RETRACTED: Zipper Is Necessary for Branching Morphogenesis of the Terminal Cells in the Drosophila melanogaster’s Tracheal System
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. D. melanogaster Strain and Crosses
2.2. Heat Kill
2.3. Immunohistochemistry (Dissection/Fixation and Immunofluorescence/Mounting)
2.4. Qualitative and Quantitative Analysis
2.5. Image Processing
3. Results
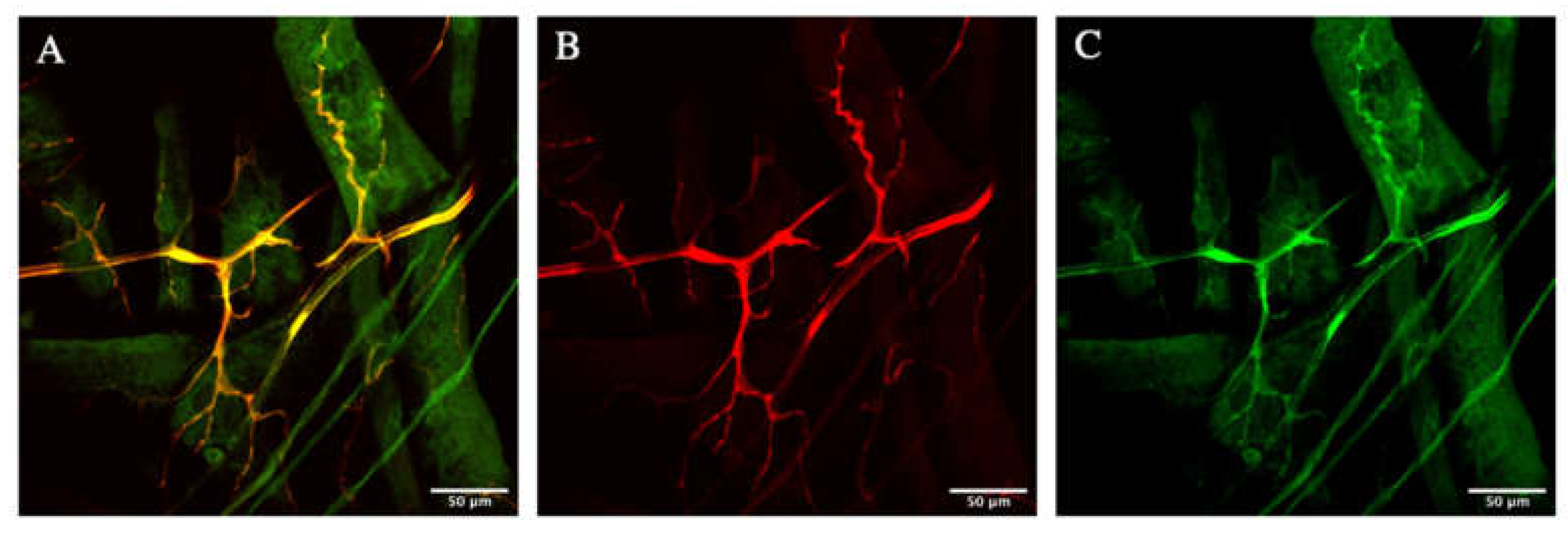
3.1. Zipper Is Localized in the Cytoplasm of the D. melanogaster’s Terminal Cells
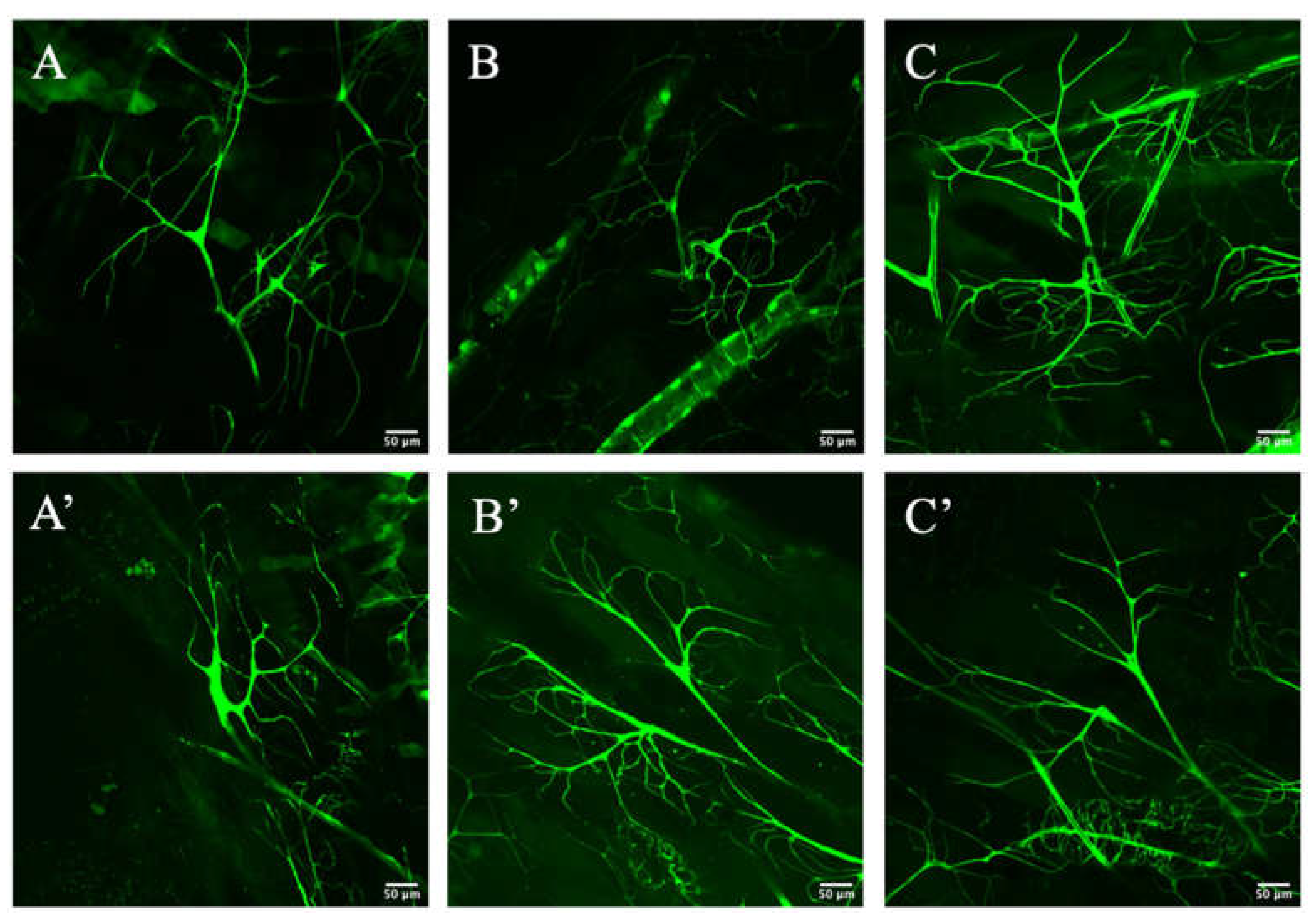
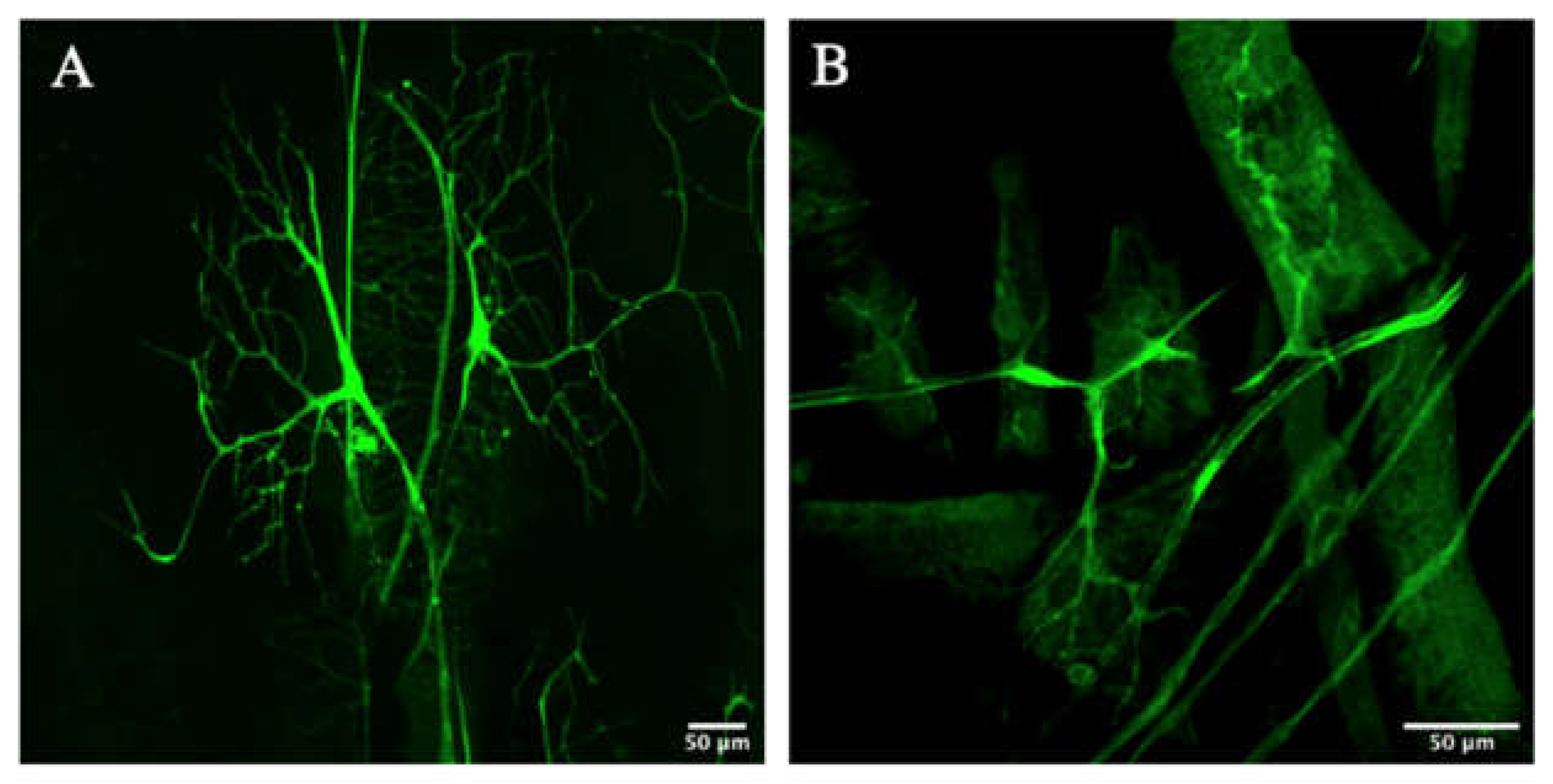
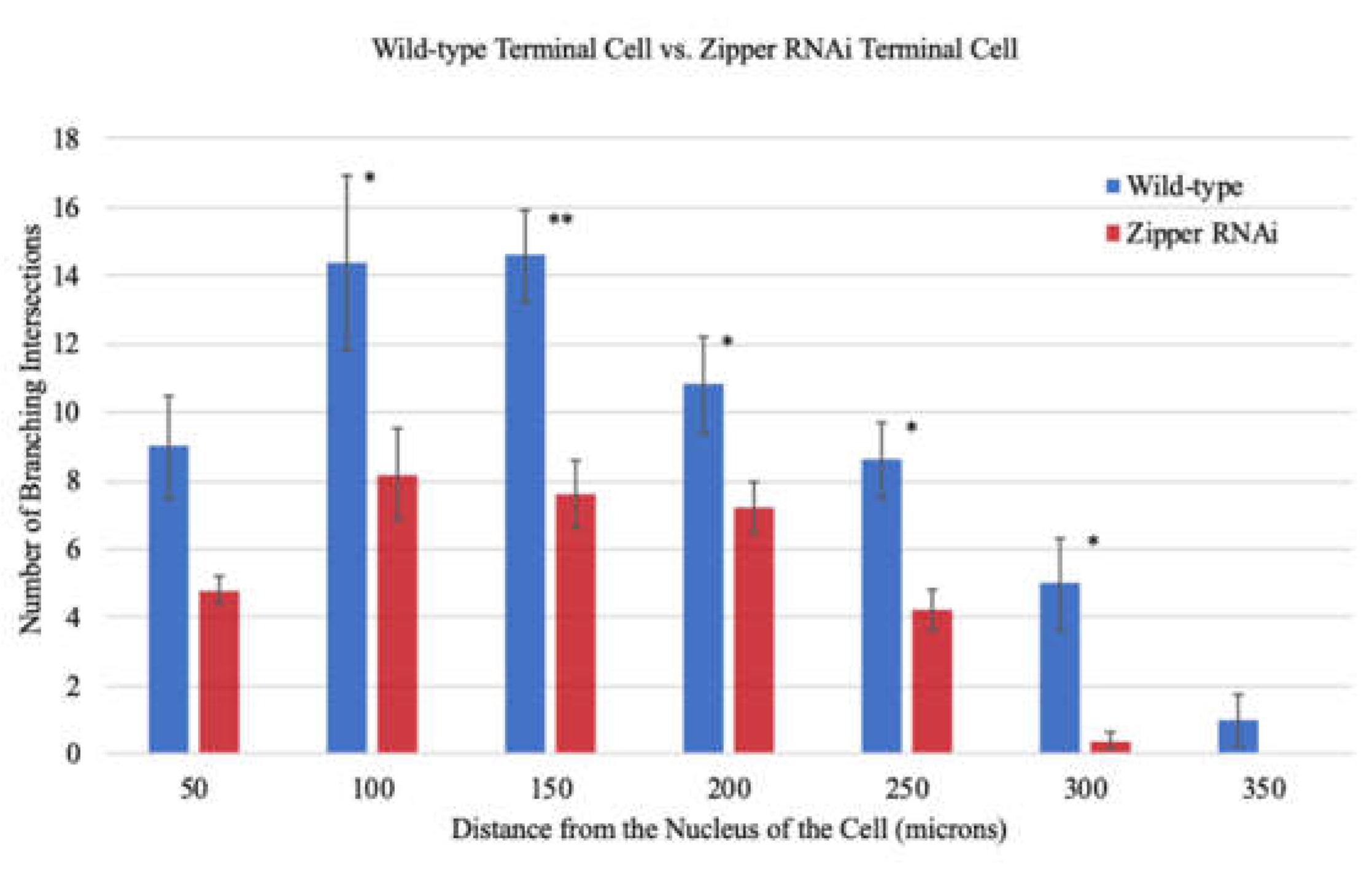
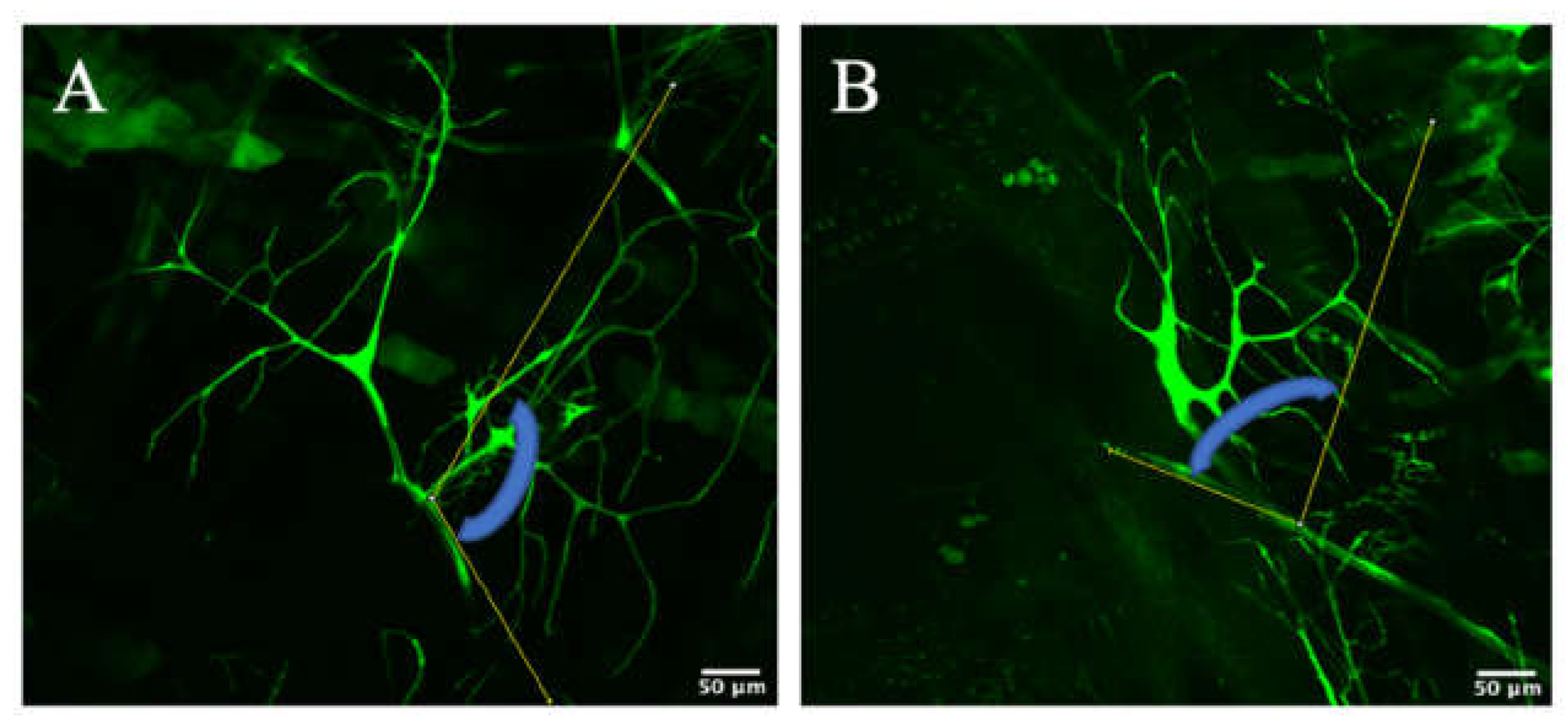
3.2. Zipper Is Necessary for Branching Morphogenesis in D. melanogaster’s Terminal Cells
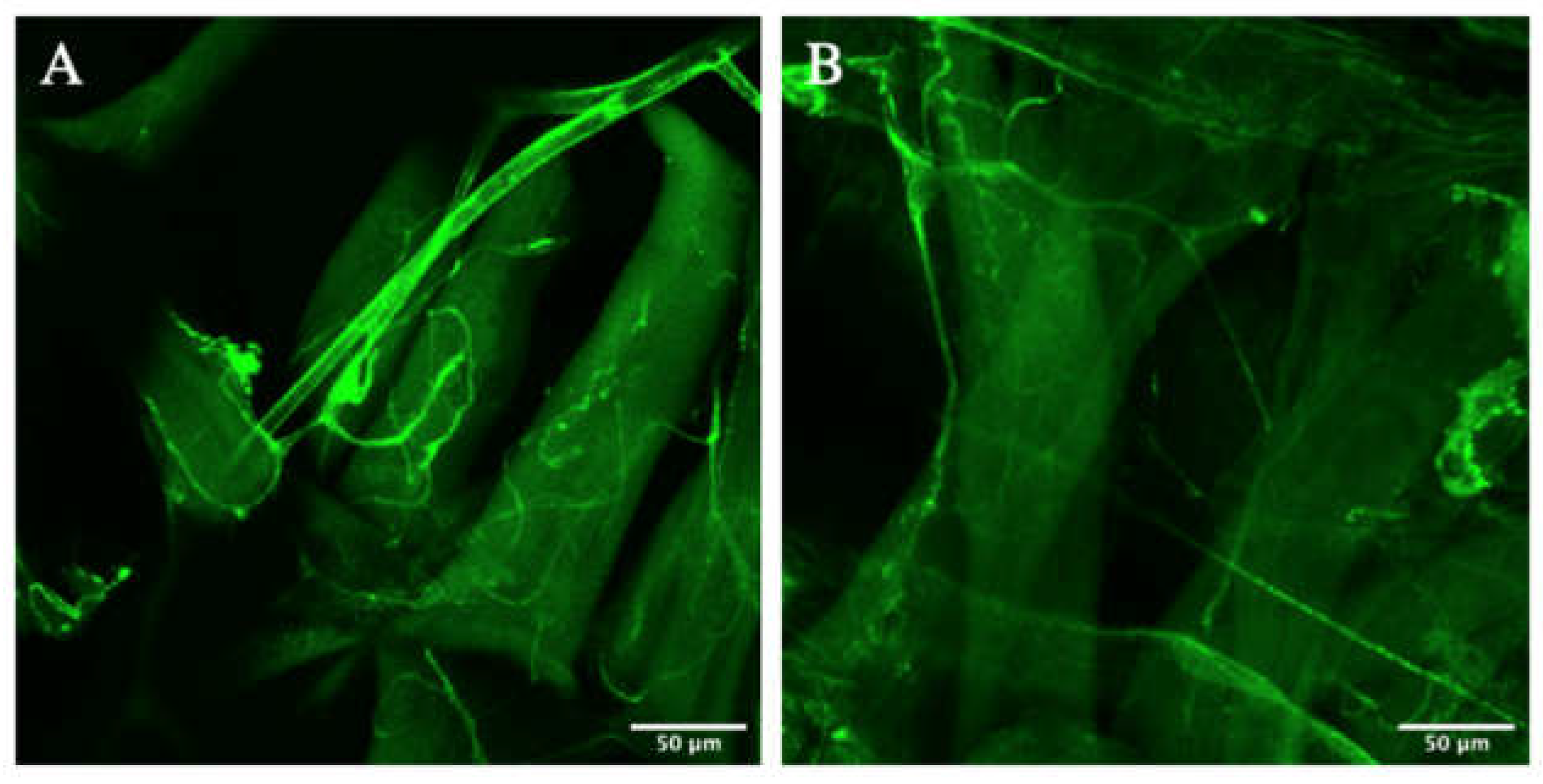
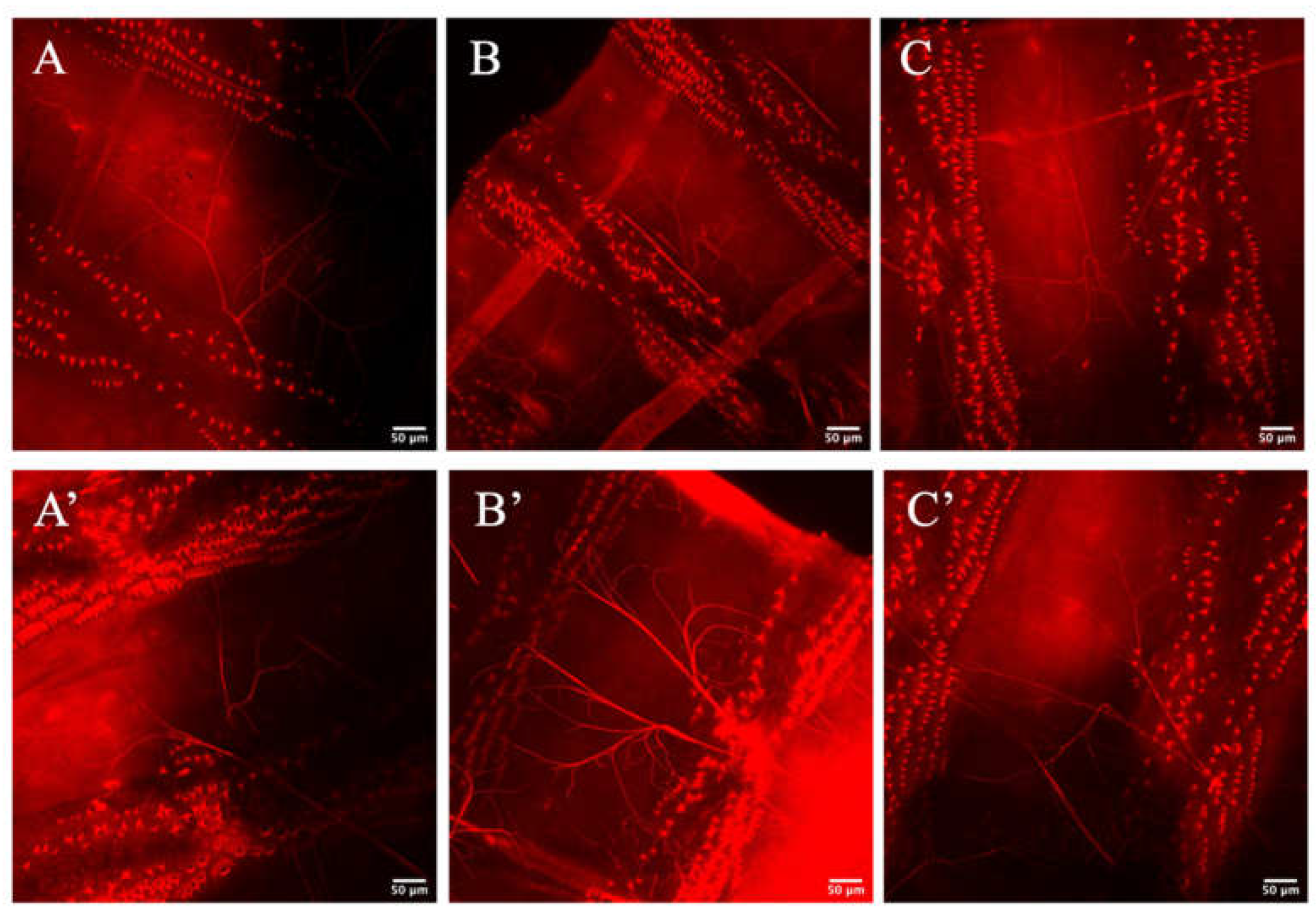
3.3. Zipper Is Not Necessary for Lumenogenesis in D. melanogaster’s Terminal Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Best, B.T. Single-cell branching morphogenesis in the Drosophila trachea. Dev. Biol. 2019, 451, 5–15. [Google Scholar] [CrossRef] [PubMed]
- Franke, J.D.; Montague, R.A.; Kiehart, D.P. Nonmuscle myosin II is required for cell proliferation, cell sheet adhesion and wing hair morphology during wing morphogenesis. Dev. Biol. 2010, 345, 117–132. [Google Scholar] [CrossRef] [PubMed]
- Ochoa-Espinosa, A.; Affolter, M. Branching Morphogenesis: From Cells to Organs and Back. Cold Spring Harb. Perspect. Biol. 2012, 4, a008243. [Google Scholar] [CrossRef] [PubMed]
- Pichel, J.; Shen, L.; Sheng, H.Z.; Granholm, A.-C.; Drago, J.; Grinberg, A.; Lee, E.J.; Huang, S.P.; Saarma, M.; Hoffer, B.J.; et al. Defects in enteric innervation and kidney development in mice lacking GDNF. Nat. Cell Biol. 1996, 382, 73–76. [Google Scholar] [CrossRef]
- Fabrowski, P.; Nećakov, A.S.; Mumbauer, S.; Loeser, E.; Reversi, A.; Streichan, S.; Briggs, J.A.G.; De Renzis, S. Tubular endocytosis drives remodelling of the apical surface during epithelial morphogenesis in Drosophila. Nat. Commun. 2013, 4, 2244. [Google Scholar] [CrossRef] [PubMed]
- Gervais, L.; Casanova, J. In Vivo Coupling of Cell Elongation and Lumen Formation in a Single Cell. Curr. Biol. 2010, 20, 359–366. [Google Scholar] [CrossRef]
- Zhou, L.; Dey, C.R.; Wert, S.E.; Whitsett, J.A. Arrested Lung Morphogenesis in Transgenic Mice Bearing an SP-C–TGF-β1 Chimeric Gene. Dev. Biol. 1996, 175, 227–238. [Google Scholar] [CrossRef]
- Costantini, F.; Shakya, R. GDNF/Ret signaling and the development of the kidney. BioEssays 2006, 28, 117–127. [Google Scholar] [CrossRef]
- Villasenor, A.; Chong, D.C.; Henkemeyer, M.; Cleaver, O. Epithelial dynamics of pancreatic branching morphogenesis. Development 2010, 137, 4295–4305. [Google Scholar] [CrossRef]
- Klämbt, C.; Glazer, L.; Shilo, B.Z. breathless, a Drosophila FGF receptor homolog, is essential for migration of tracheal and specific midline glial cells. Genes Dev. 1992, 6, 1668–1678. [Google Scholar] [CrossRef]
- Kadam, S.; McMahon, A.; Tzou, P.; Stathopoulos, A. FGF ligands in Drosophila have distinct activities required to support cell migration and differentiation. Development 2009, 136, 739–747. [Google Scholar] [CrossRef]
- Isaac, D.D.; Andrew, D.J. Tubulogenesis in Drosophila: A requirement for the trachealess gene product. Genes Dev. 1996, 10, 103–117. [Google Scholar] [CrossRef]
- Nikolova, L.S.; Metzstein, M.M. Intracellular lumen formation in Drosophila proceeds via a novel subcellular compartment. Development 2015, 142, 3964–3973. [Google Scholar] [CrossRef]
- Baer, M.; Palm, W.; Eaton, S.; Leptin, M.; Affolter, M. Microsomal triacylglycerol transfer protein (MTP) is required cell autonomously to expand tracheal lumen in Drosophila in a cell-autonomous manner. J. Cell Sci. 2012, 125, 6038–6048. [Google Scholar] [CrossRef] [PubMed]
- Samakovlis, C. Development of the Drosophila tracheal system occurs by a series of morphologically distinct but genetically coupled branching events. Development 1996, 122, 1395–1407. [Google Scholar] [CrossRef]
- Ghabrial, A.S.; Levi, B.P.; Krasnow, M.A. A Systematic Screen for Tube Morphogenesis and Branching Genes in the Drosophila Tracheal System. PLoS Genet. 2011, 7, e1002087. [Google Scholar] [CrossRef] [PubMed]
- Bar, T.; Guldner, F.H.; Wolff, J.R. “Seamless” endothelial cells of blood capillaries. Cell Tissue Res. 1984, 235, 99–106. [Google Scholar] [CrossRef]
- Berry, K.L.; Bülow, H.E.; Hall, D.H.; Hobert, O. A C. elegans CLIC-like Protein Required for Intracellular Tube Formation and Maintenance. Science 2003, 302, 2134–2137. [Google Scholar] [CrossRef]
- Jewett, C.; Vanderleest, T.E.; Miao, H.; Xie, Y.; Madhu, R.; Loerke, D.; Blankenship, J.T. Planar polarized Rab35 functions as an oscillatory ratchet during cell intercalation in the Drosophila epithelium. Nat. Commun. 2017, 8, 1–16. [Google Scholar] [CrossRef]
- Rosa, J.B.; Metzstein, M.M.; Ghabrial, A.S. An Ichor-dependent apical extracellular matrix regulates seamless tube shape and integrity. PLoS Genet. 2018, 14, e1007146. [Google Scholar] [CrossRef]
- Adler, P.N.; Krasnow, R.E.; Liu, J. Tissue polarity points from cells that have higher Frizzled levels towards cells that have lower Frizzled levels. Curr. Biol. 1997, 7, 940–949. [Google Scholar] [CrossRef]
- Young, P.E.; Richman, A.M.; Ketchum, A.S.; Kiehart, D.P. Morphogenesis in Drosophila requires nonmuscle myosin heavy chain function. Genes Dev. 1993, 7, 29–41. [Google Scholar] [CrossRef]
- Bloor, J.W.; Kiehart, D.P. zipper Nonmuscle Myosin-II Functions Downstream of PS2 Integrin in Drosophila Myogenesis and Is Necessary for Myofibril Formation. Dev. Biol. 2001, 239, 215–228. [Google Scholar] [CrossRef] [PubMed]
- Morin, X.; Daneman, R.; Zavortink, M.; Chia, W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 15050–15055. [Google Scholar] [CrossRef]
- Katz, B.-Z.; Krylov, D.; Aota, S.-I.; Olive, M.; Vinson, C.; Yamada, K. Green Fluorescent Protein Labeling of Cytoskeletal Structures—Novel Targeting Approach Based on Leucine Zippers. Biotechniques 1998, 25, 298–304. [Google Scholar] [CrossRef]
- Spudich, J.A.; Rice, S.E.; Rock, R.S.; Purcell, T.J.; Warrick, H.M. Attachment of Anti-GFP Antibodies to Microspheres for Optical Trapping Experiments. Cold Spring Harb. Protoc. 2011, 2011, 1370–1371. [Google Scholar] [CrossRef]
- Greenland, S.; Senn, S.J.; Rothman, K.J.; Carlin, J.B.; Poole, C.; Goodman, S.N.; Altman, D.G. Statistical tests, P values, confidence intervals, and power: A guide to misinterpretations. Eur. J. Epidemiol. 2016, 31, 337–350. [Google Scholar] [CrossRef] [PubMed]
- Mutterer, J.; Zinck, E. Quck-and-clean article figures with FigureJ. J. Microsc. 2013, 252, 89–91. [Google Scholar] [CrossRef]
- Jones, T.A.; Metzstein, M.M. A Novel Function for the PAR Complex in Subcellular Morphogenesis of Tracheal Terminal Cells in Drosophila melanogaster. Genetics 2011, 189, 153–164. [Google Scholar] [CrossRef] [PubMed]
- Choi, C.K.; Vicente-Manzanares, M.; Zareno, J.; Whitmore, L.A.; Mogilner, A.; Horwitz, A.R. Actin and α-actinin orchestrate the assembly and maturation of nascent adhesions in a myosin II motor-independent manner. Nat. Cell Biol. 2008, 10, 1039–1050. [Google Scholar] [CrossRef]
- Conti, M.A.; Even-Ram, S.; Liu, C.; Yamada, K.; Adelstein, R. Defects in Cell Adhesion and the Visceral Endoderm following Ablation of Nonmuscle Myosin Heavy Chain II-A in Mice. J. Biol. Chem. 2004, 279, 41263–41266. [Google Scholar] [CrossRef] [PubMed]
- Nobes, C.D.; Hall, A. Rho, Rac, and Cdc42 GTPases regulate the assembly of multimolecular focal complexes associated with actin stress fibers, lamellipodia, and filopodia. Cell 1995, 81, 53–62. [Google Scholar] [CrossRef]
- Escudero, L.M.; Bischoff, M.; Freeman, M. Myosin II Regulates Complex Cellular Arrangement and Epithelial Architecture in Drosophila. Dev. Cell 2007, 13, 717–729. [Google Scholar] [CrossRef] [PubMed]
- JayaNandanan, N.; Mathew, R.; Leptin, M. Guidance of subcellular tubulogenesis by actin under the control of a synaptotagmin-like protein and Moesin. Nat. Commun. 2014, 5, 3036. [Google Scholar] [CrossRef]
- Levi, B.P.; Ghabrial, A.S.; Krasnow, M.A. Drosophila talin and integrin genes are required for maintenance of tracheal terminal branches and luminal organization. Development 2006, 133, 2383–2393. [Google Scholar] [CrossRef]
| Cell 1 | Cell 2 | Cell 3 | Cell 4 | Cell 5 | Cell 6 | |
|---|---|---|---|---|---|---|
| Control | 78.68 | 124.64 | 93.40 | 102.59 | 146.30 | 112.30 |
| Mutant | 0.00 | 83.22 | 43.32 | 39.12 | 28.80 | 46.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shin, J.-H.; Jeong, C.-W. RETRACTED: Zipper Is Necessary for Branching Morphogenesis of the Terminal Cells in the Drosophila melanogaster’s Tracheal System. Biology 2021, 10, 729. https://doi.org/10.3390/biology10080729
Shin J-H, Jeong C-W. RETRACTED: Zipper Is Necessary for Branching Morphogenesis of the Terminal Cells in the Drosophila melanogaster’s Tracheal System. Biology. 2021; 10(8):729. https://doi.org/10.3390/biology10080729
Chicago/Turabian StyleShin, Jong-Hyeon, and Chan-Woo Jeong. 2021. "RETRACTED: Zipper Is Necessary for Branching Morphogenesis of the Terminal Cells in the Drosophila melanogaster’s Tracheal System" Biology 10, no. 8: 729. https://doi.org/10.3390/biology10080729
APA StyleShin, J.-H., & Jeong, C.-W. (2021). RETRACTED: Zipper Is Necessary for Branching Morphogenesis of the Terminal Cells in the Drosophila melanogaster’s Tracheal System. Biology, 10(8), 729. https://doi.org/10.3390/biology10080729






