Profiling Human CD55 Transgene Performance Assist in Selecting Best Suited Specimens and Tissues for Swine Organ Xenotransplantation
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Human Volunteers
2.2. Swine Samples
2.3. RNA Isolation, cDNA Synthesis and Gene Expression Analysis
2.4. Protein Extraction and Western Blot Analysis
2.5. Hemolysis Assay
2.6. Real-Time Cytotoxicity Assay
2.7. RNA Isolation from Fibroblast, cDNA Synthesis and Gene Expression Analysis
2.8. Statistical Analysis
3. Results
3.1. Human CD55 Transgene Protects from Complement-Mediated Hemolysis
3.2. Human CD55 mRNA Shows NO Relatable Expression Pattern across Collected Tissues
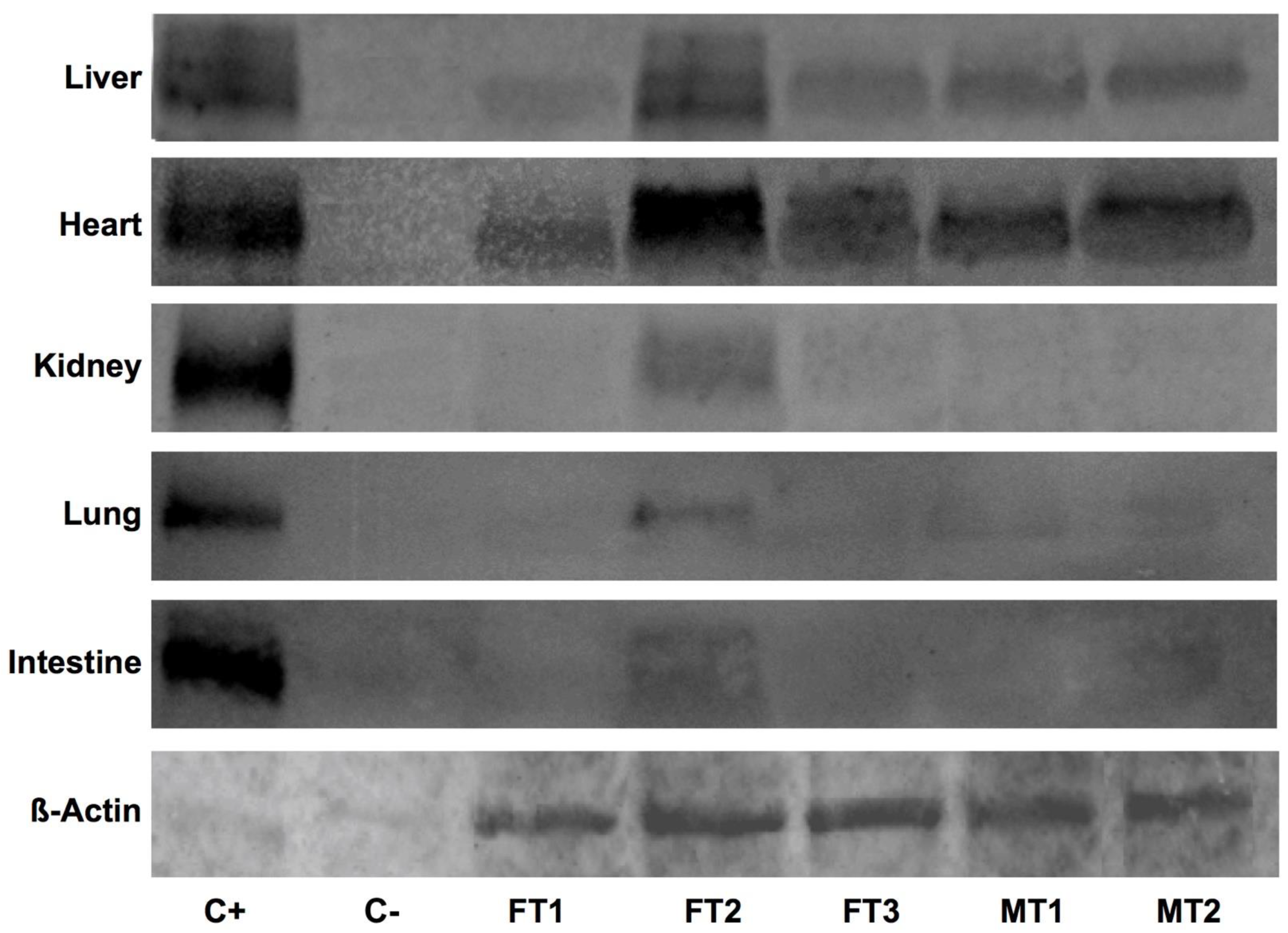
3.3. Human DAF Levels Fluctuate among Specimens and Collected Organs
3.4. Human CD55 and Human DAF Levels Can Be Correlated between Several Tissues
3.5. Fibroblast Cultures Expressing hCD55 Show Variable Responses in the RTCA Assay
3.6. Transgenic Fibroblast Viability after Serum Challenge Relates to Human CD55 Transcript Levels
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Samdani, T. Xenotransplantation. Available online: https://emedicine.medscape.com/article/432418-overview#showallTitleofSite (accessed on 13 April 2021).
- Cozzi, E.; Tucker, A.W.; Langford, G.A.; Pino-Chavez, G.; Wright, L.; O’Connell, M.-J.; Young, V.J.; Lancaster, R.; McLaughlin, M.; Hunt, K.; et al. Characterization of pigs transgenic for human decay-accelerating factor. Transplantation 1997, 64, 1383–1392. [Google Scholar] [CrossRef] [PubMed]
- Cowan, P.; Aminian, A.; Barlow, H.; Brown, A.; Chen, C.G.; Fisicaro, N.; Francis, D.M.; Goodman, D.J.; Han, W.; Kurek, M.; et al. Renal xenografts from triple transgenic pigs are not hyperacutely rejected but cause coagulopathy in non-immunosuppressed baboons. Transplantation 2000, 69, 2504–2515. [Google Scholar] [CrossRef]
- Adams, D.H.; Kadner, A.; Chen, R.H.; Farivar, R.S. Human membrane cofactor protein (MCP, CD 46) protects transgenic pig hearts from hyperacute rejection in primates. Xenotransplantation 2001, 8, 36–40. [Google Scholar] [CrossRef]
- Lavitrano, M.; Bacci, M.L.; Forni, M.; Lazzereschi, D.; Di Stefano, C.; Fioretti, D.; Giancotti, P.; Marfé, G.; Pucci, L.; Renzi, L.; et al. Efficient production by sperm-mediated gene transfer of human decay accelerating factor (hDAF) transgenic pigs for xenotransplantation. Proc. Natl. Acad. Sci. USA 2002, 99, 14230–14235. [Google Scholar] [CrossRef] [Green Version]
- Waterworth, P.D.; Dunning, J.; Tolan, M.; Cozzi, E.; Langford, G.; Chavez, G.; White, D.; Wallwork, J. Life-supporting pig-to baboon heart xenotransplantation. J. Heart Lung Transplant. 1998, 17, 1201–1207. [Google Scholar] [PubMed]
- Cozzi, E.; Bhatti, F.; Schmoeckel, M.; Chavez, G.; Smith, K.; Zaidi, A.; Bradley, J.; Thiru, S.; Goddard, M.; Vial, C.; et al. Long-term survival of nonhuman primates receiving life-supporting transgenic porcine kidney xenografts. Transplantation 2000, 70, 15–21. [Google Scholar]
- Ramirez, P.; Chavez, R.; Majado, M.; Munitiz, V.; Muñoz, A.; Hernandez, Q.; G-Palenciano, C.; Pino-Chavez, G.; Loba, M.; Loba, M.; et al. Life-supporting human complement regulator decay accelerating factor transgenic pig liver xenograft maintains the metabolic function and coagulation in the nonhuman primate for up to 8 days. Transplantation 2000, 70, 989–998. [Google Scholar] [CrossRef] [PubMed]
- Ramírez, P.; Montoya, M.J.; Ríos, A.; García Palenciano, C.; Majado, M.; Chavez, R.; Muñoz, A.; Fernández, O.M.; Sánchez, A.; Segura, B.; et al. Prevention of hyperacute rejection in a modelo of orthotopic liver xenotransplantation from pig to baboon using polytransgenic pig livers (CD55, CD59, and H-transferase). Transplant. Proc. 2005, 37, 4103–4106. [Google Scholar] [CrossRef]
- Whyte, J.J.; Prather, R.S. Genetic modifications of pigs for medicine and agriculture. Mol. Reprod. Dev. 2011, 78, 879–891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, T.; Pipkin, P.A.; Clarkson, N.A.; Stone, D.M.; Minor, P.D.; Almond, J.W. Decay-accelerating factor CD55 is identified as the receptor for echovirus 7 using CELICS, a rapid immuno-focal cloning method. EMBO J. 1994, 13, 5070–5074. [Google Scholar] [CrossRef] [PubMed]
- Ozen, A.; Comrie, W.A.; Ardy, R.C.; Domínguez Conde, C.; Dalgic, B.; Beser, Ö.F.; Morawski, A.R.; Karakoc-Aydiner, E.; Tutar, E.; Baris, S.; et al. CD55 Deficiency, Early-Onset Protein-Losing Enteropathy, and Thrombosis. N. Engl. J. Med. 2017, 377, 52–61. [Google Scholar] [CrossRef]
- Christy, J.M.; Toomey, C.B.; Cauvi, D.M.; Pollard, K.M. Chapter 25—Decay-Accelerating Factor. In The Complement Facts Book, 2nd ed.; Barnum, S., Schein, T., Eds.; Academic Press: Cambridge, MA, USA, 2018; pp. 261–270. [Google Scholar]
- Caras, I.W.; Davitz, M.A.; Rhee, L.; Weddell, G.; Martin, D.W., Jr.; Nussenzweig, V. Cloning of decay-accelerating factor suggests novel use of splicing to generate two proteins. Nature 1987, 325, 545–549. [Google Scholar] [CrossRef]
- Gelderman, K.A.; Zijlmans, H.J.; Vonk, M.J.; Gorter, A. CD55 expression patterns on intestinal neuronal tissue are divergent from the brain. Gut 2004, 53, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Nicholson-Weller, A.; March, J.P.; Rosen, C.E.; Spicer, D.B.; Austen, K.F. Surface membrane expression by human blood leukocytes and platelets of decay-accelerating factor, a regulatory protein of the complement system. Blood 1985, 65, 1237–1244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medof, M.E.; Walter, E.I.; Rutgers, J.L.; Knowles, D.M.; Nussenzweig, V. Identification of the complement decay-accelerating factor (DAF) on epithelium and glandular cells and in body fluids. J. Exp. Med. 1987, 165, 848–864. [Google Scholar] [CrossRef]
- Lublin, D.M.; Atkinson, J.P. Decay-accelerating factor: Biochemistry, molecular biology, and function. Annu. Rev. Immunol. 1989, 7, 35–58. [Google Scholar] [CrossRef] [PubMed]
- Nicholson-Weller, A.; Wang, C.E. Structure and function of decay accelerating factor CD55. J. Lab. Clin. Med. 1994, 123, 485–491. [Google Scholar]
- Miwa, T.; Sun, X.; Ohta, R.; Okada, N.; Harris, C.L.; Morgan, B.P.; Song, W.-C. Characterization of glycosylphosphatidylinositol-anchored decay accelerating factor (GPI-DAF) and transmembrane DAF gene expression in wild-type and GPI-DAF gene knockout mice using polyclonal and monoclonal antibodies with dual or single specificity. Immunology 2001, 104, 207–214. [Google Scholar] [CrossRef]
- Werth, V.P.; Ivanov, I.E.; Nussenzweig, V. Decay-accelerating factor in human skin is associated with elastic fibers. J. Investig. Dermatol. 1988, 91, 511–516. [Google Scholar] [CrossRef] [Green Version]
- Sayama, K.; Shiraishi, S.; Shirakata, Y.; Kobayashi, Y.; Miki, Y. Characterization of decay-accelerating factor (DAF) in human skin. J. Investig. Dermatol. 1991, 96, 61–64. [Google Scholar] [CrossRef] [Green Version]
- Ting, A.; Morris, P.J. A technique for lymphocyte preparation from stored heparinized blood. Vox Sang. 1971, 20, 561–563. [Google Scholar] [CrossRef]
- Martínez-Alarcón, L.; Quereda, J.J.; Herrero-Medrano, J.M.; Majado, M.J.; Mendoça, L.; Pallarés, F.J.; Ríos, A.; Ramírez, P.; Muñoz, A.; Ramis, G. Design of a real-time quantitative polymerase chain reaction to assess human complement regulatory protein gene expression in polytransgenic xenograft pigs. Transplant. Proc. 2010, 42, 3235–3238. [Google Scholar] [CrossRef] [PubMed]
- Duvigneau, J.C.; Hartl, R.T.; Groiss, S.; Gemeiner, M. Quantitative simultaneous multiplex real-time PCR for the detection of porcine cytokines. J. Immunol. Methods 2005, 306, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Erkens, T.; Van Poucke, M.; Vandesompele, J.; Goossens, K.; Van Zeveren, A.; Peelman, L.J. Development of a new set of reference genes for normalization of real-time RT-PCR data of porcine backfat and longissimus dorsi muscle, and evaluation with PPARGC1A. BMC Biotechnol. 2006, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Vandesompele, J.; De Preter, K.; Pattyn, F.; Poppe, B.; Van Roy, N.; De Paepe, A.; Speleman, F. Accurate normalization of real-time quantative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002, 3, research0034.1. [Google Scholar] [CrossRef] [Green Version]
- Ramis, G.; Martínez-Alarcón, L.; Quereda, J.J.; Mendonça, L.; Majado, M.J.; Gomez-Coelho, K.; Mrowiec, A.; Herrero-Medrano, J.M.; Abella-neda, J.M.; Pallares, F.J.; et al. Optimization of cytotoxicity assay by real-time, impedance-based cell analysis. Biomed. Microdevices 2013, 15, 985–995. [Google Scholar] [CrossRef]
- Quereda, J.J.; Martinez-Alarcon, L.; Mendoça, L.; Majado, M.; Herrero-Medrano, J.; Pallarés, F.; Ríos, A.; Ramírez, P.; Muñoz, A.; Ramis, G. Validation of xCELLigence real-time cell analyzer to assess compatibility in xenotransplantation with pig-to-baboon model. Transplant. Proc. 2010, 42, 3239–3243. [Google Scholar] [CrossRef]
- Ramis, G.; Martinez-Alarcon, L.; Majado, M.; Quereda, J.J.; Mendonça, L.; Herrero-Medrano, J.; Abellaneda, J.; Coelho, K.; López-Navas, A.; Ríos, A.; et al. Donor-Graft Compatibility Tests in Pig-to-Primate Xenotransplantation Model: Serum Versus Plasma in Real-Time Cell Analyzer Trials. Transplant. Proc. 2011, 43, 243–259. [Google Scholar] [CrossRef] [PubMed]
- Walpen, A.J.; Mohacsi, P.; Frey, C.; Roos, A.; Daha, M.R.; Rieben, R. Activation of complement pathways in xenotransplantation: An in vitro study. Transpl. Immunol. 2002, 9, 271–280. [Google Scholar] [CrossRef]
- Lee, J.-M.; Tu, C.-F.; Tai, H.-C.; Chou, N.-K.; Yang, T.-S.; Weng, C.-N.; Lee, Y.-C.; Lee, C.-J.; Lee, P.-H. The hDAF exogene protects swine endothelial and peripheral blood mononuclear cells from xenoreactive antibody mediated cytotoxicity in hDAF transgenic pigs. Transplant. Proc. 2006, 38, 2270–2272. [Google Scholar] [CrossRef]
- Smolenski, R.T.; Forni, M.; Maccherini, M.; Bacci, M.; Slominska, E.M.; Wang, H.; Fornasari, P.; Giovannoni, R.; Simeone, F.; Zannoni, A.; et al. Reduction of hyperacute rejection and protection of metabolism and function in hearts of human decay accelerating factor (hDAF)-expressing pigs. Cardiovasc. Res. 2007, 73, 143–152. [Google Scholar] [CrossRef]
- Díaz-Román, T.M.; Mañez, R.; López-Pelaez, E.; Centeno, A.; Moscoso, I.; Pértegaz, S.; Doménech, N. Human DAF on pig cells protects against human and non-human primate sera cytotoxicity mediated by exogenous or endogenous complement, as determined by flow cytometry. Transpl. Immunol. 2006, 16, 125–130. [Google Scholar] [CrossRef]
- Available online: https://www.ebi.ac.uk/gxa/genes/ensg00000196352 (accessed on 11 March 2012).
- Uhlén, M.; Fagerberg, L.; Hallström, B.M.; Lindskog, C.; Oksvold, P.; Mardinoglu, A.; Sivertsson, Å.; Kampf, C.; Sjöstedt, E.; Asplund, A.; et al. Proteomics. Tissue-based map of the human proteome. Science 2015, 23, 347. [Google Scholar]
- Jeong, Y.H.; Park, C.H.; Jang, G.H.; Jeong, Y.I.; Hwang, I.S.; Jeong, Y.; Kim, Y.-K.; Shin, T.; Kim, N.-H.; Hyun, S.-H.; et al. Production of multiple transgenic Yucatan miniature pigs expressing human complement regulatory factors, human CD55, CD59, and H-Transferase genes. PLoS ONE 2012, 8, e63241. [Google Scholar] [CrossRef]
- Pérez de la Lastra, J.M.; Harris, C.L.; Hinchliffe, S.J.; Holt, D.S.; Rushmere, N.K.; Morgan, B.P. Pigs express multiple forms of decay-accelerating factor (CD55), all of which contain only three short consensus repeats. J. Immunol. 2000, 165, 2563–2573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ichida, S.; Yuzawa, Y.; Okada, H.; Yoshioka, K.; Matsuo, S. Localization of the complement regulatory proteins in the normal human kidney. Kidney Int. 1994, 46, 89–96. [Google Scholar] [CrossRef] [Green Version]
- Di Domenico, G.; Del Vecchio, L.; Postiglione, L.; Ramaglia, L. Immunophenotypic analysis of human gingival fibroblasts and its regulation by Granulocyte-Macrophage Colony-Stimulating Factor (GM-CSF). Minerva Stomatol. 2003, 52, 81–87, 87–91. [Google Scholar]
- Iwase, H.; Jagdale, A.; Yamamoto, T.; Bikhet, M.H.; Nguyen, H.Q.; Ezzelarab, M.; Ayares, D.; Anderson, D.J.; Eckhoff, D.E.; Foote, J.B.; et al. Evidence suggesting that deletion of expression of N-glycolylneuraminic acid (Neu5Gc) in the organ-source pig is associated with increased antibody-mediated rejection of kidney transplants in baboons. Xenotransplantation 2021, 25, e12700. [Google Scholar]
- Watanabe, H.; Ariyoshi, Y.; Pomposelli, T.; Takeuchi, K.; Ekanayake-Alper, D.K.; Boyd, L.K.; Arn, S.J.; Sahara, H.; Shimizu, A.; Ayares, D.; et al. Intra-bone bone marrow transplantation from hCD47 transgenic pigs to baboons prolongs chimerism to >60 days and promotes increased porcine lung transplant survival. Xenotransplantation 2020, 27, e12552. [Google Scholar] [CrossRef]
- Abicht, J.M.; Sfriso, R.; Reichart, B.; Längin, M.; Gahle, K.; Puga Yung, G.L.; Seebach, J.D.; Rieben, R.; Ayares, D.; Wolf, E.; et al. Multiple genetically modified GTKO/hCD46/HLA-E/hβ2-mg porcine hearts are protected from complement activation and natural killer cell infiltration during ex vivo perfusion with human blood. Xenotransplantation 2018, 25, e12390. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, H.; Sahara, H.; Nomura, S.; Tanabe, T.; Ekanayake-Alper, D.K.; Boyd, L.K.; Louras, N.J.; Asfour, A.; Danton, M.A.; Ho, S.H.; et al. GalT-KO pig lungs are highly susceptible to acute vascular rejection in baboons, which may be mitigated by transgenic expression of hCD47 on porcine blood vessels. Xenotransplantation 2018, 25, e12391. [Google Scholar] [CrossRef] [PubMed]
- Cimeno, A.; Hassanein, W.; French, B.M.; Powell, J.M.; Burdorf, L.; Goloubeva, O.; Cheng, X.; Parsell, D.M.; Ramsoondar, J.; Kuravi, K.; et al. N-glycolylneuraminic acid knockout reduces erythrocyte sequestration and thromboxane elaboration in an ex vivo pig-to-human xenoperfusion model. Xenotransplantation 2017, 24, e12339. [Google Scholar] [CrossRef] [PubMed]
- Bottino, R.; Wijkstrom, M.; van der Windt, D.J.; Hara, H.; Ezzelarab, M.; Murase, N.; Bertera, S.; He, J.; Phelps, C.; Ayares, D.; et al. Pig-to-monkey islet xenotransplantation using multi-transgenic pigs. Am. J. Transplant. 2014, 14, 2275–2287. [Google Scholar] [CrossRef] [PubMed] [Green Version]





| GENE | Forward Sequence 5′-3′ | Reverse Sequence 5′-3′ |
|---|---|---|
| hCD55 | CAGCACCACCACAAATTGAC | TGCTCTCCAATCATGGTGAA |
| CYCL | TGCTTTCACAGAATAATTCCAGGATTTA | GACTTGCCACCAGTGCCATTA |
| BACT | TCTGGCACCACACCTTCT | CTCGATCATGAAGTGCGACGT |
| GAPDH | ACATGGCCTCCAAGGAGTAAGA | GATCGAGTTGGGGCTGTGACT |
| Specimen | Liver | Heart | Kidney |
|---|---|---|---|
| FT1 | 0.1064 ± 0.012 | 2.61 ± 0.17 | 0 |
| FT2 | 0.6902 ± 0.11 | 13.56 ± 0.81 | 0.10793 ± 0.03 |
| FT3 | 0.1923 ± 0.014 | 16.67 ± 0.92 | 0 |
| MT1 | 0.1913 ± 0.007 | 21.08 ± 1.34 | 0 |
| MT2 | 0.1964 ± 0.014 | 10.85 ± 0.72 | 0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martínez-Alarcón, L.; Liarte, S.; Quereda, J.J.; Sáez-Acosta, A.; Torre-Minguela, C.d.; Mendonça, L.; Abellaneda, J.M.; Majado, M.J.; Ríos, A.; Ramírez, P.; et al. Profiling Human CD55 Transgene Performance Assist in Selecting Best Suited Specimens and Tissues for Swine Organ Xenotransplantation. Biology 2021, 10, 747. https://doi.org/10.3390/biology10080747
Martínez-Alarcón L, Liarte S, Quereda JJ, Sáez-Acosta A, Torre-Minguela Cd, Mendonça L, Abellaneda JM, Majado MJ, Ríos A, Ramírez P, et al. Profiling Human CD55 Transgene Performance Assist in Selecting Best Suited Specimens and Tissues for Swine Organ Xenotransplantation. Biology. 2021; 10(8):747. https://doi.org/10.3390/biology10080747
Chicago/Turabian StyleMartínez-Alarcón, Laura, Sergio Liarte, Juan J. Quereda, Aida Sáez-Acosta, Carlos de Torre-Minguela, Livia Mendonça, Juana M. Abellaneda, María J. Majado, Antonio Ríos, Pablo Ramírez, and et al. 2021. "Profiling Human CD55 Transgene Performance Assist in Selecting Best Suited Specimens and Tissues for Swine Organ Xenotransplantation" Biology 10, no. 8: 747. https://doi.org/10.3390/biology10080747
APA StyleMartínez-Alarcón, L., Liarte, S., Quereda, J. J., Sáez-Acosta, A., Torre-Minguela, C. d., Mendonça, L., Abellaneda, J. M., Majado, M. J., Ríos, A., Ramírez, P., Muñoz, A., & Ramis, G. (2021). Profiling Human CD55 Transgene Performance Assist in Selecting Best Suited Specimens and Tissues for Swine Organ Xenotransplantation. Biology, 10(8), 747. https://doi.org/10.3390/biology10080747








