Assessment of Associations between Malaria Parasites and Avian Hosts—A Combination of Classic System and Modern Molecular Approach
Abstract
Simple Summary
Abstract
1. Introduction
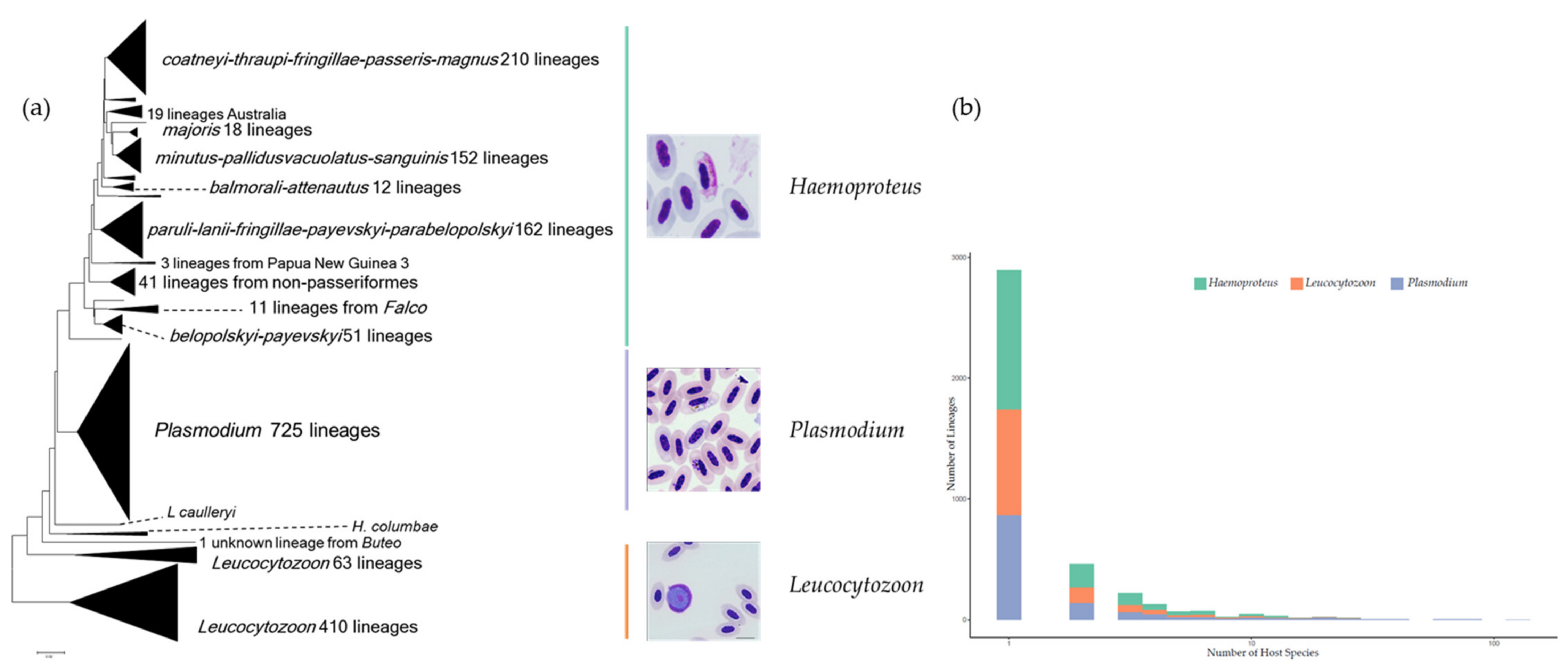
2. Avian Haemosporidian Parasites as a Classical Model System
3. Advances in Molecular Era
3.1. Molecular Approaches for Taxonomic Identification
3.2. The Milestone in Molecular Quantification
3.3. From Relative to Absolute Quantification
3.4. The Emergence of New Technologies for Future Exploration
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Combes, C. Parasitism: The Ecology and Evolution of Intimate Interactions; University of Chicago Press: Chicago, IL, USA, 2001. [Google Scholar]
- Hahn, B.H.; Shaw, G.M.; De, K.M.; Sharp, P.M. AIDS as a zoonosis: Scientific and public health implications. Science 2000, 287, 607–614. [Google Scholar] [CrossRef]
- LaDeau, S.L.; Kilpatrick, A.M.; Marra, P.P. West Nile virus emergence and large-scale declines of North American bird populations. Nature 2007, 447, 710–713. [Google Scholar] [CrossRef]
- Gortázar, C.; Ferroglio, E.; Höfle, U.; Frölich, K.; Vicente, J. Diseases shared between wildlife and livestock: A European perspective. Eur. J. Wildl. Res. 2007, 53, 241–256. [Google Scholar] [CrossRef]
- Woodworth, B.L.; Atkinson, C.T.; LaPointe, D.A.; Hart, P.J.; Spiegel, C.S.; Tweed, E.J.; Henneman, C.; LeBrun, J.; Denette, T.; DeMots, R.; et al. Host population persistence in the face of introduced vector-borne diseases: Hawaii amakihi and avian malaria. Proc. Natl. Acad. Sci. USA 2005, 102, 1531–1536. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.R.; Hoberg, E.P.; Boeger, W.A.; Gardner, S.L.; Galbreath, K.E.; Herczeg, D.; Mejía-Madrid, H.H.; Rácz, S.E.; Dursahinhan, A.T. Finding them before they find us: Informatics, parasites, and environments in accelerating climate change. Comp. Parasitol. 2014, 81, 155–164. [Google Scholar] [CrossRef]
- De Roode, J.C.; Pansini, R.; Cheesman, S.J.; Helinski, M.E.H.; Huijben, S.; Wargo, A.R.; Bell, A.S.; Chan, B.H.K.; Walliker, D.; Read, A.F. Virulence and competitive ability in genetically diverse malaria infections. Proc. Natl. Acad. Sci. USA 2005, 102, 7624–7628. [Google Scholar] [CrossRef]
- Graumans, W.; Jacobs, E.; Bousema, T.; Sinnis, P. When is a Plasmodium-infected mosquito an infectious mosquito? Trends Parasitol. 2020, 36, 705–716. [Google Scholar] [CrossRef] [PubMed]
- Poulin, R. Evolutionary Ecology of Parasites; Princeton University Press: Princeton, NJ, USA, 2011. [Google Scholar]
- Antonovics, J.; Boots, M.; Ebert, D.; Koskella, B.; Poss, M.; Sadd, B.M. The origin of specificity by means of natural selection: Evolved and nonhost resistance in host–pathogen interactions. Evolution 2013, 67, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Martin, M.J.; Rayner, J.C.; Gagneux, P.; Barnwell, J.W.; Varki, A. Evolution of human-chimpanzee differences in malaria susceptibility: Relationship to human genetic loss of N-glycolylneuraminic acid. Proc. Natl. Acad. Sci. USA 2005, 102. [Google Scholar] [CrossRef]
- Rohde, K. Ecology and biogeography of marine parasites. In Advances in Marine Biology; Academic Press: Cambridge, MA, USA, 2002; Volume 43, pp. 1–83. [Google Scholar]
- Hellgren, O.; Pérez-Tris, J.; Bensch, S. A jack-of-all-trades and still a master of some: Prevalence and host range in avian malaria and related blood parasites. Ecology 2009, 90, 2840–2849. [Google Scholar] [CrossRef]
- Cooper, N.; Griffin, R.; Franz, M.; Omotayo, M.; Nunn, C.L. Phylogenetic host specificity and understanding parasite sharing in primates. Ecol. Lett. 2012, 15, 1370–1377. [Google Scholar] [CrossRef]
- Beldomenico, P.M.; Begon, M. Disease spread, susceptibility and infection intensity: Vicious circles? Trends Ecol. Evol. 2010, 25, 21–27. [Google Scholar] [CrossRef]
- Huang, X.; Ellis, V.A.; Jönsson, J.; Bensch, S. Generalist haemosporidian parasites are better adapted to a subset of host species in a multiple host community. Mol. Ecol. 2018, 27, 4336–4346. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, C.T.; Thomas, N.J.; Hunter, D.B. Parasitic Diseases of Wild Birds; John Wiley & Sons: Hoboken, NJ, USA, 2009. [Google Scholar]
- Asghar, M.; Hasselquist, D.; Hansson, B.; Zehtindjiev, P.; Westerdahl, H.; Bensch, S. Hidden costs of infection: Chronic malaria accelerates telomere degradation and senescence in wild birds. Science 2015, 347, 436–438. [Google Scholar] [CrossRef]
- Ilgūnas, M.; Bukauskaitė, D.; Palinauskas, V.; Iezhova, A.T.; Dinhopl, N.; Nedorost, N.; Weissenbacher-Lang, C.; Weissenböck, H.; Valkiūnas, G. Mortality and pathology in birds due to Plasmodium (Giovannolaia) homocircumflexum infection, with emphasis on the exoerythrocytic development of avian malaria parasites. Malar. J. 2016, 15, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Perkins, S.L. Malaria’s many mates: Past, present, and future of the systematics of the order Haemosporida. J. Parasitol. 2014, 100, 11–25. [Google Scholar] [CrossRef] [PubMed]
- Pigeault, R.; Vézilier, J.; Cornet, S.; Zélé, F.; Nicot, A.; Perret, P.; Gandon, S.; Rivero, A. Avian malaria: A new lease of life for an old experimental model to study the evolutionary ecology of Plasmodium. Phil. Trans. R. Soc. B 2015, 370, 20140300. [Google Scholar] [CrossRef]
- Bukauskaitė, D.; Žiegytė, R.; Palinauskas, V.; Iezhova, T.A.; Dimitrov, D.; Ilgūnas, M.; Bernotienė, R.; Markovets, M.Y.; Valkiūnas, G. Biting midges (Culicoides, Diptera) transmit Haemoproteus parasites of owls: Evidence from sporogony and molecular phylogeny. Parasites Vectors 2015, 8, 303. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G. Avian Malaria Parasites and Other Haemosporidia; CRC Press: Boca Raton, FL, USA, 2005. [Google Scholar]
- Godfrey, R.D., Jr.; Fedynich, A.M.; Pence, D.B. Quantification of hematozoa in blood smears. J. Wildl. Dis. 1987, 23, 558–565. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G.; Bensch, S.; Iezhova, T.A.; Križanauskienė, A.; Hellgren, O.; Bolshakov, C.V. Nested cytochrome b polymerase chain reaction diagnostics underestimate mixed infections of avian blood haemosporidian parasites: Microscopy is still essential. J. Parasitol. 2006, 92, 418–422. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G.; Iezhova, T.A.; Križanauskienė, A.; Palinauskas, V.; Sehgal, R.N.M.; Bensch, S. A comparative analysis of microscopy and PCR-based detection methods for blood parasites. J. Parasitol. 2008, 94, 1395–1401. [Google Scholar] [CrossRef] [PubMed]
- Mukhin, A.; Palinauskas, V.; Platonova, E.; Kobylkov, D.; Vakoliuk, I.; Valkiūnas, G. The strategy to survive primary malaria infection: An experimental study on behavioural changes in parasitized birds. PloS ONE 2016, 11, e0159216. [Google Scholar] [CrossRef] [PubMed]
- Bruce, M.C.; Day, K.P. Cross-species regulation of malaria parasitaemia in the human host. Curr. Opin. Microbiol. 2002, 5, 431–437. [Google Scholar] [CrossRef]
- Nilsson, E.; Taubert, H.; Hellgren, O.; Huang, X.; Palinauskas, V.; Markovets, M.; Valkiūnas, G.; Bensch, S. Multiple cryptic species of sympatric generalists within the avian blood parasite Haemoproteus majoris. J. Evol. Biol. 2016, 29, 1812–1826. [Google Scholar] [CrossRef]
- Palinauskas, V.; Žiegytė, R.; Ilgūnas, M.; Iezhova, T.A.; Bernotienė, A.; Bolshakov, C. Description of the first cryptic avian malaria parasite, Plasmodium homocircumflexum n. sp. with experimental data on its virulence and development in avian hosts and mosquitoes. Int. J. Parasitol. 2015, 45. [Google Scholar] [CrossRef]
- Valkiūnas, G.; Iezhova, T.A. Keys to the avian malaria parasites. Malar. J. 2018, 17, 212. [Google Scholar] [CrossRef]
- Clark, N.J.; Clegg, S.M. Integrating phylogenetic and ecological distances reveals new insights into parasite host specificity. Mol. Ecol. 2017, 26, 3074–3086. [Google Scholar] [CrossRef]
- Fecchio, A.; Wells, K.; Bell, J.A.; Tkach, V.V.; Lutz, H.L.; Weckstein, J.D.; Clegg, S.M.; Clark, N.J. Climate variation influences host specificity in avian malaria parasites. Ecol. Lett. 2019, 22, 547–557. [Google Scholar] [CrossRef]
- Bensch, S.; Stjernman, M.; Hasselquist, D.; Örjan, Ö.; Hannson, B.; Westerdahl, H.; Pinheiro, R.T. Host specificity in avian blood parasites: A study of Plasmodium and Haemoproteus mitochondrial DNA amplified from birds. Proc. Royal Soc. B. 2000, 267, 1583–1589. [Google Scholar] [CrossRef]
- Perkins, S.L.; Schall, J.J. A molecular phylogeny of malarial parasites recovered from cytochrome b gene sequences. J. Parasitol. 2002, 88, 972–978. [Google Scholar] [CrossRef]
- Fallon, S.M.; Ricklefs, R.E.; Swanson, B.L.; Bermingham, E. Detecting avian malaria: An improved polymerase chain reaction diagnostic. J. Parasitol. 2003, 89, 1044–1047. [Google Scholar] [CrossRef] [PubMed]
- Waldenström, J.; Bensch, S.; Hasselquist, D.; Ostman, O. A new nested polymerase chain reaction method very efficient in detecting Plasmodium and Haemoproteus infections from avian blood. J. Parasitol. 2004, 90, 191–194. [Google Scholar] [CrossRef]
- Beadell, J.S.; Fleischer, R.C. A restriction enzyme-based assay to distinguish between avian hemosporidians. J. Parasitol. 2005, 91, 683–685. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tris, J.; Hasselquist, D.; Hellgren, O.; Krizanauskiene, A.; Waldenström, J.; Bensch, S. What are malaria parasites? Trends Parasitol. 2005, 21, 209–211. [Google Scholar] [CrossRef] [PubMed]
- Bensch, S.; Hellgren, O.; Pérez-Tris, J. MalAvi: A public database of malaria parasites and related haemosporidians in avian hosts based on mitochondrial cytochrome b lineages. Mol. Ecol. Resour. 2009, 9, 1353–1358. [Google Scholar] [CrossRef] [PubMed]
- Dunn, J.C.; Outlaw, D.C. Flying into the future: Avian haemosporidians and the advancement of understanding host–parasite systems. Parasitology 2019, 146, 1487–1489. [Google Scholar] [CrossRef]
- Hellgren, O.; Atkinson, C.T.; Bensch, S.; Albayrak, T.; Dimitrov, D.; Ewen, J.G.; Kim, K.S.; Lima, M.R.; Martin, L.; Palinauskas, V. Global phylogeography of the avian malaria pathogen Plasmodium relictum based on MSP1 allelic diversity. Ecography 2015, 38, 842–850. [Google Scholar] [CrossRef]
- Clark, N.J.; Clegg, S.M.; Lima, M.R. A review of global diversity in avian haemosporidians (Plasmodium and Haemoproteus: Haemosporida): New insights from molecular data. Int. J. Parasitol. 2014, 44, 329–338. [Google Scholar] [CrossRef]
- Rivero, A.; Gandon, S. Evolutionary ecology of avian malaria: Past to present. Trends Parasitol. 2018, 34, 712–726. [Google Scholar] [CrossRef]
- Ellis, V.A.; Bensch, S. Host specificity of avian haemosporidian parasites is unrelated among sister lineages but shows phylogenetic signal across larger clades. Int. J. Parasitol. 2018, 48, 897–902. [Google Scholar] [CrossRef]
- Ricklefs, R.E.; Outlaw, D.C.; Svensson-Coelho, M.; Medeiros, M.C.I.; Ellis, V.A.; Latta, S. Species formation by host shifting in avian malaria parasites. Proc. Natl. Acad. Sci. USA 2014, 111, 14816–14821. [Google Scholar] [CrossRef]
- Alcala, N.; Jenkins, T.; Christe, P.; Vuilleumier, S. Host shift and cospeciation rate estimation from co-phylogenies. Ecol. Lett. 2017, 20, 1014–1024. [Google Scholar] [CrossRef]
- Ciloglu, A.; Ellis, V.A.; Duc, M.; Downing, P.A.; Inci, A.; Bensch, S. Evolution of vector transmitted parasites by host switching revealed through sequencing of Haemoproteus parasite mitochondrial genomes. Mol. Phylogen. Evol. 2020, 153, 106947. [Google Scholar] [CrossRef]
- Bernotienė, R.; Palinauskas, V.; Iezhova, T.; Murauskaitė, D.; Valkiūnas, G. Avian haemosporidian parasites (Haemosporida): A comparative analysis of different polymerase chain reaction assays in detection of mixed infections. Exp. Parasitol. 2016, 163, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Martinez, J.; Martinez-De La Puente, J.; Herrero, J.; Del Cerro, S.; Lobato, E.; Rivero-De Aguilar, J.; Vasquez, R.A.; Merino, S. A restriction site to differentiate Plasmodium and Haemoproteus infections in birds: On the inefficiency of general primers for detection of mixed infections. Parasitology 2009, 136, 713–722. [Google Scholar] [CrossRef] [PubMed]
- Ciloglu, A.; Ellis, V.A.; Bernotienė, R.; Valkiūnas, G.; Bensch, S. A new one-step multiplex PCR assay for simultaneous detection and identification of avian haemosporidian parasites. Parasitol. Res. 2019, 118, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Valkiūnas, G.; Iezhova, T.A. Exo-erythrocytic development of avian malaria and related haemosporidian parasites. Malar. J. 2017, 16, 101. [Google Scholar] [CrossRef] [PubMed]
- Keesing, F.; Holt, R.D.; Ostfeld, R.S. Effects of species diversity on disease risk. Ecol. Lett. 2006, 9, 485–498. [Google Scholar] [CrossRef] [PubMed]
- Fenton, A.; Streicker, D.G.; Petchey, O.L.; Pedersen, A.B. Are all hosts created equal? Partitioning host species contributions to parasite persistence in multihost communities. Am. Nat. 2015, 186, 610–622. [Google Scholar] [CrossRef] [PubMed]
- Moens, M.A.; Valkiūnas, G.; Paca, A.; Bonaccorso, E.; Aguirre, N.; Pérez-Tris, J. Parasite specialization in a unique habitat: Hummingbirds as reservoirs of generalist blood parasites of Andean birds. J. Anim. Ecol. 2016, 85, 1234–1245. [Google Scholar] [CrossRef]
- Bentz, S.; Rigaud, T.; Barroca, M.; Martin-Laurent, F.; Bru, D.; Moreau, J.; Faivre, B. Sensitive measure of prevalence and parasitaemia of haemosporidia from European blackbird (Turdus merula) populations: Value of PCR-RFLP and quantitative PCR. Parasitology 2006, 133, 685–692. [Google Scholar] [CrossRef] [PubMed]
- Fallon, S.M.; Ricklefs, R.E. Parasitemia in PCR-detected Plasmodium and Haemoproteus infections in birds. J. Avian Biol. 2008, 39, 514–522. [Google Scholar] [CrossRef]
- Ishtiaq, F.; Rao, M.; Huang, X.; Bensch, S. Estimating prevalence of avian haemosporidians in natural populations: A comparative study on screening protocols. Parasites Vectors 2017, 10, 127. [Google Scholar] [CrossRef] [PubMed]
- Asghar, M.; Hasselquist, D.; Bensch, S. Are chronic avian haemosporidian infections costly in wild birds? J. Avian Biol. 2011, 42, 530–537. [Google Scholar] [CrossRef]
- Logan, J.; Logan, J.M.; Edwards, K.J.; Saunders, N.A. Real-Time PCR: Current Technology and Applications; Horizon Scientific Press: Poole, UK, 2009. [Google Scholar]
- Kubista, M.; Andrade, J.M.; Bengtsson, M.; Forootan, A.; Jonák, J.; Lind, K.; Sindelka, R.; Sjöback, R.; Sjögreen, B.; Strömbom, L.; et al. The real-time polymerase chain reaction. Mol. Asp. Med. 2006, 27, 95–125. [Google Scholar] [CrossRef]
- DeBarry, J.D.; Kissinger, J.C. Jumbled Genomes: Missing Apicomplexan Synteny. Mol. Biol. Evol. 2011, 28, 2855–2871. [Google Scholar] [CrossRef]
- Nikbakht, H.; Xia, X.; Hickey, D.A. The evolution of genomic GC content undergoes a rapid reversal within the genus Plasmodium. Genome 2014, 57, 507–511. [Google Scholar] [CrossRef]
- Videvall, E. Genomic advances in avian malaria research. Trends Parasitol. 2019, 35, 254–266. [Google Scholar] [CrossRef]
- Knowles, S.C.L.; Wood, M.J.; Alves, R.; Wilkin, T.A.; Bensch, S.; Sheldon, B.C. Molecular epidemiology of malaria prevalence and parasitaemia in a wild bird population. Mol. Ecol. 2011, 20, 1062–1076. [Google Scholar] [CrossRef] [PubMed]
- Friedl, T.W.P.; Groscurth, E. A real-time PCR protocol for simple and fast quantification of blood parasite infections in evolutionary and ecological studies and some data on intensities of blood parasite infections in a subtropical weaverbird. J. Ornithol. 2012, 153, 239–247. [Google Scholar] [CrossRef]
- Bell, J.; Weckstein, J.; Fecchio, A.; Tkach, V. A new real-time PCR protocol for detection of avian haemosporidians. Parasites Vectors 2015, 8, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Lauron, E.J.; Oakgrove, K.S.; Tell, L.A.; Biskar, K.; Roy, S.W.; Sehgal, R.N. Transcriptome sequencing and analysis of Plasmodium gallinaceum reveals polymorphisms and selection on the apical membrane antigen-1. Malar. J. 2014, 13, 382. [Google Scholar] [CrossRef][Green Version]
- Videvall, E.; Cornwallis, C.K.; Ahrén, D.; Palinauskas, V.; Valkiūnas, G.; Hellgren, O. The transcriptome of the avian malaria parasite Plasmodium ashfordi displays host-specific gene expression. Mol. Ecol. 2017, 26, 2939–2958. [Google Scholar] [CrossRef] [PubMed]
- Videvall, E.; Paxton, K.L.; Campana, M.G.; Cassin-Sackett, L.; Atkinson, C.T.; Fleischer, R.C. Transcriptome assembly and differential gene expression of the invasive avian malaria parasite Plasmodium relictum in Hawai’i. Ecol. Evol. 2021, 11, 4935–4944. [Google Scholar] [CrossRef]
- Bensch, S.; Canbäck, B.; DeBarry, J.D.; Johansson, T.; Hellgren, O.; Kissinger, J.C.; Palinauskas, V.; Videvall, E.; Valkiūnas, G. The genome of Haemoproteus tartakovskyi and its relationship to human malaria parasites. Genome Biol. Evol. 2016, 8, 1361–1373. [Google Scholar] [CrossRef]
- Böhme, U.; Otto, T.D.; Cotton, J.; Steinbiss, S.; Sanders, M.; Oyola, S.O.; Nicot, A.; Gandon, S.; Patra, K.P.; Herd, C. Complete avian malaria parasite genomes reveal features associated with lineage specific evolution in birds and mammals. Genome Res. 2018, 28, 547–560. [Google Scholar] [CrossRef]
- Zehtindjiev, P.; Ilieva, M.; Westerdahl, H.; Hansson, B.; Valkiūnas, G.; Bensch, S. Dynamics of parasitemia of malaria parasites in a naturally and experimentally infected migratory songbird, the great reed warbler Acrocephalus arundinaceus. Exp. Parasitol. 2008, 119, 99–110. [Google Scholar] [CrossRef]
- Huang, X.; Jönsson, J.; Bensch, S. Persistence of avian haemosporidians in the wild: A case study to illustrate seasonal infection patterns in relation to host life stages. Int. J. Parasitol. 2020, 50, 611–619. [Google Scholar] [CrossRef] [PubMed]
- Podmokła, E.; Dubiec, A.; Drobniak, S.M.; Arct, A.; Gustafsson, L.; Cichoń, M. Determinants of prevalence and intensity of infection with malaria parasites in the Blue Tit. J. Ornithol. 2014, 155, 721–727. [Google Scholar] [CrossRef]
- Dubiec, A.; Podmokła, E.; Gustafsson, L. Intra-individual changes in haemosporidian infections over the nesting period in great tit females. Parasitol. Res. 2017, 116, 2385–2392. [Google Scholar] [CrossRef] [PubMed]
- Ferraguti, M.; Martinez-de la Puente, J.; Munoz, J.; Roiz, D.; Ruiz, S.; Soriguer, R.; Figuerola, J. Avian Plasmodium in Culex and Ochlerotatus mosquitoes from Southern Spain: Effects of season and host-feeding source on parasite dynamics. PLoS ONE 2013, 8. [Google Scholar] [CrossRef]
- Marzal, A.; Bensch, S.; Reviriego, M.; Balbontin, J.; De Lope, F. Effects of malaria double infection in birds: One plus one is not two. J. Evol. Biol. 2008, 21, 979–987. [Google Scholar] [CrossRef]
- Huang, X.; Huang, D.; Liang, Y.; Zhang, L.; Yang, G.; Liu, B.; Peng, Y.; Deng, W.; Dong, L. A new protocol for absolute quantification of haemosporidian parasites in raptors and comparison with current assays. Parasites Vectors 2020, 13, 354. [Google Scholar] [CrossRef]
- Beaver, J.A.; Jelovac, D.; Balukrishna, S.; Cochran, R.L.; Croessmann, S.; Zabransky, D.J.; Wong, H.Y.; Valda Toro, P.; Cidado, J.; Blair, B.G.; et al. Detection of Cancer DNA in Plasma of Patients with Early-Stage Breast Cancer. Clin. Cancer Res. 2014, 20, 2643–2650. [Google Scholar] [CrossRef]
- Dodd, D.W.; Gagnon, K.T.; Corey, D.R. Digital Quantitation of Potential Therapeutic Target RNAs. Nucleic Acid Ther. 2013, 23, 188–194. [Google Scholar] [CrossRef]
- Yang, R.; Paparini, A.; Monis, P.; Ryan, U. Comparison of next-generation droplet digital PCR (ddPCR) with quantitative PCR (qPCR) for enumeration of Cryptosporidium oocysts in faecal samples. Int. J. Parasitol. 2014, 44, 1105–1113. [Google Scholar] [CrossRef]
- Koepfli, C.; Nguitragool, W.; Hofmann, N.E.; Robinson, L.J.; Ome-Kaius, M.; Sattabongkot, J.; Felger, I.; Mueller, I. Sensitive and accurate quantification of human malaria parasites using droplet digital PCR (ddPCR). Sci. Rep. 2016, 6, 39183. [Google Scholar] [CrossRef]
- Murzin, D.; Mapps, D.J.; Levada, K.; Belyaev, V.; Omelyanchik, A.; Panina, L.; Rodionova, V. Ultrasensitive Magnetic Field Sensors for Biomedical Applications. Sensors 2020, 20, 1569. [Google Scholar] [CrossRef] [PubMed]
- Peng, W.K.; Kong, T.F.; Ng, C.S.; Chen, L.; Huang, Y.; Bhagat, A.A.S.; Nguyen, N.-T.; Preiser, P.R.; Han, J. Micromagnetic resonance relaxometry for rapid label-free malaria diagnosis. Nat. Med. 2014, 20, 1069–1073. [Google Scholar] [CrossRef] [PubMed]
- Arndt, L.; Koleala, T.; Orbán, Á.; Ibam, C.; Lufele, E.; Timinao, L.; Lorry, L.; Butykai, Á.; Kaman, P.; Molnár, A.P.; et al. Magneto-optical diagnosis of symptomatic malaria in Papua New Guinea. Nat.Commun. 2021, 12, 969. [Google Scholar] [CrossRef] [PubMed]
- Toh, R.J.; Peng, W.K.; Han, J.; Pumera, M. Haemoglobin electrochemical detection on various reduced graphene surfaces: Well-defined glassy carbon electrode outperforms the graphenoids. RSC Adv. 2014, 4, 8050–8054. [Google Scholar] [CrossRef]
- Toh, R.J.; Peng, W.K.; Han, J.; Pumera, M. Direct In Vivo Electrochemical Detection of Haemoglobin in Red Blood Cells. Sci. Rep. 2014, 4, 6209. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Yang, C.; Preiser, P.; Zheng, Y. A Photoacoustic-Surface-Acoustic-Wave Sensor for Ring-Stage Malaria Parasite Detection. IEEE Trans. Circuits Syst. II Express Briefs 2020, 67, 881–885. [Google Scholar] [CrossRef]


| Method | Summary | Potentials | Restriction | Ref. |
|---|---|---|---|---|
| photoacoustic PA-SAW | Diagnosis of malaria at early stage based on photoacoustics signal. | Applicable in Plasmodium | Applicable at ring-stage which inly present in Plasmodium | [89] |
| Magneto-optical diagnosis | Detect hemozoin in very small concentrations based on crystal structure. | Worth trying | Nucleoid red blood cell may be inhibitor. | [86] |
| Magnetic Field Sensors | Detect infection based on the increased magnetic susceptibility of infected red blood cells. | Potentially useful | [84] | |
| Haemoglobin electrochemical | Detect infection based on transfer characteristics of haemoglobin. | Potentially useful | Unfit for Leucocytozoon due to lack of pigment | [87,88] |
| Method | Scope | Fields of Application | Advantages | Limitations | Ref. |
|---|---|---|---|---|---|
| Microscopy | Diversity Infection intensity | Morphological identification Life stage description | Hardly any false positive; Identify mixed infections | Time-consuming Low sensitivity | [23] |
| Conventional PCR | Diversity | Molecular identification Phylogenetic analysis | Taxonomic classification; Cost-effective | Underestimate mixed and abortive infections | [34,36,37,38] |
| qPCR | Infection intensity | Relative quantification Infection pattern in small scale | Rapid diagnosis; Eliminate false-positive | Taxonomy undefinable; Rely on standard | [16,59,66,67] |
| ddPCR | Infection intensity | Absolute quantification Infection pattern in large scale | Sensitive and repeatable; Low demand in samples | Taxonomy undefinable; Costly | [79] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, X. Assessment of Associations between Malaria Parasites and Avian Hosts—A Combination of Classic System and Modern Molecular Approach. Biology 2021, 10, 636. https://doi.org/10.3390/biology10070636
Huang X. Assessment of Associations between Malaria Parasites and Avian Hosts—A Combination of Classic System and Modern Molecular Approach. Biology. 2021; 10(7):636. https://doi.org/10.3390/biology10070636
Chicago/Turabian StyleHuang, Xi. 2021. "Assessment of Associations between Malaria Parasites and Avian Hosts—A Combination of Classic System and Modern Molecular Approach" Biology 10, no. 7: 636. https://doi.org/10.3390/biology10070636
APA StyleHuang, X. (2021). Assessment of Associations between Malaria Parasites and Avian Hosts—A Combination of Classic System and Modern Molecular Approach. Biology, 10(7), 636. https://doi.org/10.3390/biology10070636






