus with close relatives widespread in southern Arabia. Virol. J. 2014, 11, 103. [Google Scholar] [CrossRef] [Green Version]
- Kon, T.; Dolores, L.M.; Murayama, A.; Bajet, N.B.; Hase, S.; Takahashi, H.; Ikegami, M. Genome organization of an infectious clone of Tomato leaf curl virus (Philippines), a new monopartite begomovirus. J. Phytopathol. 2002, 150, 587–591. [Google Scholar] [CrossRef]
- Ueda, S.; Kimura, T.; Onuki, M.; Hanada, K.; Iwanami, T. Three distinct groups of isolates of Tomato yellow leaf curl virus in Japan and construction of an infectious clone. J. Gen. Plant Pathol. 2004, 70, 232–238. [Google Scholar] [CrossRef]
- Romay, G.; Geraud-Pouey, F.; Chirinos, D.T.; Mahillon, M.; Gillis, A.; Mahillon, J.; Bragard, C. Tomato Twisted Leaf Virus: A Novel Indigenous New World Monopartite Begomovirus Infecting Tomato in Venezuela. Viruses 2019, 11, 327. [Google Scholar] [CrossRef] [Green Version]
- Davino, S.; Napoli, C.; Dellacroce, C.; Miozzi, L.; Noris, E.; Davino, M.; Accotto, G.P. Two new natural begomovirus recombinants associated with the tomato yellow leaf curl disease co-exist with parental viruses in tomato epidemics in Italy. Virus Res. 2009, 143, 15–23. [Google Scholar] [CrossRef]
- Heydarnejad, J.; Kamali, M.; Hassanvand, V.; Massumi, H.; Shamshiri, M.; Varsani, A. Turnip leaf curl disease associated with two begomoviruses in south-eastern Iran. Trop. Plant Pathol. 2018, 43, 165–169. [Google Scholar] [CrossRef]
- Just, K.; Leke, W.N.; Sattar, M.N.; Luik, A.; Kvarnheden, A. Detection of tomato yellow leaf curl virus in imported tomato fruit in northern europe. Plant Pathol. 2014, 63, 1454–1460. [Google Scholar] [CrossRef]
- Salati, R.; Nahkla, M.K.; Rojas, M.R.; Guzman, P.; Jaquez, J.; Maxwell, D.P.; Gilbertson, R.L. Tomato yellow leaf curl virus in the Dominican Republic: Characterization of an Infectious Clone, Virus Monitoring in Whiteflies, and Identification of Reservoir Hosts. Phytopathology 2002, 92, 487–496. [Google Scholar] [CrossRef] [Green Version]
- Urbino, C.; Thébaud, G.; Granier, M.; Blanc, S.; Peterschmitt, M. A novel cloning strategy for isolating, genotyping and phenotyping genetic variants of geminiviruses. Virol. J. 2008, 5, 135. [Google Scholar] [CrossRef] [Green Version]
- García-Andrés, S.; Monci, F.; Navas-Castillo, J.; Moriones, E. Begomovirus genetic diversity in the native plant reservoir Solanum nigrum: Evidence for the presence of a new virus species of recombinant nature. Virology 2006, 350, 433–442. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Gong, H.; Zhou, X. Molecular characterization and pathogenicity of tomato yellow leaf curl virus in China. Virus Genes 2009, 39, 249–255. [Google Scholar] [CrossRef]
- Packialakshmi, R.M.; Usha, R. A simple and efficient method for agroinfection of Vernonia cinerea with infectious clones of Vernonia yellow vein virus. Virus Genes 2011, 43, 465–470. [Google Scholar] [CrossRef]
- Garrido-Ramirez, E.R.; Sudarshana, M.R.; Gilbertson, R.L. Bean golden yellow mosaic virus from Chiapas, Mexico: Characterization, Pseudorecombination with Other Bean-Infecting Geminiviruses and Germ Plasm Screening. Phytopathology 2000, 90, 1224–1232. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhou, X. Molecular characterization and experimental host-range of two begomoviruses infecting Clerodendrum cyrtophyllum in China. Virus Genes 2010, 41, 250–259. [Google Scholar] [CrossRef]
- Blawid, R.; Fontenele, R.S.; Lacorte, C.; Ribeiro, S.G. Molecular and biological characterization of corchorus mottle virus, a new begomovirus from Brazil. Arch. Virol. 2013, 158, 2603–2609. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Kulshreshtha, A.; Roshan, P.; Hallan, V. Molecular characterization and infectivity analysis of a bipartite begomovirus associated with cotton leaf curl Multan betasatellite naturally infecting Rumex nepalensis in northern India. J. Plant Pathol. 2019, 101, 935–941. [Google Scholar] [CrossRef]
- Jailani, A.; Kumar, A.; Mandal, B.; Sivasudha, T.; Roy, A. Agroinfection of tobacco by croton yellow vein mosaic virus and designing of a replicon vector for expression of foreign gene in plant. Virus Dis. 2016, 27, 277–286. [Google Scholar] [CrossRef] [Green Version]
- Hagen, C.; Rojas, M.R.; Sudarshana, M.R.; Xoconostle-Cazares, B.; Natwick, E.T.; Turini, T.A.; Gilbertson, R.L. Biology and Molecular Characterization of Cucurbit leaf crumple virus, an Emergent Cucurbit-Infecting Begomovirus in the Imperial Valley of California. Plant Dis. 2008, 92, 781–793. [Google Scholar] [CrossRef] [Green Version]
- Ndunguru, J.; Legg, J.; Fofana, B.; Aveling, T.; Thompson, G.; Fauquet, C. Identification of a defective molecule derived from DNA-A of the bipartite begomovirus of East African Cassava Mosaic Virus. Plant Pathol. 2006, 55, 2–10. [Google Scholar] [CrossRef]
- Gao, S.; Qu, J.; Chua, N.H.; Ye, J. A new strain of Indian cassava mosaic virus causes a mosaic disease in the biodiesel crop Jatropha curcas. Arch. Virol. 2010, 155, 607–612. [Google Scholar] [CrossRef]
- Polston, J.E.; Londoño, M.A.; Capobianco, H. The complete genome sequence of New World jatropha mosaic virus. Arch. Virol. 2014, 159, 3131–3136. [Google Scholar] [CrossRef]
- Ilyas, M.; Qazi, J.; Mansoor, S.; Briddon, R.W. Molecular characterisation and infectivity of a "Legumovirus" (genus Begomovirus: Family Geminiviridae) infecting the leguminous weed Rhynchosia minima in Pakistan. Virus Res. 2009, 145, 279–284. [Google Scholar] [CrossRef]
- Kumar, S.; Tanti, B.; Patil, B.L.; Mukherjee, S.K.; Sahoo, L. RNAi-derived transgenic resistance to Mungbean yellow mosaic India virus in cowpea. PLoS ONE 2017, 12, e0186786. [Google Scholar] [CrossRef]
- Haq, Q.M.; Rouhibakhsh, A.; Ali, A.; Malathi, V.G. Infectivity analysis of a blackgram isolate of Mungbean yellow mosaic virus and genetic assortment with MYMIV in selective hosts. Virus Genes 2011, 42, 429–439. [Google Scholar] [CrossRef] [PubMed]
- Shakir, S.; Nawaz-ul-Rehman, M.S.; Mubin, M.; Ali, Z. Characterization, phylogeny and recombination analysis of Pedilanthus leaf curl virus-Petunia isolate and its associated betasatellite. Virol. J. 2018, 15. [Google Scholar] [CrossRef] [PubMed]
- Koeda, S.; Homma, K.; Tanaka, Y.; Onizaki, D.; Kesumawati, E.; Zakaria, S. Inoculation of capsicums with pepper yellow leaf curl indonesia virus by combining agroinoculation and grafting. Hortic. J. 2018, 87, 364–371. [Google Scholar] [CrossRef] [Green Version]
- Urbino, C.; Polston, J.E.; Patte, C.P.; Caruana, M.L. Characterization and genetic diversity of potato yellow mosaic virus from the Caribbean. Arch. Virol. 2004, 149, 417–424. [Google Scholar] [CrossRef]
- Al-Aqeel, H.A.; Iqbal, Z.; Polston, J.E. Characterization of sida golden mottle virus isolated from Sida santaremensis Monteiro in Florida. Arch. Virol. 2018, 163, 2907–2911. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Tovar, R.; Navas-Castillo, J. Deciphering the biology of deltasatellites from the New World: Maintenance by New World begomoviruses and whitefly transmission. New Phytol. 2016, 212, 680–692. [Google Scholar] [CrossRef] [Green Version]
- Kon, T.; Dolores, L.M.; Bajet, N.B.; Hase, S.; Takahashi, H.; Ikegami, M. Molecular characterisation of a strain of Squash leaf curl China virus from the Philippines. J. Phytopathol. 2003, 151, 535–539. [Google Scholar] [CrossRef]
- Ferreira, P.T.O.; Lemos, T.O.; Nagata, T.; Inoue-Nagata, A.K. One-step cloning approach for construction of agroinfectious begomovirus clones. J. Virol. Methods 2008, 147, 351–354. [Google Scholar] [CrossRef]
- Malik, A.H.; Briddon, R.W.; Mansoor, S. Infectious clones of Tomato leaf curl Palampur virus with a defective DNA B and their pseudo-recombination with Tomato leaf curl New Delhi virus. Virol. J. 2011, 8. [Google Scholar] [CrossRef] [Green Version]
- Guo, W.; Yang, X.; Xie, Y.; Cui, X.; Zhou, X. Tomato yellow leaf curl Thailand virus-[Y72] from Yunnan is a monopartite begomovirus associated with DNAbeta. Virus Genes 2009, 38, 328–333. [Google Scholar] [CrossRef]
- Fiallo-Olivé, E.; Hernández-Zepeda, C.; Trejo-Saavedra, D.; Carillo-Tripp, J.; Rivera-Bustamante, R.F.; Martinez-Zubiaur, Y. Complete genome and pathogenicity of Tomato yellow leaf distortion virus, a bipartite begomovirus infecting tomato in Cuba. Eur. J. Plant Pathol. 2012, 134, 13–21. [Google Scholar] [CrossRef]
- Zaim, M.; Kumar, Y.; Hallan, V.; Zaidi, A.A. Velvet bean severe mosaic virus: A distinct begomovirus species causing severe mosaic in Mucuna pruriens (L.) DC. Virus Genes 2011, 43, 138–146. [Google Scholar] [CrossRef]
- Al-Musa, A.; Anfoka, G.; Al-Abdulat, A.; Misbeh, S.; Haj Ahmed, F.; Otri, I. Watermelon chlorotic stunt virus (WmCSV): A serious disease threatening watermelon production in Jordan. Virus Genes 2011, 43, 79–89. [Google Scholar] [CrossRef]
- Collins, A.M.; Mujaddad-ur-Rehman, M.; Brown, J.K.; Reddy, C.; Wang, A.; Fondong, V.; Roye, M.E. Molecular characterization and experimental host range of an isolate of Wissadula golden mosaic St. Thomas virus. Virus Genes 2009, 39, 387–395. [Google Scholar] [CrossRef]
- Boulton, M.I. Construction of infectious clones for DNA viruses: Mastreviruses. Methods Mol. Biol. 2008, 451, 503–523. [Google Scholar] [CrossRef]
- Bunawan, H.; Dusik, L.; Bunawan, S.N.; Amin, N.M. Rice Tungro Disease: From Identification to Disease Control. World Appl. Sci. J. 2014, 31, 1221–1226. [Google Scholar] [CrossRef]
- Biswas, K.K.; Varma, A. Agroinoculation: A method of screening germplasm resistance to mungbean yellow mosaic Geminivirus. Indian Phytopathol. 2001, 54, 240–245. [Google Scholar]
- Picó, B.; Díez, M.J.; Nuez, F. Viral diseases causing the greatest economic losses to the tomato crop. II. The Tomato yellow leaf curl virus—A review. Sci. Hortic. 1996, 67, 151–196. [Google Scholar] [CrossRef]
- Brewer, H.C.; Hird, D.L.; Bailey, A.M.; Seal, S.E.; Foster, G.D. A guide to the contained use of plant virus infectious clones. Plant Biotechnol. J. 2018, 16, 832–843. [Google Scholar] [CrossRef]
- Lange, M.; Yellina, A.L.; Orashakova, S.; Becker, A. Virus-induced gene silencing (VIGS) in plants: An overview of target species and the virus-derived vector systems. Methods Mol. Biol. 2013, 975, 1–14. [Google Scholar] [CrossRef]
- Lee, W.S.; Hammond-Kosack, K.E.; Kanyuka, K. Barley stripe mosaic virus-mediated tools for investigating gene function in cereal plants and their pathogens: Virus-induced gene silencing, host-mediated gene silencing, and virus-mediated overexpression of heterologous protein. Plant Physiol. 2012, 160, 582–590. [Google Scholar] [CrossRef] [Green Version]
- Moury, B.; Morel, C.; Johansen, E.; Guilbaud, L.; Souche, S.; Ayme, V.; Caranta, C.; Palloix, A.; Jacquemond, M. Mutations in potato virus Y genome-linked protein determine virulence toward recessive resistances in Capsicum annuum and Lycopersicon hirsutum. Mol. Plant-Microbe Interact. Mpmi 2004, 17, 322–329. [Google Scholar] [CrossRef] [Green Version]
- Roshan, P.; Kulshreshtha, A.; Kumar, S.; Purohit, R.; Hallan, V. AV2 protein of tomato leaf curl Palampur virus promotes systemic necrosis in Nicotiana benthamiana and interacts with host Catalase2. Sci. Rep. 2018, 8, 1273. [Google Scholar] [CrossRef] [Green Version]
- Saeed, F.; Naeem, M.; Hameed, U.; Ilyas, M.; Saleem, M.; Hamza, M.; Mansoor, S.; Amin, I. Infectivity of okra enation leaf curl virus and the role of its V2 protein in pathogenicity. Virus Res. 2018, 255, 90–94. [Google Scholar] [CrossRef]
- Hipp, K.; Rau, P.; Schäfer, B.; Gronenborn, B.; Jeske, H. The RXL motif of the African cassava mosaic virus Rep protein is necessary for rereplication of yeast DNA and viral infection in plants. Virology 2014, 462–463, 189–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stanley, J. Analysis of African cassava mosaic virus recombinants suggests strand nicking occurs within the conserved nonanucleotide motif during the initiation of rolling circle DNA replication. Virology 1995, 206, 707–712. [Google Scholar] [CrossRef] [Green Version]
- Chandran, S.A.; Jeyabharathy, C.; Usha, R. The C2 protein of Bhendi yellow vein mosaic virus plays an important role in symptom determination and virus replication. Virus Genes 2014, 48, 203–207. [Google Scholar] [CrossRef] [PubMed]
- Stanley, J.; Latham, J.R.; Pinner, M.S.; Bedford, I.; Markham, P.G. Mutational analysis of the monopartite geminivirus beet curly top virus. Virology 1992, 191, 396–405. [Google Scholar] [CrossRef]
- Baliji, S.; Sunter, J.; Sunter, G. Transcriptional analysis of complementary sense genes in Spinach curly top virus and functional role of C2 in pathogenesis. Mol. Plant-Microbe Interact. Mpmi 2007, 20, 194–206. [Google Scholar] [CrossRef] [Green Version]
- Iqbal, Z.; Sattar, M.N.; Kvarnheden, A.; Mansoor, S.; Briddon, R.W. Effects of the mutation of selected genes of Cotton leaf curl Kokhran virus on infectivity, symptoms and the maintenance of Cotton leaf curl Multan betasatellite. Virus Res. 2012, 169, 107–116. [Google Scholar] [CrossRef]
- Jupin, I.; De Kouchkovsky, F.; Jouanneau, F.; Gronenborn, B. Movement of tomato yellow leaf curl geminivirus (TYLCV): Involvement of the protein encoded by ORF C4. Virology 1994, 204, 82–90. [Google Scholar] [CrossRef]
- Piroux, N.; Saunders, K.; Page, A.; Stanley, J. Geminivirus pathogenicity protein C4 interacts with Arabidopsis thaliana shaggy-related protein kinase AtSKeta, a component of the brassinosteroid signalling pathway. Virology 2007, 362, 428–440. [Google Scholar] [CrossRef]
- Rojas, M.R.; Jiang, H.; Salati, R.; Xoconostle-Cázares, B.; Sudarshana, M.R.; Lucas, W.J.; Gilbertson, R.L. Functional analysis of proteins involved in movement of the monopartite begomovirus, Tomato yellow leaf curl virus. Virology 2001, 291, 110–125. [Google Scholar] [CrossRef] [Green Version]
- Etessami, P.; Saunders, K.; Watts, J.; Stanley, J. Mutational analysis of complementary-sense genes of African cassava mosaic virus DNA A. J. Gen. Virol. 1991, 72 Pt 5, 1005–1012. [Google Scholar] [CrossRef]
- Hong, Y.; Stanley, J. Regulation of African cassava mosaic virus complementary-sense gene expression by N-terminal sequences of the replication-associated protein AC1. J. Gen. Virol. 1995, 76 Pt 10, 2415–2422. [Google Scholar] [CrossRef]
- Pooma, W.; Petty, I.T. Tomato golden mosaic virus open reading frame AL4 is genetically distinct from its C4 analogue in monopartite geminiviruses. J. Gen. Virol. 1996, 77 Pt 8, 1947–1951. [Google Scholar] [CrossRef]
- Chellappan, P.; Vanitharani, R.; Fauquet, C.M. MicroRNA-binding viral protein interferes with Arabidopsis development. Proc. Natl. Acad. Sci. USA 2005, 102, 10381–10386. [Google Scholar] [CrossRef] [Green Version]
- Vanitharani, R.; Chellappan, P.; Pita, J.S.; Fauquet, C.M. Differential roles of AC2 and AC4 of cassava geminiviruses in mediating synergism and suppression of posttranscriptional gene silencing. J. Virol. 2004, 78, 9487–9498. [Google Scholar] [CrossRef] [Green Version]
- Mansoor, S.; Zafar, Y.; Briddon, R.W. Geminivirus disease complexes: The threat is spreading. Trends Plant Sci. 2006, 11, 209–212. [Google Scholar] [CrossRef]
- Jose, J.; Usha, R. Bhendi yellow vein mosaic disease in India is caused by association of a DNA Beta satellite with a begomovirus. Virology 2003, 305, 310–317. [Google Scholar] [CrossRef] [Green Version]
- Cui, X.; Li, G.; Wang, D.; Hu, D.; Zhou, X. A Begomovirus DNAbeta-encoded protein binds DNA, functions as a suppressor of RNA silencing, and targets the cell nucleus. J. Virol. 2005, 79, 10764–10775. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Guo, W.; Ma, X.; An, Q.; Zhou, X. Molecular characterization of Tomato leaf curl China virus, infecting tomato plants in China, and functional analyses of its associated betasatellite. Appl. Environ. Microbiol. 2011, 77, 3092–3101. [Google Scholar] [CrossRef] [Green Version]
- Ariyo, O.A.; Atiri, G.I.; Dixon, A.G.; Winter, S. The use of biolistic inoculation of cassava mosaic begomoviruses in screening cassava for resistance to cassava mosaic disease. J. Virol. Methods 2006, 137, 43–50. [Google Scholar] [CrossRef]
- Briddon, R.W.; Liu, S.; Pinner, M.S.; Markham, P.G. Infectivity of African cassava mosaic virus clones to cassava by biolistic inoculation. Arch. Virol. 1998, 143, 2487–2492. [Google Scholar] [CrossRef]
- Berrie, L.C.; Rybicki, E.P.; Rey, M. Complete nucleotide sequence and host range of South African cassava mosaic virus: Further evidence for recombination amongst begomoviruses. J. Gen. Virol. 2001, 82 Pt 1, 53–58. [Google Scholar] [CrossRef]
- Ariyo, O.A.; Dixon, A.G.; Atiri, G.I. Whitefly Bemisia tabaci (Homoptera: Aleyrodidae) infestation on cassava genotypes grown at different ecozones in Nigeria. J. Econ. Entomol. 2005, 98, 611–617. [Google Scholar] [CrossRef]
- Lapidot, M.; Friedmann, M. Breeding for resistance to whitefly-transmitted geminviruses. Ann. Appl. Biol. 2005, 140, 109–127. [Google Scholar] [CrossRef]
- Ayeh, K.O.; Ramsell, J.N.E. The use of biolistic inoculation of cassava mosaic begomoviruses in screening cassava cultivars from Ghana for resistance/susceptibility to cassava mosaic disease. Afr. J. Biotechnol. 2008, 7, 2353–2358. [Google Scholar]
- Ferro, M.M.M.; Ramos-Sobrinho, R.; Xavier, C.A.D.; Zerbini, F.M.; Lima, G.S.A.; Nagata, T.; Assunção, I.P. New approach for the construction of infectious clones of a circular DNA plant virus using Gibson Assembly. J. Virol. Methods 2019, 263, 20–23. [Google Scholar] [CrossRef] [PubMed]
- Cooper, B. Proof by synthesis of Tobacco mosaic virus. Genome Biol. 2014, 15, 67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lovato, A.; Faoro, F.; Gambino, G.; Maffi, D.; Bracale, M.; Polverari, A.; Santi, L. Construction of a synthetic infectious cDNA clone of Grapevine Algerian latent virus (GALV-Nf) and its biological activity in Nicotiana benthamiana and grapevine plants. Virol. J. 2014, 11, 186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pasin, F.; Menzel, W.; Daròs, J.-A. Harnessed viruses in the age of metagenomics and synthetic biology: An update on infectious clone assembly and biotechnologies of plant viruses. Plant Biotechnol. J. 2019, 17, 1010–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bornancini, V.A.; Irazoqui, J.M.; Flores, C.R.; Vaghi Medina, C.G.; Amadio, A.F.; López Lambertini, P.M. Reconstruction and characterization of full-length begomovirus and alphasatellite genomes infecting pepper through metagenomics. Viruses 2020, 12, 202. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fakhfakh, H.; Vilaine, F.; Makni, M.; Robaglia, C. Cell-free cloning and biolistic inoculation of an infectious cDNA of potato virus Y. J. Gen. Virol. 1996, 77, 519–523. [Google Scholar] [CrossRef]
- Jailani, A.A.K.; Shilpi, S.; Mandal, B. Rapid demonstration of infectivity of a hybrid strain of potato virus Y occurring in India through overlapping extension PCR. Physiol. Mol. Plant Pathol. 2017, 98, 62–68. [Google Scholar] [CrossRef]
- Youssef, F.; Marais, A.; Faure, C.; Gentit, P.; Candresse, T. Strategies to facilitate the development of uncloned or cloned infectious full-length viral cDNAs: Apple chlorotic leaf spot virus as a case study. Virol. J. 2011, 8. [Google Scholar] [CrossRef] [Green Version]
- Roy, A.; Zhai, Y.; Ortiz, J.; Neff, M.; Mandal, B.; Mukherjee, S.K.; Pappu, H.R. Multiplexed editing of a begomovirus genome restricts escape mutant formation and disease development. PLoS ONE 2019, 14. [Google Scholar] [CrossRef] [Green Version]
- Khan, S.; Mahmood, M.S.; Ur Rahman, S.; Rizvi, F.; Ahmad, A. Evaluation of the CRISPR/Cas9 system for the development of resistance against Cotton leaf curl virus in model plants. Plant Protect. Sci. 2020, 56, 154–162. [Google Scholar] [CrossRef]
| Genome Structure | Virus | Genome Amplification | Cloning Vector | Transformation Method | Plant (Description) | Reference |
|---|---|---|---|---|---|---|
| Monopartite | Ageratum enation virus (AgEV) | RCA | pCambia2301 | Agroinoculation | Amaranthus cruentus (severe leaf curl, enation and yellow vein symptom) | [41] |
| N. benthamiana (severe leaf curl, enation and yellow vein symptom) | ||||||
| N. glutinosa (severe leaf curl, enation and yellow vein symptom) | ||||||
| Monopartite | Ageratum yellow vein China virus (AYVCNV) | RCA | pBin19 | Agroinoculation | N. benthamiana (severe downward leaf curl, leaf crinkle, stunting and vein swelling) | [42] |
| N. glutinosa (leaf curling and distortion) | ||||||
| P. hybrida (leaf curling and distortion) | ||||||
| L. esculentum (leaf curling and distortion) | ||||||
| Ageratum conyzoides (yellow vein) | ||||||
| Monopartite | Ageratum yellow vein Taiwan virus (AYVTV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (vein yellowing, leaf curling) | [30] |
| Monopartite | Cotton leaf curl virus (CLCuV) | PCR | pBin19 | Biolistic | Cotton (vein swelling, vein darkening, leaf curling, and enations) | [43] |
| N. benthamiana (mild stunting and yellowing) | ||||||
| Monopartite | Euphorbia leaf curl virus (EuLCV) | PCR | pBinPLUS | Agroinoculation | P. hybrida (severe leaf curling and crinkling, vein swelling and stunting) | [44] |
| N. benthamiana (severe upward leaf curling, vein swelling, and stunting) | ||||||
| N. glutinosa (vein swelling, stunting and downward leaf curling) | ||||||
| Solanum lycopersicum (severe leaf crinkling, vein swelling and stunting) | ||||||
| Monopartite | Grapevine red blotch virus (GRBV) | RCA | pCambia2301 | Agroinoculation | Chenopodium album (symptomless) | [45] |
| Lycopersicon esculentum (symptomless) | ||||||
| Monopartite | Malvastrum leaf curl Guandong virus (MLCuGdV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (yellow vein, leaf curling, vein swelling and stunting symptoms) | [46] |
| N. glutinosa (leaf curling, vein swelling and stunting) | ||||||
| P. hybrida (leaf curling, vein swelling and stunting) | ||||||
| Monopartite | Malvastrum yellow vein virus (MYVV) | PCR | pLH9000 | Agroinoculation | N. benthamiana (downward curling of the leaves, yellow vein and leaf crinkling) | [47] |
| N. glutinosa (downward curling of the leaves, yellow vein and leaf crinkling) | ||||||
| P. hybrida (downward curling of the leaves, yellow vein and leaf crinkling) | ||||||
| M. coromandelianum (yellow vein) | ||||||
| Monopartite | Malvastrum yellow vein Yunnan virus (MYVYNV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (yellow vein, vein thickening and upward leaf curl symptoms) | [48] |
| Monopartite | Papaya leaf curl China virus (PaLCuCNV) | PCR | pBinPLUS | Agroinoculation | S. lycopersicum (leaf downward curling and crinkling) | [49] |
| N. benthamiana (leaf upward curling and crinkling) | ||||||
| N. tabacum (leaf curling and crinkling) | ||||||
| N. glutinosa (leaf downward curling and crinkling) | ||||||
| P. hybrida (leaf upward curling and crinkling) | ||||||
| Monopartite | Passion fruit chlorotic mottle virus (PCMoV) | RCA | pJETt 1.2 | Agroinoculation | N. benthamiana (chlorotic spots, mottle and growth impairment) | [50] |
| Arabidopsis thaliana (mild symptom) | ||||||
| Passiflora edulis (mild symptom) | ||||||
| Monopartite | Stachytarpheta leaf curl virus (StaLCV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (upward leaf curling, swelling of the veins and stunting) | [51] |
| N. tabacum (upward curling and vein swelling) | ||||||
| Lycopersicon esculentum (mild leaf distortion, puckering and leaf crinkling) | ||||||
| P. hybrida (severe upward leaf curling and vein swelling) | ||||||
| Monopartite | Sweet potato leaf curl virus (SPLCV) | PCR | pGEM-T easy | Biolistic | Sweet potatoes (mild leaf curl) | [52] |
| Monopartite | Sweet potato leaf curl virus-Jiangsu (SPLCV-JS) & | RCA | pPZP100 | Agroinoculation | Sweet potato: golden vein, mosaic and leaf curling symptoms | [53] |
| I. setosa: mosaic, chlorosis and severe downward leaf curling | ||||||
| N. benthamiana (downward leaf curling) | ||||||
| Monopartite | Tobacco curly shoot virus (TbCSV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (severe upward leaf curl, vein swelling and stunting) | [54] |
| N. glutinosa (downward leaves curling) | ||||||
| N. tabacum (mild downward curling, puckering of leaves and vein thickening) | ||||||
| Monopartite | Tobacco leaf curl Yunnan virus (TbLCYNV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (severe upward leaf curling and vein thickening) | [55] |
| P. hybrida (severe upward leaf curling and stunting) | ||||||
| N. glutinosa (slight leaf curling and vein thickening) | ||||||
| Monopartite | Tomato leaf curl Al-Batinah virus (ToLCABV) | RCA | pCambia1301 | Agroinoculation | N. benthamiana (downward leaf curl, leaf crumpling, vein swelling, interveinal yellowing, a reduced leaf size for young leaves and stunting of plants) | [56] |
| Tomato (interveinal chlorosis, leaf curling or crumpling) | ||||||
| Monopartite | Tomato leaf curl Sudan virus (ToLCSDV) | RCA | pGreen0029 | Agroinoculation | N. benthamiana (vein swelling and severe upward leaf curling) | [57] |
| Tomato (mild downward curling, crumpling and interveinal yellowing) | ||||||
| Monopartite | Tomato leaf curl virus (ToLCV) | RCA | Pg-Gez3 | Agroinoculation | N. benthamiana (stunting, leaf curling, vein thickening, stem deformation | [58] |
| Tomato (upward curling, reduced size of new leaflets and mild vein yellowing) | ||||||
| Monopartite | Tomato leaf curl virus (Philippines) (ToLCV-Ph) | PCR | pBI121 | Agroinoculation | Tomato (curling) | [59] |
| N. benthamiana (wrinkling) | ||||||
| Monopartite | Tomato leaf curl virus (TYLCV) | PCR | pBISzYI | Agroinoculation | N. benthamiana (upward leaf curl and plant stunting) | [60] |
| N. glutinosa (stunting and mild leaf curling) | ||||||
| Tomato (mild leaf curl) | ||||||
| Monopartite | Tomato twisted leaf virus (ToTLV) | RCA | pCAMBIA1300 | Agroinoculation | Tomato (size reduction of young leaves, chlorosis and leaf deformation) | [61] |
| N. benthamiana (mosaic symptoms) | ||||||
| Monopartite | Tomato yellow leaf curl Sardinia virus (TYLCSV) | PCR | pBin19 | Agroinoculation | Tomato (downward leaf curl and rugosity) | [62] |
| N. benthamiana (downward curling, bubbling and interveinal yellowing) | ||||||
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | PCR | pGreen0.24 | Agroinoculation | Tomato seedlings (downward leaf curling, reduced leaf size and marginal leaf chlorosis) | [63] |
| Turnip seedling (outward leaf curling, swelling of veins, thick and brittle leaves) | ||||||
| Watermelon seedlings (severe stunting, severe leaf curling, leaf necrosis) | ||||||
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | PCR | pLH7000 | Agroinoculation | Tomato (leaf curling and distortion, yellowing of leaves and reduction in plant growth) | [64] |
| N. benthamiana (leaf curling and distortion, yellowing of leaves and reduction in plant growth) | ||||||
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | PCR | pGEM-5Zf+ | Agroinoculation | Tomato (stunted and erect, leaves curl upwards, crumple and distinctive interveinal chlorosis) | [65] |
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | PCR | pGreen | Agroinoculation | Tomato (yellow leaf curl and stunting) | [66] |
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | PCR | pBBR1MCS-5 | Agroinoculation | Solanum nigrum (yellowing, leaf distortion, dwarfing) | [67] |
| tomato (yellowing and distortion) | ||||||
| Common bean (yellowing) | ||||||
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (downward leaf curling and vein yellowing) | [68] |
| S. lycopersicum (downward leaf curling, yellowing and vein yellowing) | ||||||
| N. tabacum (downward leaf curling) | ||||||
| N. glutinosa (downward leaf curling) | ||||||
| P. hybrida (upward leaf curling) | ||||||
| Monopartite | Tomato yellow leaf curl virus (TYLCV) | RCA | pCambia0390 | Agroinoculation | Tomato (leaf curling, yellowing, and stunting) | [31] |
| Monopartite | Vernonia yellow vein virus (VeYVV) | RCA | pCambia2301 | Agroinoculation | Vernonia cinerea (vein yellowing) | [69] |
| Bipartite | Bean golden yellow mosaic virus (BGYMV) | PCR | pCGN1547 | Agroinoculation | Bean plants (moderate vein clearing and epinasty symptoms) | [70] |
| N. benthamiana (mild epinasty and leaf crumpling) | ||||||
| Bipartite | Clerodendrum golden mosaic China virus (ClGMCNV) | RCA | pBinPLUS | Agroinoculation | N. benthamiana (leaf crumpling and stunting) | [71] |
| N. glutinosa (chlorotic spots and leaf distortion) | ||||||
| N. tabacum (leaf distortion and stunting) | ||||||
| Petunia hybrida (symptomless) | ||||||
| Solanum lycopersicum (no infection) | ||||||
| Bipartite | Corchorus mottle virus (CoMoV) | RCA | pBluescript SK+ | Biolistic | Sida rhombifolia (systemic vein chlorosis, mottling and leaf deformation) | [72] |
| N. benthamiana and tomato (symptomless) | ||||||
| Bipartite | Cotton leaf curl Multan betasatellite (CLCuMB) | RCA | pCambia1300 | Agroinoculation | N. benthamiana (downward leaf curling, leaf puckering, chlorosis, reduction of the leaf lamina and shortening of internodes) Rumex nepalensis (no symptom) | [73] |
| Bipartite | Croton yellow vein mosaic virus (CYVMV) | PCR | pCambia2300 | Agroinoculation | Nicotiana tabacum (mild puckering and vein clearing) N. benthamiana (leaf curling, leaf rolling, and vein thickening) N. glutinosa (mild puckering on leaves) | [74] |
| Bipartite | Cucurbit leaf crumple virus (CuLCrV) | PCR | pZeroBlunt | Agroinoculation | Common bean (stunted growth, leaf curl and chlorosis) N. benthamiana (stunted growth, leaf curl and chlorosis) Pumpkin (stunted growth, leaf curl, leaf yellowing and mottling) Zucchini (stunted growth, leaf curl, leaf yellowing and mottling) | [75] |
| Bipartite | East African cassava mosaic Cameroon virus (EACMCV) | PCR | pSKDf | Biolistic | N.benthamiana (mosaic, leaf distortion and yellowing) | [76] |
| Bipartite | Indian cassava mosaic virus (ICMV) | PCR | pCambia1300 | Agroinoculation | N. benthamiana (plant stunting, downward leaf curling, yellow-green mosaic leaves) J. curcas (downward leaf curling, yellow-green mosaic, serration, leaf size reduction and plant stunting) | [77] |
| Bipartite | Jatropha mosaic virus (JaMV) | RCA | pBluescript KS (−) | Biolistic | Jatropha multifida (distortion, mosaic and necrosis) N. tabacum (chlorotic mottle and mild leaf curling) Phaseolus vulgaris (mosaic, leaf deformation and stunting) | [78] |
| Bipartite | Legume yellow mosaic virus (LYMV) | RCA | pBin19 | Agroinoculation | N. bentahamiana (no infection) Soybean (mild chlorotic spots) | [79] |
| Bipartite | Mungbean yellow mosaic India virus (MYMIV) | RCA | pCambia3300 | Agroinoculation | Cowpea (stunted growth, severe leaf curling, reduced leaf size, yellow patches and distortion of leaf lamina) Mungbean (stunted growth, severe leaf curling, reduced leaf size, yellow patches and distortion of leaf lamina) | [80] |
| Bipartite | Mungbean yellow mosaic virus (MYMV) | PCR | pBin19 | Agroinoculation | Blackgram (mosaic pattern) Mungbean (mosaic pattern) French bean (backward leaf curling and stunting) Cowpea (severe leaf curl and leaf distortion) | [81] |
| Bipartite | Pedilanthus leaf curl virus (PeLCV) | RCA | pCambia2300 | Agroinoculation | N. benthamiana (severe downward curling, swelling of leaves and vein yellowing) Petunia atkinsiana (upward curling and swelling of leaves) | [82] |
| Bipartite | Pepper yellow leaf curl Indonesia virus (PepYLCIV) | RCA | pGreen11 | Agroinoculation | N. benthamiana (leaf curling, yellowing, mottling and stunting) Tomato (no symptom) | [83] |
| Bipartite | Potato yellow mosaic virus (PYMV) | PCR | pBluescript SK (+) | Agroinoculation | Capsicum annuum (mild yellow mosaic and distortion) Datura stramonium (yellow spots and leaf distortion) N. benthamiana (mild chlorosis and curling) N. tabacum (mild chlorosis and curling) Petunia hybrida (mild chlorosis and leaf curling) Solanum tuberosum (bright yellow mosaic and distortion) | [84] |
| Bipartite | Sida golden mosaic virus (SiGMoV) | PCR | pBluescript KS (−) pLitmus28 | Biolistic | Common bean (mild mosaic and stunting) Cotton (asymptomatic) N. bethamiana (mosaic, leaf distortion and stunting N. tabacum (asymptomatic) Sida santaremensis (chlorotic mottle) Tomato (asymptomatic) | [85] |
| Bipartite | Sida golden yellow vein virus (SiGYVV) | RCA | pBluescript II SK (+) | Agroinoculation | N. benthamiana (chlorosis, leaf curling and severe stunting) Malvastrum coromandelianum (no symptom) Sidastrum micranthum (mild yellow mosaics) | [86] |
| Bipartite | Squash leaf curl China virus (SLCCNV) | PCR | pBI121 | Agroinoculation | Pumpkin (stunting and leaf curling) | [87] |
| Bipartite | Tomato golden vein virus (TGVV) | RCA | pCambia0380 | Agroinoculation | Tomato (vein yellowing) N. benthamiana (vein yellowing) | [88] |
| Bipartite | Tomato leaf curl Palampur virus (ToLCHnV) | PCR | pGreen 0029 | Biolistic | N. benthamiana (upward leaf curling and vein swelling) Muskmelon (leaves necrosis near the vein) | [89] |
| Bipartite | Tomato yellow leaf curl Thailand virus (TYLCTHV) | PCR | pBinPLUS | Agroinoculation | N. benthamiana (leaf curling and vein thickening) N. glutinosa (downward leaf curling) Solanum lycopersicum (downward leaf curling and mild yellowing) | [90] |
| Bipartite | Tomato yellow leaf distortion virus (ToYLDV) | RCA | pBluescript SK pGEM-T easy | Biolistic | Tomato (yellowing and distortion) Soybean (no symptom) N. tabacum (yellowing and distortion) N. benthamiana (yellowing and distortion) | [91] |
| Bipartite | Velvet bean severe mosaic virus (VbSMV) | RCA | pCambia1300 | Agroinoculation | N. benthamiana (downward leaf curling. Mottling and puckering) N. glutinosa (downward leaf curling, mottling and puckering) | [92] |
| Bipartite | Watermelon chlorotic stunt virus (WmCSV) | RCA | pCambia2300 | Agroinoculation | N. benthamiana (mild disease symptoms) Watermelon (mottling, yellowing and severe leaf curling) | [93] |
| Bipartite | Wissadula golden mosaic St Thomas virus (WGMSTV) | PCR | pCR2.1-TOPO | Biolistic | Red pea (yellowing) Wissadula amplissima (blistering, yellow patches) N. benthamiana (blistering, yellowing) Lycopersicon esculentum (chlorosis) | [94] |
| Gene Study | Virus | Strategy | Impact | Reference |
|---|---|---|---|---|
| AV2 | Tomato leaf curl Palampur virus (ToLCPalV) | AV2 mutant infectious clones | Systematic infection occurs in N. benthamiana and S. lycopersicum | [103] |
| Okra enation leaf virus (OELV) | AV2 mutant infectious clones | No necrotic and leaf curling symptom is observed in N. benthamiana | [104] | |
| AC1 | African Cassava mosaic virus (ACMV) | RXL mutant infectious clones | No symptom induction in N. benthamiana. | [105] |
| C2 | Bhendi yellow mosaic virus (BYMV) | C2 mutant infectious clones | Abolished symptom and reduced viral load in N. benthamiana. | [107] |
| AC4 | African Cassava mosaic virus (ACMV) | AC4 mutant infectious clones | Full symptom and infectivity on the infected plants. | [114,115] |
| Tomato golden mosaic virus (TGMV) | AC4 mutant infectious clones | No infection and symptomless effect on the infected N. benthamiana. | [116] | |
| BC1 | Tomato leaf curl China virus (ToLCCNV) | ToLCCNB infectious clones | Asymptomatic infection occurs in N. benthamiana. | [122] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saad, M.F.M.; Sau, A.R.; Akbar, M.A.; Baharum, S.N.; Ramzi, A.B.; Talip, N.; Bunawan, H. Construction of Infectious Clones of Begomoviruses: Strategies, Techniques and Applications. Biology 2021, 10, 604. https://doi.org/10.3390/biology10070604
Saad MFM, Sau AR, Akbar MA, Baharum SN, Ramzi AB, Talip N, Bunawan H. Construction of Infectious Clones of Begomoviruses: Strategies, Techniques and Applications. Biology. 2021; 10(7):604. https://doi.org/10.3390/biology10070604
Chicago/Turabian StyleSaad, Mohd Faiz Mat, Aziz Ramlee Sau, Muhamad Afiq Akbar, Syarul Nataqain Baharum, Ahmad Bazli Ramzi, Noraini Talip, and Hamidun Bunawan. 2021. "Construction of Infectious Clones of Begomoviruses: Strategies, Techniques and Applications" Biology 10, no. 7: 604. https://doi.org/10.3390/biology10070604
APA StyleSaad, M. F. M., Sau, A. R., Akbar, M. A., Baharum, S. N., Ramzi, A. B., Talip, N., & Bunawan, H. (2021). Construction of Infectious Clones of Begomoviruses: Strategies, Techniques and Applications. Biology, 10(7), 604. https://doi.org/10.3390/biology10070604







