A Gut-Ex-Vivo System to Study Gut Inflammation Associated to Inflammatory Bowel Disease (IBD)
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Reagents and Materials
2.2. Silicone-Based Device and Organ Culture
2.3. Colon Cultures and Treatments
2.4. Tissue Viability Assay
2.5. Quantitative PCR (qPCR)
2.6. Western Blotting Analysis
2.7. ELISA
2.8. Hematoxylin/Eosin Staining
2.9. TUNEL
2.10. Statistical Analysis
3. Results
3.1. Ex Vivo Culturing and Viability of Colon Tissues—GEVS
3.2. DNBS Induces Tissue Stress and Compromises the Permeability Barrier of Colon Cultivated in a GEVS
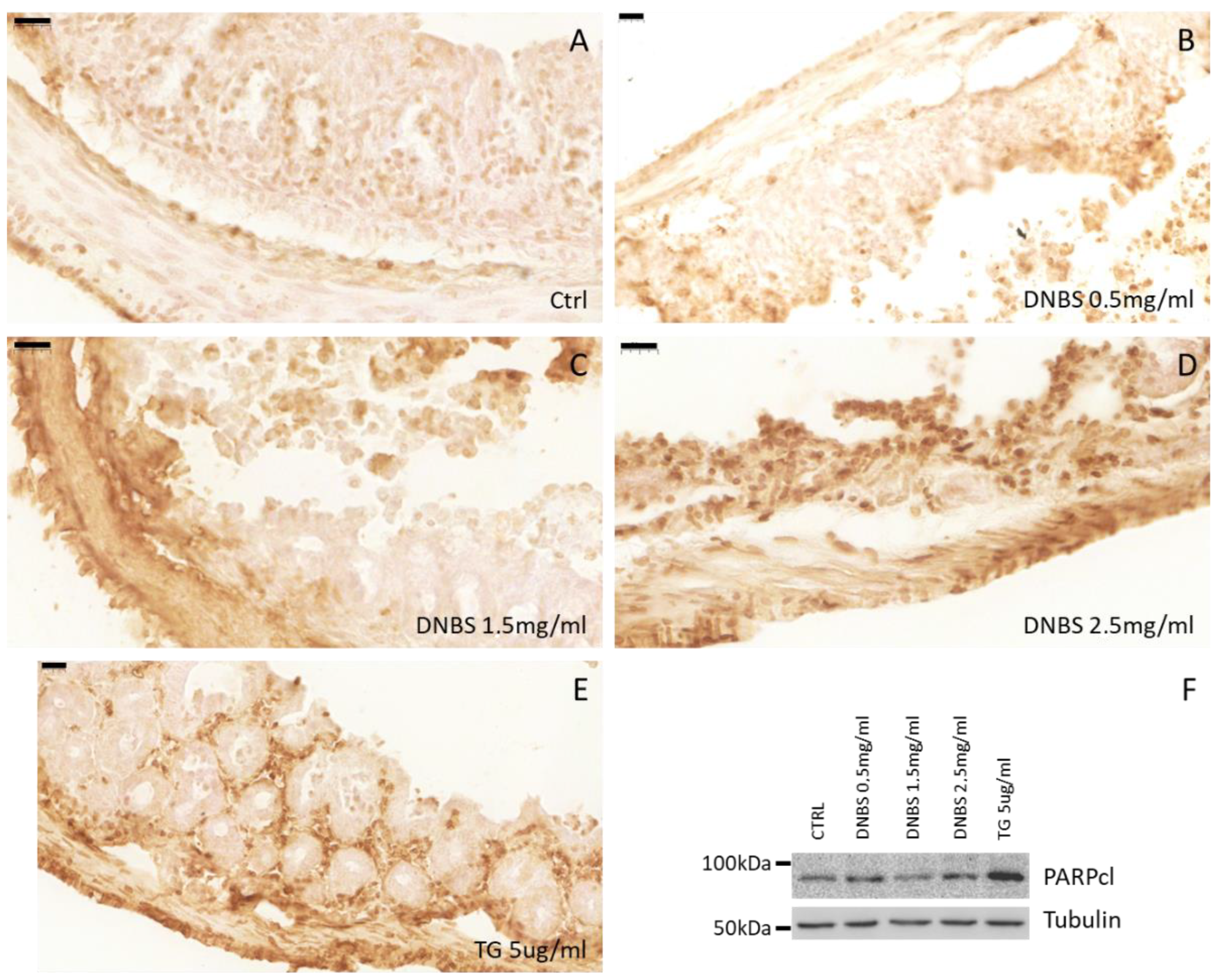
3.3. DNBS Induces ER Stress in Colon Cultivated in a GEVS
3.4. The Expression of Cytokines Associated to IBD Development Are Modulated by DNBS in Tissues Cultivated in a GEVS
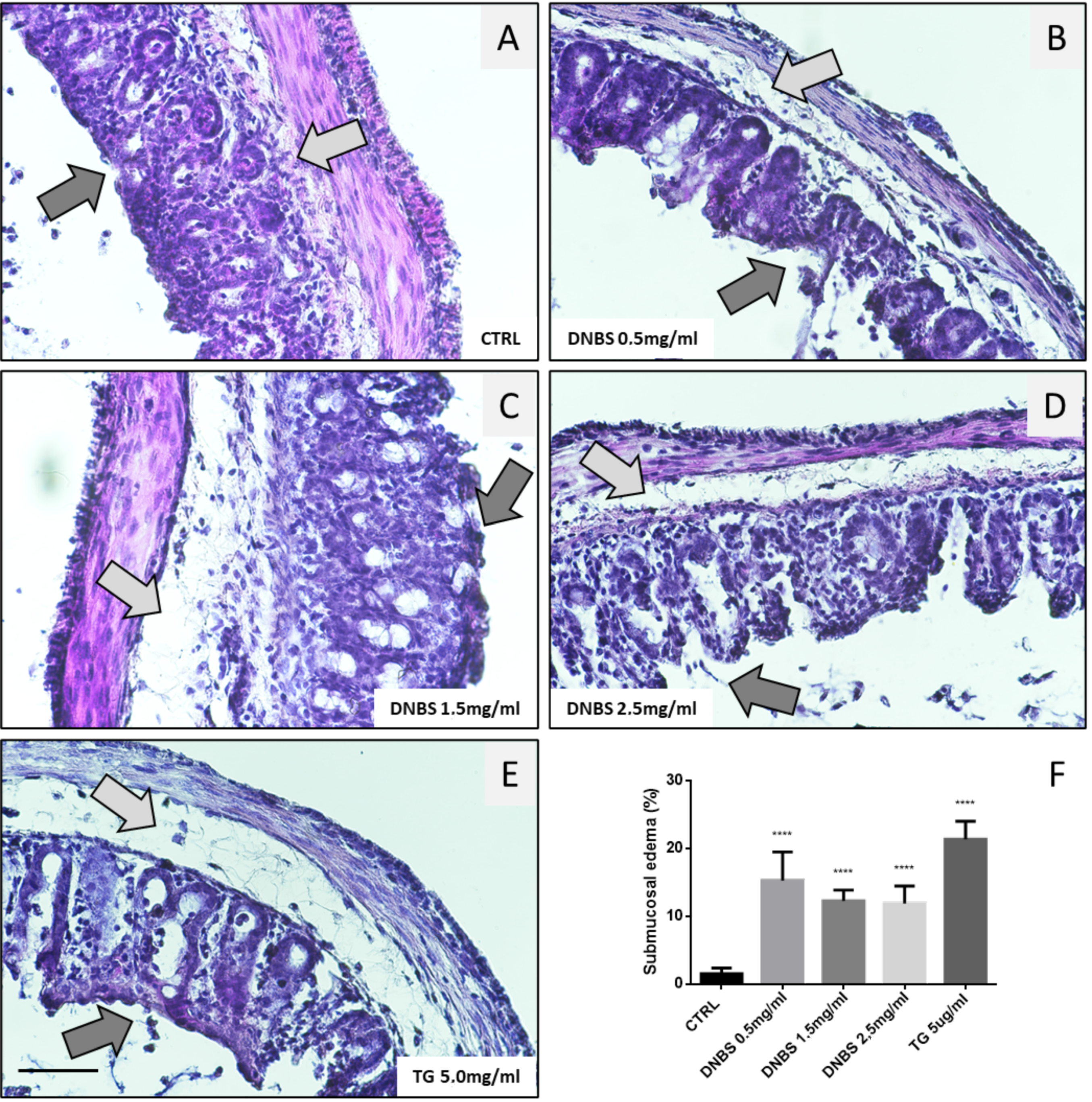
3.5. Colon Tissue Damage, upon Stimulation with DNBS
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Bischoff, S.C.; Barbara, G.; Buurman, W.; Ockhuizen, T.; Schulzke, J.-D.; Serino, M.; Tilg, H.; Watson, A.; Wells, J.M. Intestinal permeability—A new target for disease prevention and therapy. BMC Gastroenterol. 2015, 14. [Google Scholar] [CrossRef] [Green Version]
- Garcia-Carbonell, R.; Yao, S.-J.; Das, S.; Guma, M. Dysregulation of Intestinal Epithelial Cell RIPK Pathways Promotes Chronic Inflammation in the IBD Gut. Front. Immunol. 2019, 10, 1094. [Google Scholar] [CrossRef]
- Romano, M.; De Francesco, F.; Zarantonello, L.; Ruffolo, C.; Ferraro, A.G.; Zanus, G.; Giordano, A.; Bassi, N.; Cillo, U. From Inflammation to Cancer in Inflammatory Bowel Disease: Molecular Perspectives. Anticancer. Res. 2016, 36, 1447–1460. [Google Scholar]
- Fiocchi, C.; Iliopoulos, D. What’s new in IBD therapy: An “omics network” approach. Pharmacol. Res. 2020, 159, 104886. [Google Scholar] [CrossRef] [PubMed]
- Okin, D.; Medzhitov, R. Evolution of Inflammatory Diseases. Curr. Biol. 2012, 22, R733–R740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dejban, P.; Nikravangolsefid, N.; Chamanara, M.; Dehpour, A.; Rashidian, A. The role of medicinal products in the treat-ment of inflammatory bowel disease (IBD) through inhibition of TLR4/NF-kappaB pathway. Phytother. Res. 2021. [Google Scholar] [CrossRef] [PubMed]
- Gagliardi, M.; Clemente, N.; Monzani, R.; Fusaro, L.; Ferrari, E.; Saverio, V.; Grieco, G.; Pańczyszyn, E.; Carton, F.; Santoro, C.; et al. Gut-Ex-Vivo system as a model to study gluten response in celiac disease. Cell Death Discov. 2021, 7, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Yissachar, N.; Zhou, Y.; Ung, L.; Lai, N.Y.; Mohan, J.F.; Ehrlicher, A.; Weitz, D.A.; Kasper, D.L.; Chiu, I.M.; Mathis, D.; et al. An Intestinal Organ Culture System Uncovers a Role for the Nervous System in Microbe-Immune Crosstalk. Cell 2017, 168, 1135–1148.e12. [Google Scholar] [CrossRef] [Green Version]
- Gagliardi, M.; Cotella, D.; Santoro, C.; Corà, D.; Barlev, N.A.; Piacentini, M.; Corazzari, M. Aldo-keto reductases protect metastatic melanoma from ER stress-independent ferroptosis. Cell Death Dis. 2019, 10, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Ferrari, E.; Monzani, R.; Saverio, V.; Gagliardi, M.; Pańczyszyn, E.; Raia, V.; Villella, V.; Bona, G.; Pane, M.; Amoruso, A.; et al. Probiotics Supplements Reduce ER Stress and Gut Inflammation Associated with Gliadin Intake in a Mouse Model of Gluten Sensitivity. Nutrients 2021, 13, 1221. [Google Scholar] [CrossRef]
- Kühl, A.A.; Kakirman, H.; Janotta, M.; Dreher, S.; Cremer, P.; Pawlowski, N.N.; Loddenkemper, C.; Heimesaat, M.M.; Grollich, K.; Zeitz, M.; et al. Aggravation of different types of ex-perimental colitis by depletion or adhesion blockade of neutrophils. Gastroenterology 2007, 133, 1882–1892. [Google Scholar] [CrossRef]
- Elli, L.; Ciulla, M.M.; Busca, G.; Roncoroni, L.; Maioli, C.; Ferrero, S.; Bardella, M.T.; Bonura, A.; Paliotti, R.; Terrani, C.; et al. Beneficial effects of treatment with transglutaminase inhibitor cystamine on the severity of inflammation in a rat model of inflammatory bowel disease. Lab. Investig. 2010, 91, 452–461. [Google Scholar] [CrossRef] [Green Version]
- Jeong, E.M.; Son, Y.H.; Choi, Y.; Kim, J.-H.; Lee, J.-H.; Cho, S.-Y.; Kim, I.-G. Transglutaminase 2 is dispensable but required for the survival of mice in dextran sulfate sodium-induced colitis. Exp. Mol. Med. 2016, 48, e267. [Google Scholar] [CrossRef] [Green Version]
- Yamamoto-Furusho, J.K.; Mendivil, E.J.; Fonseca-Camarillo, G. Differential expression of occludin in patients with ulcera-tive colitis and healthy controls. Inflamm. Bowel Dis. 2012, 18, E1999. [Google Scholar] [CrossRef] [PubMed]
- Zeissig, S.; Bürgel, N.; Günzel, D.; Richter, J.; Mankertz, J.; Wahnschaffe, U.; Kroesen, E.J.; Zeits, M.; Fromm, M.; Schulzke, J.D. Changes in expression and distribution of claudin 2, 5 and 8 lead to discontinuous tight junctions and barrier dysfunction in active Crohn’s disease. Gut 2007, 56, 61–72. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Dai, Z.; Sun, K.; Zhang, Y.; Chen, J.; Yang, Y.; Tso, P.; Wu, G.; Wu, Z. Intestinal Epithelial Cell Endoplasmic Reticulum Stress and In-flammatory Bowel Disease Pathogenesis: An Update Review. Front Immunol. 2017, 8, 1271. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogaert, S.; De Vos, M.; Olievier, K.; Peeters, H.; Elewaut, D.; Lambrecht, B.; Pouliot, P.; Laukens, D. Involvement of Endoplasmic Reticulum Stress in Inflammatory Bowel Disease: A Different Implication for Colonic and Ileal Disease? PLoS ONE 2011, 6, e25589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rees, W.D.; Stahl, M.; Jacobson, K.; Bressler, B.; Sly, L.; Vallance, A.B.; Steiner, T.S. Enteroids Derived From Inflammatory Bowel Disease Patients Display Dysregulated Endoplasmic Reticulum Stress Pathways, Leading to Differential Inflammatory Responses and Dendritic Cell Maturation. J. Crohn’s Coliti. 2019, 14, 948–961. [Google Scholar] [CrossRef]
- Cao, S.S.; Zimmermann, E.M.; Chuang, B.; Song, B.; Nwokoye, A.; Wilkinson, J.E.; Eaton, K.A.; Kaufman, R.J. The Unfolded Protein Response and Chemical Chaperones Reduce Protein Misfolding and Colitis in Mice. Gastroenterology 2013, 144, 989–1000.e6. [Google Scholar] [CrossRef] [Green Version]
- Giglio, P.; Gagliardi, M.; Tumino, N.; Antunes, F.; Smaili, S.; Cotella, D.; Santoro, C.; Bernardini, R.; Mattei, M.; Piacentini, M.; et al. PKR and GCN2 stress kinases promote an ER stress-independent eIF2α phosphorylation responsible for calreticulin exposure in melanoma cells. OncoImmunology 2018, 7, e1466765. [Google Scholar] [CrossRef] [Green Version]
- Pagliarini, V.; Giglio, P.; Bernardoni, P.; De Zio, D.; Fimia, G.M.; Piacentini, M.; Corazzari, M. Down-regulation of E2F1 during ER stress is required to induce apoptosis. J. Cell Sci. 2015, 128, 1166–1179. [Google Scholar] [CrossRef] [Green Version]
- Neurath, M.F. Cytokines in inflammatory bowel disease. Nat. Rev. Immunol. 2014, 14, 329–342. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto-Furusho, J.K.; Sanchez-Muñoz, F.; Dominguez-Lopez, A. Role of cytokines in inflammatory bowel disease. World J. Gastroenterol. 2008, 14, 4280–4288. [Google Scholar] [CrossRef]
- Cuzzocrea, S.; McDonald, M.C.; Mazzon, E.; Mota-Filipe, H.; Centorrino, T.; Terranova, M.L.; Ciccolo, A.; Britti, A.; Caputi, A.P.; Thiemermann, C.; et al. Calpain inhibitor I reduces colon injury caused by dinitrobenzene sulphonic acid in the rat. Gut 2001, 14, 478–488. [Google Scholar] [CrossRef] [Green Version]
- Mentella, M.C.; Scaldaferri, F.; Pizzoferrato, M.; Gasbarrini, A.; Miggiano, G.A.D.; Chiara, M.M.; Franco, S.; Marco, P.; Antonio, G.; Donato, M.G.A. Nutrition, IBD and Gut Microbiota: A Review. Nutrients 2020, 12, 944. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Katz, L.H.; Kopylov, U.; Fudim, E.; Yavzori, M.; Picard, O.; Ungar, B.; Eliakim, R.; Ben-Horin, S.; Chowers, J. Expression of IL-2, IL-17 and TNF-alpha in pa-tients with Crohn’s disease treated with anti-TNF antibodies. Clin. Gastroenterol. Hepatol. 2014, 38, 491–498. [Google Scholar] [CrossRef]
- Pedersen, G. Development, validation and implementation of an in vitro model for the study of metabolic and immune function in normal and inflamed human colonic epithelium. Dan. Med. J. 2015, 62, 4973. [Google Scholar]
- Vadstrup, K.; Galsgaard, E.D.; Gerwien, J.; Vester-Andersen, M.K.; Pedersen, J.S.; Rasmussen, J.; Neermark, N.; Kiszka-Kanowitz, M.; Jensen, T.; Bendtsen, F. Validation and opti-mization of an ex vivo assay of intestinal mucosal biopsies in Crohn’s disease: Reflects inflammation and drug effects. PLoS ONE 2016. [Google Scholar] [CrossRef] [Green Version]
- Rieux, A.D.; Fievez, V.; Théate, I.; Mast, J.; Préat, V.; Schneider, Y.J. An improved in vitro model of human intestinal folli-cle-associated epithelium to study nanoparticle transport by M cells. Eur. J. Pharm. Sci. 2007, 30, 380–391. [Google Scholar] [CrossRef] [PubMed]
- Spottl, T.; Hausmann, M.; Gunckel, M.; Herfarth, H.; Herlyn, M.; Schoelmerich, J.; Rogler, G. A new organotypic model to study cell interactions in the intestinal mucosa. Eur. J. Gastroenterol. Hepatol. 2006, 18, 901–909. [Google Scholar] [CrossRef] [PubMed]
- Tanoue, T.; Nishitani, Y.; Kanazawa, K.; Hashimoto, T.; Mizuno, M. In vitro model to estimate gut inflammation using co-cultured Caco-2 and RAW264.7 cells. Biochem. Biophys. Res. Commun. 2008, 374, 565–569. [Google Scholar] [CrossRef] [Green Version]
- Motavallian-Naeini, A.; Andalib, S.; Rabbani, M.; Mahzouni, P.; Afsharipour, M.; Minaiyan, M. Validation and optimization of experimental colitis induction in rats using 2, 4, 6-trinitrobenzene sulfonic acid. Res. Pharm. Sci. 2012, 7, 159–169. [Google Scholar]
- Goyal, N.; Rana, A.; Ahlawat, A.; Bijjem, K.R.V.; Kumar, P. Animal models of inflammatory bowel disease: A review. Inflammopharmacology 2014, 22, 219–233. [Google Scholar] [CrossRef]
- Dou, W.; Zhang, J.; Sun, A.; Zhang, E.; Ding, L.; Mukherjee, S.; Wei, X.; Chou, G.; Wang, Z.-T.; Mani, S. Protective effect of naringenin against experimental colitis via suppression of Toll-like receptor 4/NK-kB signaling. Br. J. Nutr. 2013, 110, 599–608. [Google Scholar] [CrossRef] [Green Version]
- Perše, M.; Cerar, A. Dextran Sodium Sulphate Colitis Mouse Model: Traps and Tricks. J. Biomed. Biotechnol. 2012, 2012, 718617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joshi, S.V.; Vyas, B.A.; Shah, P.D.; Shah, D.R.; Shah, S.A.; Gandhi, T.R. Protective effect of aqueous extract of Oroxylum in-dicum Linn. (root bark) against DNBS-induced colitis in rats. Indian J. Pharmacol. 2011, 14, 656–661. [Google Scholar]
- Ko, J.K.S.; Lam, F.Y.L.; Cheung, A.P.L. Amelioration of experimental colitis by Astragalus membranaceus through an-ti-oxidation and inhibition of adhesion molecule synthesis. World J. Gastroenterol. 2005, 11, 5787–5794. [Google Scholar] [CrossRef] [PubMed]
- Eri, R.D.; Adams, R.J.; Tran, T.V.; Tong, H.; Das, I.; Roche, D.K.; Oancea, I.; Png, C.W.; Jeffery, P.L.; Radford-Smith, G.L.; et al. An intestinal epithelial defect conferring ER stress results in inflammation involving both innate and adaptive immunity. Mucosal Immunol. 2010, 4, 354–364. [Google Scholar] [CrossRef]
- Cao, S. Epithelial ER stress in Crohn’s disease and ulcerative colitis. Inflamm. Bowel Dis. 2016, 22, 984–993. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Lee, S.; Lee, H.; Ju, S.; Park, S.; Kwon, D.; Yo, J.-W.; Yoon, I.S.; Min, D.S.; Jung, Y.S.; et al. A colon-targeted prodrug, 4-phenylbutyric acid glutamic acid con-jugate, ameliorates 2,4-dinitrobenzene sulfonic acid-induced colitis in rats. Pharmaceutics 2020, 12, 843. [Google Scholar] [CrossRef] [PubMed]
- Michielan, A.; D’Incà, R. Intestinal Permeability in inflammation bowel disease: Pathogenesis, clinical evaluation, and therapy of leaky gut. Mediat. Inflamm. 2015, 628157. [Google Scholar] [CrossRef] [Green Version]
- Suenaert, P.; Bulteel, V.; Lemmens, L.; Noman, M.; Geypens, B.; Assche, G.V.; Geboes, K.; Ceuppens, J.L.; Rutgeerts, P. Anti-tumor necrosis factor treatment re-stores the gut barrier in Crohn’s disease. Am. J. Gastroenterol. 2002, 97, 2000–2004. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Chain, F.; Miquel, S.; Lu, J.; Gratadoux, J.-J.; Sokol, H.; Verdu, E.F.; Bercik, P.; Humaran, L.G.B.; Langella, P. The Commensal Bacterium Faecalibacterium prausnitzii Is Protective in DNBS-induced Chronic Moderate and Severe Colitis Models. Inflamm. Bowel Dis. 2014, 20, 417–430. [Google Scholar] [CrossRef] [PubMed]
- McGuckin, M.A.; Eri, R.; Simms, L.A.; Florin, T.H.; Radford-Smith, G. Intestinal barrier dysfunction in inflammatory bowel diseases. Inflamm. Bowel Dis. 2009, 15, 100–113. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, E.-J.; Lee, H.J.; Kim, W.-J.; Han, K.-I.; Iwasa, M.; Kobayashi, K.; Debnath, T.; Tang, Y.; Kwak, Y.-S.; Yoon, J.-H.; et al. Enterococcus faecalis EF-2001 protects DNBS-induced inflammatory bowel disease in mice model. PLoS ONE 2019, 14, e0210854. [Google Scholar] [CrossRef] [PubMed] [Green Version]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gagliardi, M.; Monzani, R.; Clemente, N.; Fusaro, L.; Saverio, V.; Grieco, G.; Pańczyszyn, E.; Yissachar, N.; Boccafoschi, F.; Corazzari, M. A Gut-Ex-Vivo System to Study Gut Inflammation Associated to Inflammatory Bowel Disease (IBD). Biology 2021, 10, 605. https://doi.org/10.3390/biology10070605
Gagliardi M, Monzani R, Clemente N, Fusaro L, Saverio V, Grieco G, Pańczyszyn E, Yissachar N, Boccafoschi F, Corazzari M. A Gut-Ex-Vivo System to Study Gut Inflammation Associated to Inflammatory Bowel Disease (IBD). Biology. 2021; 10(7):605. https://doi.org/10.3390/biology10070605
Chicago/Turabian StyleGagliardi, Mara, Romina Monzani, Nausicaa Clemente, Luca Fusaro, Valentina Saverio, Giovanna Grieco, Elżbieta Pańczyszyn, Nissan Yissachar, Francesca Boccafoschi, and Marco Corazzari. 2021. "A Gut-Ex-Vivo System to Study Gut Inflammation Associated to Inflammatory Bowel Disease (IBD)" Biology 10, no. 7: 605. https://doi.org/10.3390/biology10070605
APA StyleGagliardi, M., Monzani, R., Clemente, N., Fusaro, L., Saverio, V., Grieco, G., Pańczyszyn, E., Yissachar, N., Boccafoschi, F., & Corazzari, M. (2021). A Gut-Ex-Vivo System to Study Gut Inflammation Associated to Inflammatory Bowel Disease (IBD). Biology, 10(7), 605. https://doi.org/10.3390/biology10070605






