Clinical Outcome Prediction in COVID-19 Patients by Lymphocyte Subsets Analysis and Monocytes’ iTNF-α Expression
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population and Sample Collection
2.2. Flow Cytometry Analysis
2.3. Statistical Analysis
3. Results
3.1. Patient Characteristics
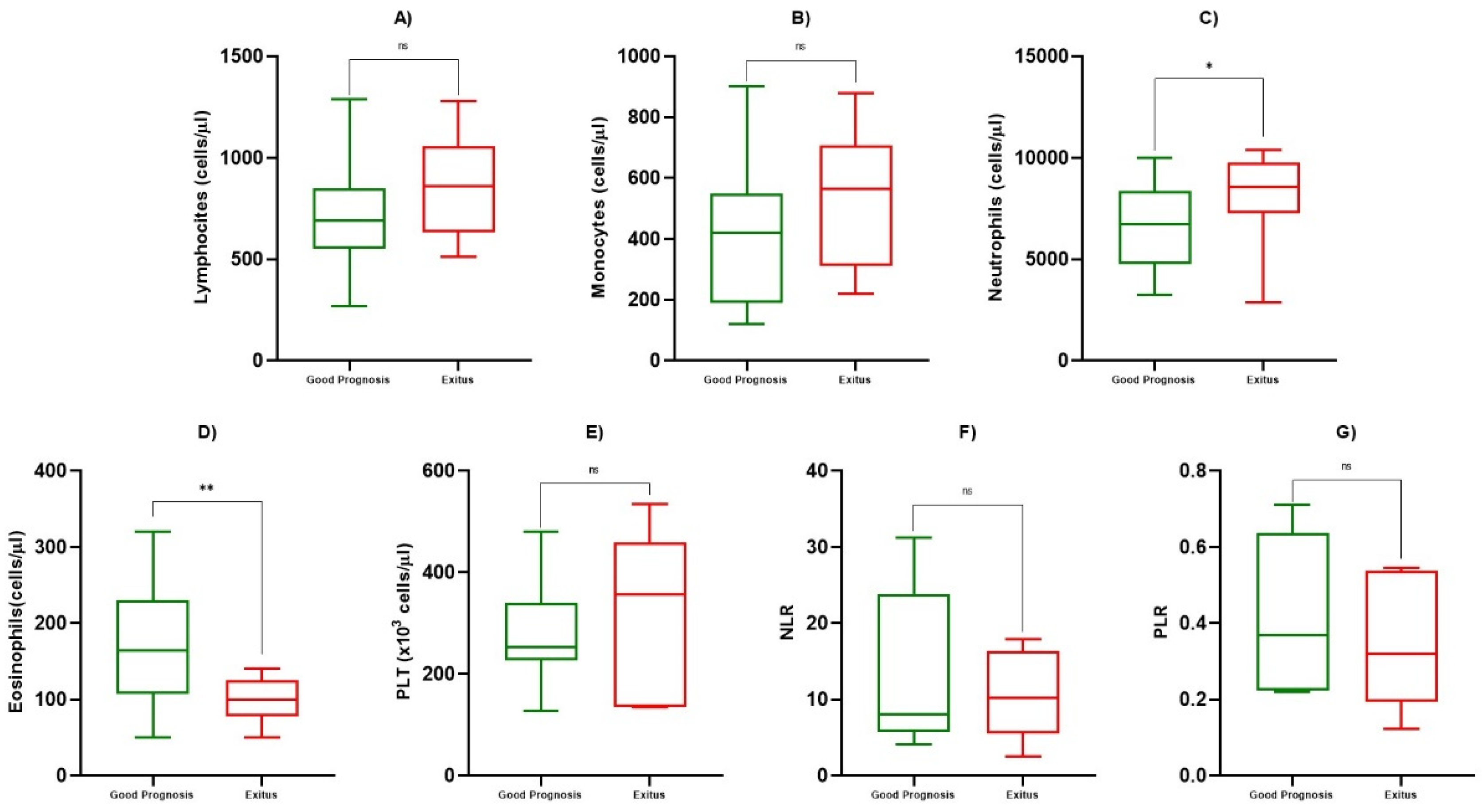
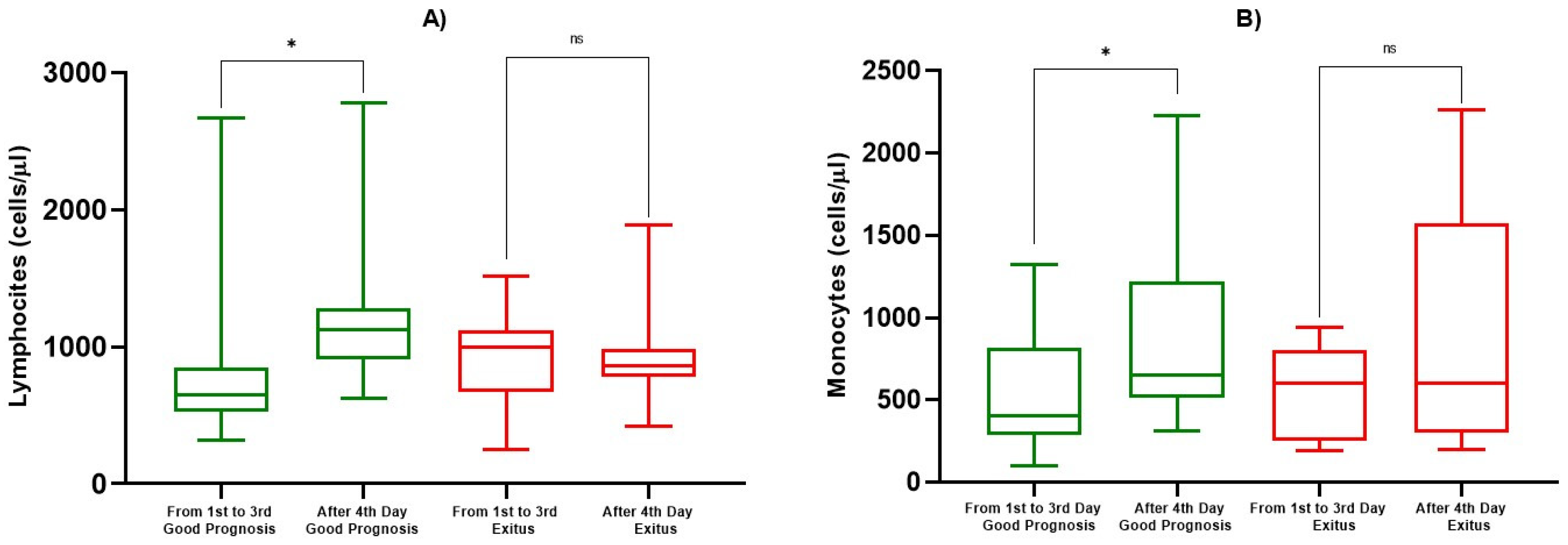
3.2. Hematological and Inflammatory Values
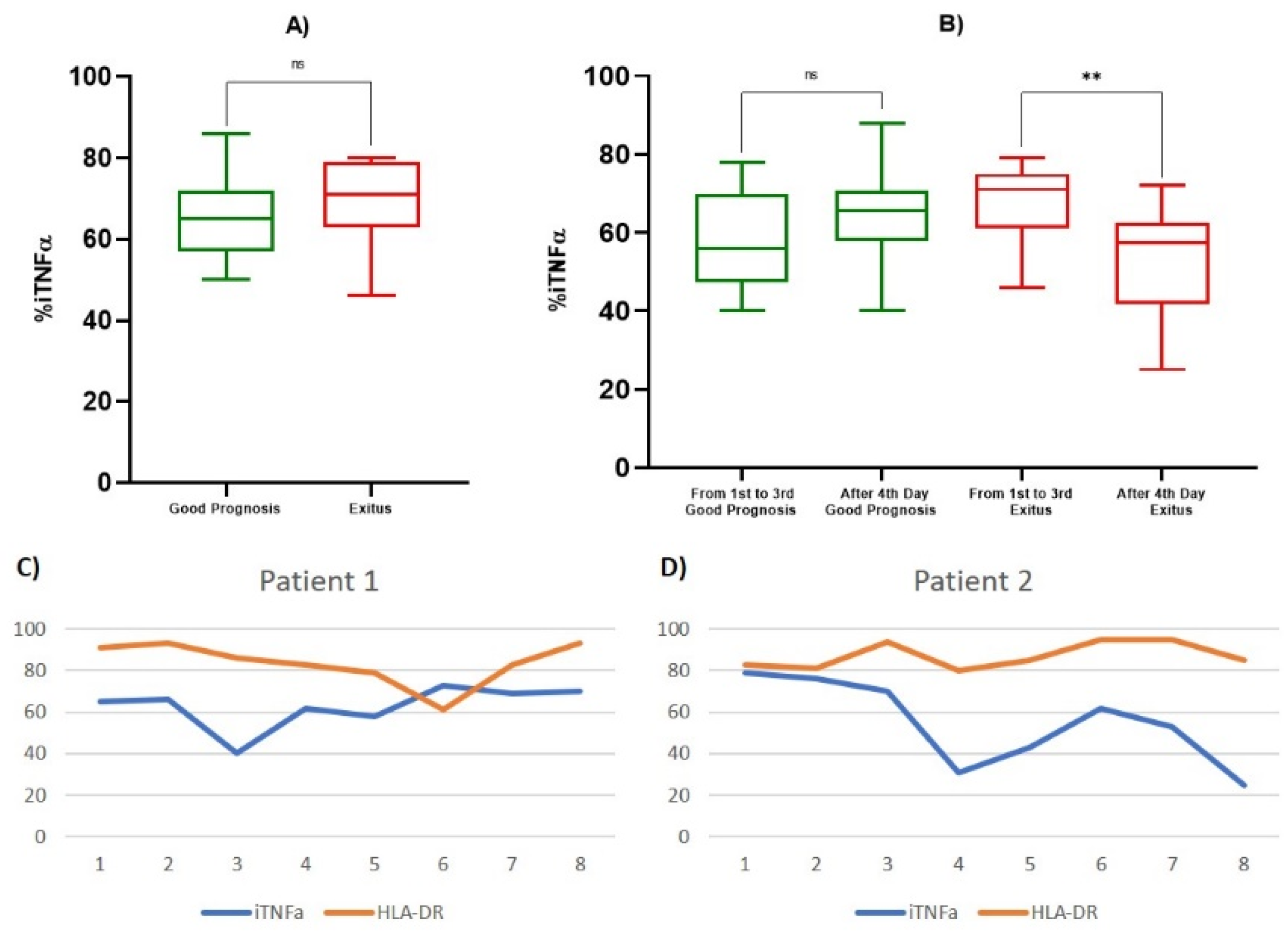
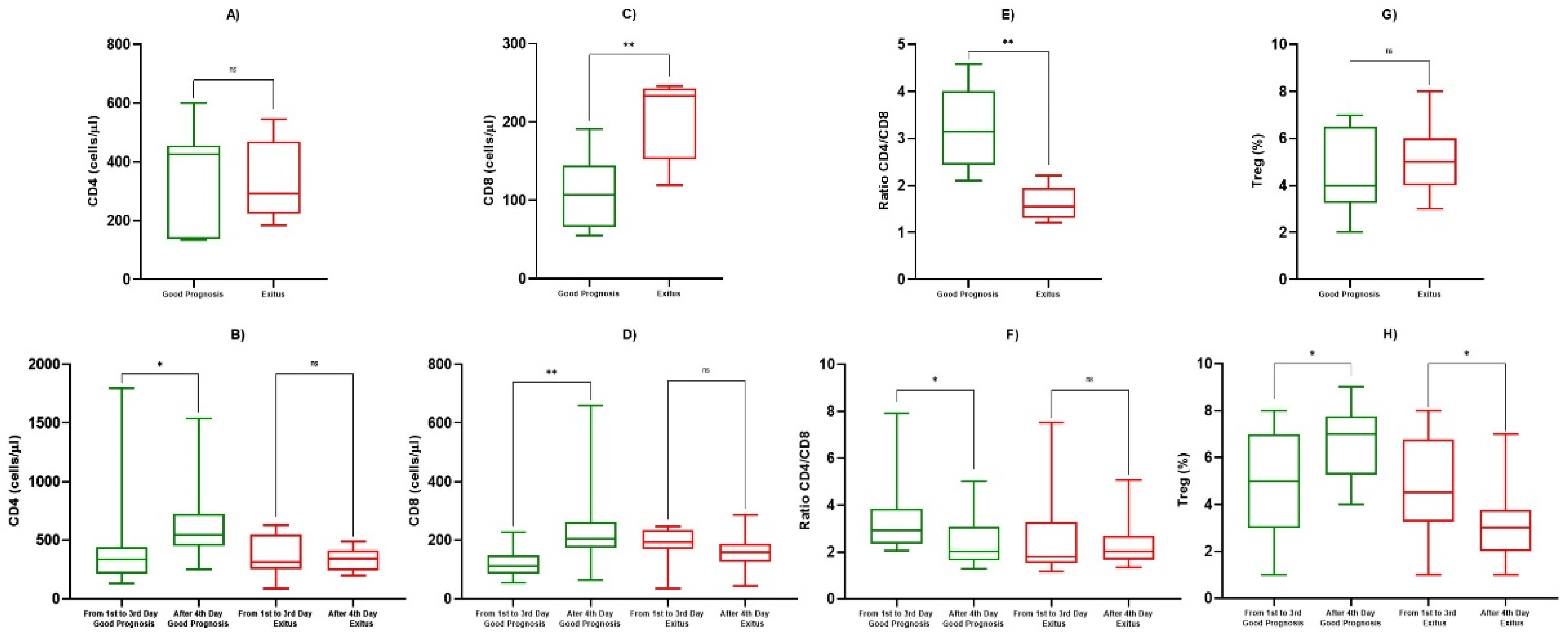
3.3. TNF-α and Lymphocyte Subpopulations Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhu, N.; Zhang, D.; Wang, W.; Li, X.; Yang, B.; Song, J.; Zhao, X.; Huang, B.; Shi, W.; Lu, R.; et al. A Novel Coronavirus from Patients with Pneumonia in China, 2019. N. Engl. J. Med. 2020, 382, 727–733. [Google Scholar] [CrossRef]
- Lu, R.; Zhao, X.; Li, J.; Niu, P.; Yang, B.; Wu, H.; Wang, W.; Song, H.; Huang, B.; Zhu, N.; et al. Genomic characterisation and epidemiology of 2019 novel coronavirus: Implications for virus origins and receptor binding. Lancet 2020, 395, 565–574. [Google Scholar] [CrossRef] [Green Version]
- Tan, W.; Zhao, X.; Ma, X.; Wang, W.; Niu, P.; Xu, W.; Gao, G.F.; Wu, G. A Novel Coronavirus Genome Identified in a Cluster of Pneumonia Cases—Wuhan, China 2019−2020. China CDC Wkly. 2020, 2, 61–62. [Google Scholar] [CrossRef]
- Huang, C.; Wang, Y.; Li, X.; Ren, L.; Zhao, J.; Hu, Y.; Zhang, L.; Fan, G.; Xu, J.; Gu, X.; et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet 2020, 395, 497–506. [Google Scholar] [CrossRef] [Green Version]
- Lechien, J.R.; Chiesa-Estomba, C.M.; De Siati, D.R.; Horoi, M.; Le Bon, S.D.; Rodriguez, A.; Dequanter, D.; Blecic, S.; El Afia, F.; Distinguin, L.; et al. Olfactory and gustatory dysfunctions as a clinical presentation of mild-to-moderate forms of the coronavirus disease (COVID-19): A multicenter European study. Eur. Arch. Oto-Rhino-Laryngol. 2020, 277, 2251–2261. [Google Scholar] [CrossRef]
- Wu, Z.; McGoogan, J.M. Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China. JAMA 2020, 323, 1239. [Google Scholar] [CrossRef]
- Matthay, M.A.; Ware, L.B.; Zimmerman, G.A. The acute respiratory distress syndrome. J. Clin. Investig. 2012, 122, 2731–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tisoncik, J.R.; Korth, M.J.; Simmons, C.P.; Farrar, J.; Martin, T.R.; Katze, M.G. Into the Eye of the Cytokine Storm. Microbiol. Mol. Biol. Rev. 2012, 76, 16–32. [Google Scholar] [CrossRef] [Green Version]
- Mehta, P.; McAuley, D.; Brown, M.; Sanchez, E.; Tattersall, R.S.; Manson, J.J. COVID-19: Consider cytokine storm syndromes and immunosuppression. Lancet 2020, 395, 1033–1034. [Google Scholar] [CrossRef]
- Chatenoud, L.; Ferran, C.; Legendre, C.; Thouard, I.; Merite, S.; Reuter, A.; Gevaert, Y.; Kreis, H.; Franchimont, P.; Bach, J.-F. In Vivo Cell Activation Following Okt3 Administration. Transplantation 1990, 49, 697–702. [Google Scholar] [CrossRef]
- Wang, H.; Ma, S. The cytokine storm and factors determining the sequence and severity of organ dysfunction in multiple organ dysfunction syndrome. Am. J. Emerg. Med. 2008, 26, 711–715. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Chen, H.; Xiaoyin, M.; Yang, G.; Hu, Y.; Xie, K.; Yu, Y. Autophagy Activation Improves Lung Injury and Inflammation in Sepsis. Inflammation 2019, 42, 426–439. [Google Scholar] [CrossRef] [PubMed]
- De Jong, M.D.; Simmons, C.P.; Thanh, T.T.; Hien, V.M.; Smith, G.J.; Chau, T.N.B.; Hoang, D.M.; Chau, N.V.V.; Khanh, T.H.; Dong, V.C.; et al. Fatal outcome of human influenza A (H5N1) is associated with high viral load and hypercytokinemia. Nat. Med. 2006, 12, 1203–1207. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Guo, R.; Lei, L.; Liu, H.; Wang, Y.; Wang, Y.; Qian, H.; Dai, T.; Zhang, T.; Lai, Y.; et al. Frontline Science: COVID-19 infection induces readily detectable morphologic and inflammation-related phenotypic changes in peripheral blood monocytes. J. Leukoc. Biol. 2020, 109, 13–22. [Google Scholar] [CrossRef] [PubMed]
- Diao, B.; Wang, C.; Tan, Y.; Chen, X.; Liu, Y.; Ning, L.; Chen, L.; Li, M.; Liu, Y.; Wang, G.; et al. Reduction and Functional Exhaustion of T Cells in Patients With Coronavirus Disease 2019 (COVID-19). Front. Immunol. 2020, 11, 827. [Google Scholar] [CrossRef]
- Guihot, A.; Litvinova, E.; Autran, B.; Debré, P.; Vieillard, V. Cell-Mediated Immune Responses to COVID-19 Infection. Front. Immunol. 2020, 11, 1662. [Google Scholar] [CrossRef]
- Ng, C.T.; Snell, L.M.; Brooks, D.; Oldstone, M.B. Networking at the Level of Host Immunity: Immune Cell Interactions during Persistent Viral Infections. Cell Host Microbe 2013, 13, 652–664. [Google Scholar] [CrossRef] [Green Version]
- Fenwick, C.; Joo, V.; Jacquier, P.; Noto, A.; Banga, R.; Perreau, M.; Pantaleo, G. T-cell exhaustion in HIV infection. Immunol. Rev. 2019, 292, 149–163. [Google Scholar] [CrossRef]
- Montesarchio, V.; Parella, R.; Iommelli, C.; Bianco, A.; Manzillo, E.; Fraganza, F.; Palumbo, C.; Rea, G.; Murino, P.; De Rosa, R.; et al. Outcomes and biomarker analyses among patients with COVID-19 treated with interleukin 6 (IL-6) receptor antagonist sarilumab at a single institution in Italy. J. Immunother. Cancer 2020, 8, e001089. [Google Scholar] [CrossRef]
- McGonagle, D.; Sharif, K.; O’Regan, A.; Bridgewood, C. The Role of Cytokines including Interleukin-6 in COVID-19 induced Pneumonia and Macrophage Activation Syndrome-Like Disease. Autoimmun. Rev. 2020, 19, 102537. [Google Scholar] [CrossRef]
- Watanabe, S.; Chan, K.W.K.; Wang, J.; Rivino, L.; Lok, S.-M.; Vasudevan, S.G. Dengue Virus Infection with Highly Neutralizing Levels of Cross-Reactive Antibodies Causes Acute Lethal Small Intestinal Pathology without a High Level of Viremia in Mice. J. Virol. 2015, 89, 5847–5861. [Google Scholar] [CrossRef] [Green Version]
- Mathison, J.C.; Wolfson, E.; Ulevitch, R.J. Participation of tumor necrosis factor in the mediation of gram negative bacterial lipopolysaccharide-induced injury in rabbits. J. Clin. Investig. 1988, 81, 1925–1937. [Google Scholar] [CrossRef] [Green Version]
- Li, C.; Gu, H.; Yu, M.; Yang, P.; Zhang, M.; Ba, H.; Yin, Y.; Wang, J.; Yin, B.; Zhou, X.; et al. Inhibition of transmembrane TNF-α shedding by a specific antibody protects against septic shock. Cell Death Dis. 2019, 10, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Lv, S.; Han, M.; Yi, R.; Kwon, S.; Dai, C.; Wang, R. Anti-TNF-α therapy for patients with sepsis: A systematic meta-analysis. Int. J. Clin. Pract. 2014, 68, 520–528. [Google Scholar] [CrossRef]
- Mehrad, B.; Strieter, R.M.; Standiford, T.J. Role of TNF-alpha in pulmonary host defense in murine invasive aspergillosis. J. Immunol. 1999, 162, 1633–1640. [Google Scholar] [PubMed]
- Qiu, P.; Cui, X.; Barochia, A.; Li, Y.; Natanson, C.; Eichacker, P.Q. The evolving experience with therapeutic TNF inhibition in sepsis: Considering the potential influence of risk of death. Expert Opin. Investig. Drugs 2011, 20, 1555–1564. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vandewalle, J.; Steeland, S.; Van Ryckeghem, S.; Eggermont, M.; Van Wonterghem, E.; Vandenbroucke, R.E.; Libert, C. A Study of Cecal Ligation and Puncture-Induced Sepsis in Tissue-Specific Tumor Necrosis Factor Receptor 1-Deficient Mice. Front. Immunol. 2019, 10, 2574. [Google Scholar] [CrossRef]
- Jin, J.-M.; Bai, P.; He, W.; Wu, F.; Liu, X.-F.; Han, D.-M.; Liu, S.; Yang, J.-K. Gender Differences in Patients with COVID-19: Focus on Severity and Mortality. Front. Public Health 2020, 8, 152. [Google Scholar] [CrossRef] [PubMed]
- Cai, H. Sex difference and smoking predisposition in patients with COVID-19. Lancet Respir. Med. 2020, 8, e20. [Google Scholar] [CrossRef]
- Henry, B.M. COVID-19, ECMO, and lymphopenia: A word of caution. Lancet Respir. Med. 2020, 8, e24. [Google Scholar] [CrossRef]
- Han, M.; Xu, M.; Zhang, Y.; Liu, Z.; Li, S.; He, T.; Li, J.; Gao, Y.; Liu, W.; Li, T.; et al. Assessing SARS-CoV-2 RNA levels and lymphocyte/T cell counts in COVID-19 patients revealed initial immune status as a major determinant of disease severity. Med. Microbiol. Immunol. 2020, 209, 657–668. [Google Scholar] [CrossRef] [PubMed]
- Swadling, L.; Maini, M.K. T cells in COVID-19—United in diversity. Nat. Immunol. 2020, 21, 1307–1308. [Google Scholar] [CrossRef] [PubMed]
- Fan, B.E.; Chong, V.C.L.; Chan, S.S.W.; Lim, G.H.; Lim, K.G.E.; Tan, G.B.; Mucheli, S.S.; Kuperan, P.; Ong, K.H. Hematologic parameters in patients with COVID-19 infection. Am. J. Hematol. 2020, 95, E131–E134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.; Zhou, J.; Zhou, Q.; Hu, L.; Long, Y. Role of eosinophils in the diagnosis and prognostic evaluation of COVID-19. J. Med. Virol. 2020, 93, 1105–1110. [Google Scholar] [CrossRef] [PubMed]
- Monneret, G.; Demaret, J.; Gossez, M.; Reverdiau, E.; Malergue, F.; Rimmelé, T.; Venet, F. Novel Approach in Monocyte Intracellular TNF Measurement. Shock 2017, 47, 318–322. [Google Scholar] [CrossRef]
- Bastug, A.; Bodur, H.; Erdogan, S.; Gokcinar, D.; Kazancioglu, S.; Kosovali, B.D.; Ozbay, B.O.; Gok, G.; Turan, I.O.; Yilmaz, G.; et al. Clinical and laboratory features of COVID-19: Predictors of severe prognosis. Int. Immunopharmacol. 2020, 88, 106950. [Google Scholar] [CrossRef]
- Zhou, F.; Yu, T.; Du, R.; Fan, G.; Liu, Y.; Liu, Z.; Xiang, J.; Wang, Y.; Song, B.; Gu, X.; et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020, 395, 1054–1062. [Google Scholar] [CrossRef]
- Ortiz-Prado, E.; Simbaña-Rivera, K.; Barreno, L.G.; Rubio-Neira, M.; Guaman, L.P.; Kyriakidis, N.C.; Muslin, C.; Jaramillo, A.M.G.; Barba-Ostria, C.; Cevallos-Robalino, D.; et al. Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the Coronavirus Disease 2019 (COVID-19), a comprehensive literature review. Diagn. Microbiol. Infect. Dis. 2020, 98, 115094. [Google Scholar] [CrossRef]
- Qin, C.; Zhou, L.; Hu, Z.; Zhang, S.; Yang, S.; Tao, Y.; Xie, C.; Ma, K.; Shang, K.; Wang, W.; et al. Dysregulation of Immune Response in Patients With Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. 2020, 71, 762–768. [Google Scholar] [CrossRef]
- Boyette, L.B.; Macedo, C.; Hadi, K.; Elinoff, B.D.; Walters, J.; Ramaswami, B.; Chalasani, G.; Taboas, J.M.; Lakkis, F.G.; Metes, D.M. Phenotype, function, and differentiation potential of human monocyte subsets. PLoS ONE 2017, 12, e0176460. [Google Scholar] [CrossRef]
- Ingersoll, M.A.; Spanbroek, R.; Lottaz, C.; Gautier, E.; Frankenberger, M.; Hoffmann, R.; Lang, R.; Haniffa, M.; Collin, M.; Tacke, F.; et al. Comparison of gene expression profiles between human and mouse monocyte subsets. Blood 2010, 115, e10–e19. [Google Scholar] [CrossRef] [PubMed]
- Pence, B.D. Severe COVID-19 and aging: Are monocytes the key? GeroScience 2020, 42, 1051–1061. [Google Scholar] [CrossRef]
- Mousavi, S.A.; Rad, S.; Rostami, T.; Rostami, M.; Mousavi, S.A.; Mirhoseini, S.A.; Kiumarsi, A. Hematologic predictors of mortality in hospitalized patients with COVID-19: A comparative study. Hematology 2020, 25, 383–388. [Google Scholar] [CrossRef] [PubMed]
- Keicho, N.; Itoyama, S.; Kashiwase, K.; Phi, N.C.; Long, H.T.; Ha, L.D.; Van Ban, V.; Hoa, B.K.; Le Hang, N.T.; Hijikata, M.; et al. Association of human leukocyte antigen class II alleles with severe acute respiratory syndrome in the Vietnamese population. Hum. Immunol. 2009, 70, 527–531. [Google Scholar] [CrossRef]
- Chan, J.F.-W.; Kok, K.-H.; Zhu, Z.; Chu, H.; To, K.K.-W.; Yuan, S.; Yuen, K.-Y. Genomic characterization of the 2019 novel human-pathogenic coronavirus isolated from a patient with atypical pneumonia after visiting Wuhan. Emerg. Microbes Infect. 2020, 9, 221–236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, Y.; Huang, Z.; Yin, G.; Zhang, X.; Ye, W.; Hu, Z.; Hu, C.; Wei, H.; Zeng, Y.; Chi, Y.; et al. Study of the lymphocyte change between COVID-19 and non-COVID-19 pneumonia cases suggesting other besides uncontrolled inflammation contributed to multi-organ injury. medRxiv 2020. [Google Scholar] [CrossRef]
- Tan, L.; Wang, Q.; Zhang, D.; Ding, J.; Huang, Q.; Tang, Y.; Wang, Q.; Miao, H. Lymphopenia predicts disease severity of COVID-19: A descriptive and predictive study. Signal. Transduct. Target. Ther. 2020, 5, 1–3. [Google Scholar] [CrossRef] [PubMed]
- Ziadi, A.; Hachimi, A.; Admou, B.; Hazime, R.; Brahim, I.; Douirek, F.; Zarrouki, Y.; El Adib, A.R.; Younous, S.; Samkaoui, A.M. Lymphopenia in critically ill COVID-19 patients: A predictor factor of severity and mortality. Int. J. Lab. Hematol. 2020, 43. [Google Scholar] [CrossRef]
- Scapini, P.; Lapinet-Vera, J.A.; Gasperini, S.; Calzetti, F.; Bazzoni, F.; Cassatella, M.A. The neutrophil as a cellular source of chemokines. Immunol. Rev. 2000, 177, 195–203. [Google Scholar] [CrossRef] [PubMed]
- Cassatella, M.A. The production of cytokines by polymorphonuclear neutrophils. Immunol. Today 1995, 16, 21–26. [Google Scholar] [CrossRef]
- Bordon, J.; Aliberti, S.; Fernandez-Botran, R.; Uriarte, S.M.; Rane, M.J.; Duvvuri, P.; Peyrani, P.; Morlacchi, L.C.; Blasi, F.; Ramirez, J. Understanding the roles of cytokines and neutrophil activity and neutrophil apoptosis in the protective versus deleterious inflammatory response in pneumonia. Int. J. Infect. Dis. 2013, 17, e76–e83. [Google Scholar] [CrossRef] [Green Version]
- Zou, L.; Dai, L.; Zhang, Y.; Fu, W.; Gao, Y.; Zhang, Z.; Zhang, Z. Clinical Characteristics and Risk Factors for Disease Severity and Death in Patients With Coronavirus Disease 2019 in Wuhan, China. Front. Med. 2020, 7, 532. [Google Scholar] [CrossRef] [PubMed]
- Martinez, F.O.; Combes, T.W.; Orsenigo, F.; Gordon, S. Monocyte activation in systemic Covid-19 infection: Assay and rationale. EBioMedicine 2020, 59, 102964. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Rial, J.; Rivero-Calle, I.; Salas, A.; Martinón-Torres, F. Role of Monocytes/Macrophages in Covid-19 Pathogenesis: Implications for Therapy. Infect. Drug Resist. 2020, 13, 2485–2493. [Google Scholar] [CrossRef]
- Mann, E.R.; Menon, M.; Knight, S.B.; Konkel, J.E.; Jagger, C.; Shaw, T.N.; Krishnan, S.; Rattray, M.; Ustianowski, A.; Bakerly, N.D.; et al. Longitudinal immune profiling reveals key myeloid signatures associated with COVID-19. Sci. Immunol. 2020, 5, eabd6197. [Google Scholar] [CrossRef] [PubMed]
- Ravin, K.A.; Loy, M. The Eosinophil in Infection. Clin. Rev. Allergy Immunol. 2015, 50, 214–227. [Google Scholar] [CrossRef] [PubMed]
- Samarasinghe, A.E.; Melo, R.C.N.; Duan, S.; LeMessurier, K.; Liedmann, S.; Surman, S.L.; Lee, J.J.; Hurwitz, J.L.; Thomas, P.G.; McCullers, J.A. Eosinophils Promote Antiviral Immunity in Mice Infected with Influenza A Virus. J. Immunol. 2017, 198, 3214–3226. [Google Scholar] [CrossRef] [PubMed]
- Mu, T.; Yi, Z.; Wang, M.; Wang, J.; Zhang, C.; Chen, H.; Bai, M.; Jiang, L.; Zhang, Y. Expression of eosinophil in peripheral blood of patients with COVID-19 and its clinical significance. J. Clin. Lab. Anal. 2020, 35, e23620. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Hu, B.; Hu, C.; Zhu, F.; Liu, X.; Zhang, J.; Wang, B.; Xiang, H.; Cheng, Z.; Xiong, Y.; et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus–Infected Pneumonia in Wuhan, China. JAMA 2020, 323, 1061. [Google Scholar] [CrossRef] [PubMed]
- Chiappelli, F. CoViD-19 Immunopathology & Immunotherapy. Bioinformation 2020, 16, 219–222. [Google Scholar] [CrossRef]
- Ruan, Q.; Yang, K.; Wang, W.; Jiang, L.; Song, J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensiv. Care Med. 2020, 46, 846–848. [Google Scholar] [CrossRef] [Green Version]
- Combadière, B. Immunité adaptative contre le virus SARS-CoV-2. Med. Sci. 2020, 36, 908–913. [Google Scholar] [CrossRef]
- Hu, D.; Li, L.; Shi, W.; Zhang, L. Less expression of CD4+ and CD8+ T cells might reflect the severity of infection and predict worse prognosis in patients with COVID-19: Evidence from a pooled analysis. Clin. Chim. Acta 2020, 510, 1–4. [Google Scholar] [CrossRef] [PubMed]
- Wong, R.; Wu, A.; To, K.F.; Lee, N.; Lam, C.W.K.; Wong, C.K.; Chan, P.; Ng, M.H.L.; Yu, L.M.; Hui, D.; et al. Haematological manifestations in patients with severe acute respiratory syndrome: Retrospective analysis. BMJ 2003, 326, 1358–1362. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Nie, J.; Wang, H.; Zhao, Q.; Xiong, Y.; Deng, L.; Song, S.; Ma, Z.; Mo, P.; Zhang, Y. Characteristics of Peripheral Lymphocyte Subset Alteration in COVID-19 Pneumonia. J. Infect. Dis. 2020, 221, 1762–1769. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Huang, J.; Huang, Y.; Chen, J.; Jiang, X.; Shi, Y. Characteristics of immune cells and cytokines in patients with coronavirus disease 2019 in Guangzhou, China. Hum. Immunol. 2020, 81, 702–708. [Google Scholar] [CrossRef]
- Sadeghi, A.; Tahmasebi, S.; Mahmood, A.; Kuznetsova, M.; Valizadeh, H.; Taghizadieh, A.; Nazemiyeh, M.; Aghebati-Maleki, L.; Jadidi-Niaragh, F.; Abbaspour-Aghdam, S.; et al. Th17 and Treg cells function in SARS-CoV2 patients compared with healthy controls. J. Cell. Physiol. 2020, 236, 2829–2839. [Google Scholar] [CrossRef]
- Meidaninikjeh, S.; Sabouni, N.; Marzouni, H.Z.; Bengar, S.; Khalili, A.; Jafari, R. Monocytes and macrophages in COVID-19: Friends and foes. Life Sci. 2021, 269, 119010. [Google Scholar] [CrossRef] [PubMed]
- Schulte-Schrepping, J.; Reusch, N.; Paclik, D.; Baßler, K.; Schlickeiser, S.; Zhang, B.; Krämer, B.; Krammer, T.; Brumhard, S.; Bonaguro, L.; et al. Severe COVID-19 Is Marked by a Dysregulated Myeloid Cell Compartment. Cell 2020, 182, 1419–1440.e23. [Google Scholar] [CrossRef] [PubMed]
- Chaudhry, H.; Zhou, J.; Zhong, Y.; Ali, M.M.; McGuire, F.; Nagarkatti, P.S.; Nagarkatti, M. Role of cytokines as a double-edged sword in sepsis. In Vivo 2013, 27, 669–684. [Google Scholar] [PubMed]
| Study Population | ALL Patients N. 25 (100%) | Good Prognosis N. 16 (76%) | Exitus N. 9 (24%) |
|---|---|---|---|
| Female/Male, n (%) | 5 (20)/20 (80) | 3 (19)/13 (81)/ | 2 (22)/7 (78) |
| Median Age (Range) | 61 (34–78) | 64 (34–71) | 59 (48–67) |
| Median Days Oxygen Therapy (Range) | 10 (5–17) | 7 (5–12) | 13 (8–17) |
| Median P/F (Range) | 165 (60–338) | 139 (60–276) | 200 (80–338) |
| Subjects Without Comorbidity, n (%) | 14 (56) | 11 (78) | 3 (22) |
| Subjects With ONE Comorbidity, n (%) | 7 (28) | 3 (42) | 4 (58) |
| Subjects With TWO Comorbidities, n (%) | 4 (16) | 1 (25) | 3 (75) |
| Diabetes, n | 2 | 0 | 2 |
| Hypertension, n | 10 | 4 | 6 |
| Chronic Liver Disease, n | 1 | 0 | 1 |
| Chronic Pulmonary Disease, n | 1 | 0 | 1 |
| Chronic Renal Disease, n | 1 | 1 | 0 |
| Parameter at First Observation | Good Prognosis Mean ± SD | Exitus Mean ± SD | p-Value |
|---|---|---|---|
| ALC (cells/µL) | 767 ± 262 | 846 ± 241 | 0.684 |
| AMC (cells/µL) | 438 ± 255 | 540 ± 208 | 0.469 |
| ANC (cells/µL) | 7149 ± 2499 | 8422 ± 3163 | 0.0161 |
| AEC (cells/µL) | 179 ± 82 | 108 ± 25 | 0.0033 |
| PLT (cells/µL) | 276 ± 109 | 308 ± 172 | 0.569 |
| NLR | 13.16 ± 10.33 | 10.77 ± 5.89 | 0.935 |
| PLR | 0.440 ± 0.206 | 0.357 ± 0.180 | 0.464 |
| Parameter | Determination Points | Good Prognosis Mean ± SD | p-Value | Exitus Mean ± SD | p-Value |
|---|---|---|---|---|---|
| ALC (cells/µL) | 1st–3rd Day | 814 ± 546 | 0.0193 | 925 ± 268 | 0.725 |
| After 4th Day | 1184 ± 449 | 881 ± 250 | |||
| AMC (cells/µL) | 1st–3rd Day | 551 ± 367 | 0.0339 | 550 ± 282 | 0.290 |
| After 4th Day | 847 ± 471 | 908 ± 699 |
| Parameter | Determination Points | Mean ± SD | p-Value |
|---|---|---|---|
| iTNF-α % | 1st Observation (Good Prognosis) | 65.429 ± 11.914 | 0.469 |
| 1st Observation (Exitus) | 69.200 ± 13.570 | ||
| 1st–3rd Day (Good Prognosis) | 58.111 ± 12.6207 | 0.432 | |
| After 4th Day (Good Prognosis) | 64.167 ± 10.9450 | ||
| 1st–3rd Day (Exitus) | 67.467 ± 10.2878 | 0.0046 | |
| After 4th Day (Exitus) | 53.357 ± 14.1073 | ||
| CD4+ (cells/µL) | 1st Observation (Good Prognosis) | 367 ± 426 | 0.758 |
| 1st Observation (Exitus) | 334 ± 291 | ||
| 1st–3rd Day (Good Prognosis) | 431 ± 383 | 0.045 | |
| After 4th Day (Good Prognosis) | 561 ± 288 | ||
| 1st–3rd Day (Exitus) | 363 ± 162 | 0.748 | |
| After 4th Day (Exitus) | 334 ± 91 | ||
| CD8+ (cells/µL) | 1st Observation (Good Prognosis) | 109 ± 47 | 0.0085 |
| 1st Observation (Exitus) | 204 ± 54 | ||
| 1st–3rd Day (Good Prognosis) | 121 ± 51 | 0.0017 | |
| After 4th day (Good Prognosis) | 228 ± 126 | ||
| 1st–3rd Day (Exitus) | 181 ± 67 | 0.151 | |
| After 4th Day (Exitus) | 160 ± 57 | ||
| Ratio CD4+/CD8+ | 1st Observation (Good Prognosis) | 3.25 ± 0.85 | 0.0026 |
| 1st Observation (Exitus) | 1.61 ± 0.37 | ||
| 1st–3rd Day (Good Prognosis) | 3.26 ± 1.39 | 0.0209 | |
| After 4th Day (Good Prognosis) | 2.87 ± 0.63 | ||
| 1st–3rd day (Exitus) | 2.16 ± 0.95 | 0.349 | |
| After 4th Day (Exitus) | 2.28 ± 0.88 | ||
| Tregs (cells/µL) | 1st Observation (Good Prognosis) | 4.83 ± 1.89 | 0.884 |
| 1st Observation (Exitus) | 4.92 ± 1.58 | ||
| 1st–3rd Day (Good Prognosis) | 4.92 ± 2.35 | 0.0429 | |
| After 4th Day (Good Prognosis) | 6.79 ± 1.55 | ||
| 1st–3rd Day (Exitus) | 4.75 ± 2.22 | 0.0426 | |
| After 4th Day (Exitus) | 3.75 ± 1.65 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madonna, G.; Sale, S.; Capone, M.; De Falco, C.; Santocchio, V.; Di Matola, T.; Fiorentino, G.; Pirozzi, C.; D’Antonio, A.; Sabatino, R.; et al. Clinical Outcome Prediction in COVID-19 Patients by Lymphocyte Subsets Analysis and Monocytes’ iTNF-α Expression. Biology 2021, 10, 735. https://doi.org/10.3390/biology10080735
Madonna G, Sale S, Capone M, De Falco C, Santocchio V, Di Matola T, Fiorentino G, Pirozzi C, D’Antonio A, Sabatino R, et al. Clinical Outcome Prediction in COVID-19 Patients by Lymphocyte Subsets Analysis and Monocytes’ iTNF-α Expression. Biology. 2021; 10(8):735. https://doi.org/10.3390/biology10080735
Chicago/Turabian StyleMadonna, Gabriele, Silvia Sale, Mariaelena Capone, Chiara De Falco, Valentina Santocchio, Tiziana Di Matola, Giuseppe Fiorentino, Caterina Pirozzi, Anna D’Antonio, Rocco Sabatino, and et al. 2021. "Clinical Outcome Prediction in COVID-19 Patients by Lymphocyte Subsets Analysis and Monocytes’ iTNF-α Expression" Biology 10, no. 8: 735. https://doi.org/10.3390/biology10080735
APA StyleMadonna, G., Sale, S., Capone, M., De Falco, C., Santocchio, V., Di Matola, T., Fiorentino, G., Pirozzi, C., D’Antonio, A., Sabatino, R., Atripaldi, L., Atripaldi, U., Raffone, M., Curvietto, M., Grimaldi, A. M., Vanella, V., Festino, L., Scarpato, L., Palla, M., ... Atripaldi, L. (2021). Clinical Outcome Prediction in COVID-19 Patients by Lymphocyte Subsets Analysis and Monocytes’ iTNF-α Expression. Biology, 10(8), 735. https://doi.org/10.3390/biology10080735







