Free Amino Acids Profile and Expression Analysis of Core Genes Involved in Branched-Chain Amino Acids Metabolism during Fruit Development of Longan (Dimocarpus longan Lour.) Cultivars with Different Aroma Types
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Chemicals and Reagents
2.3. FAAs Determination
2.4. Gene Identification and Analysis
2.5. Statistical Analysis
3. Results
3.1. Changes in Phenotypic Traits and FAAs during Fruit Development
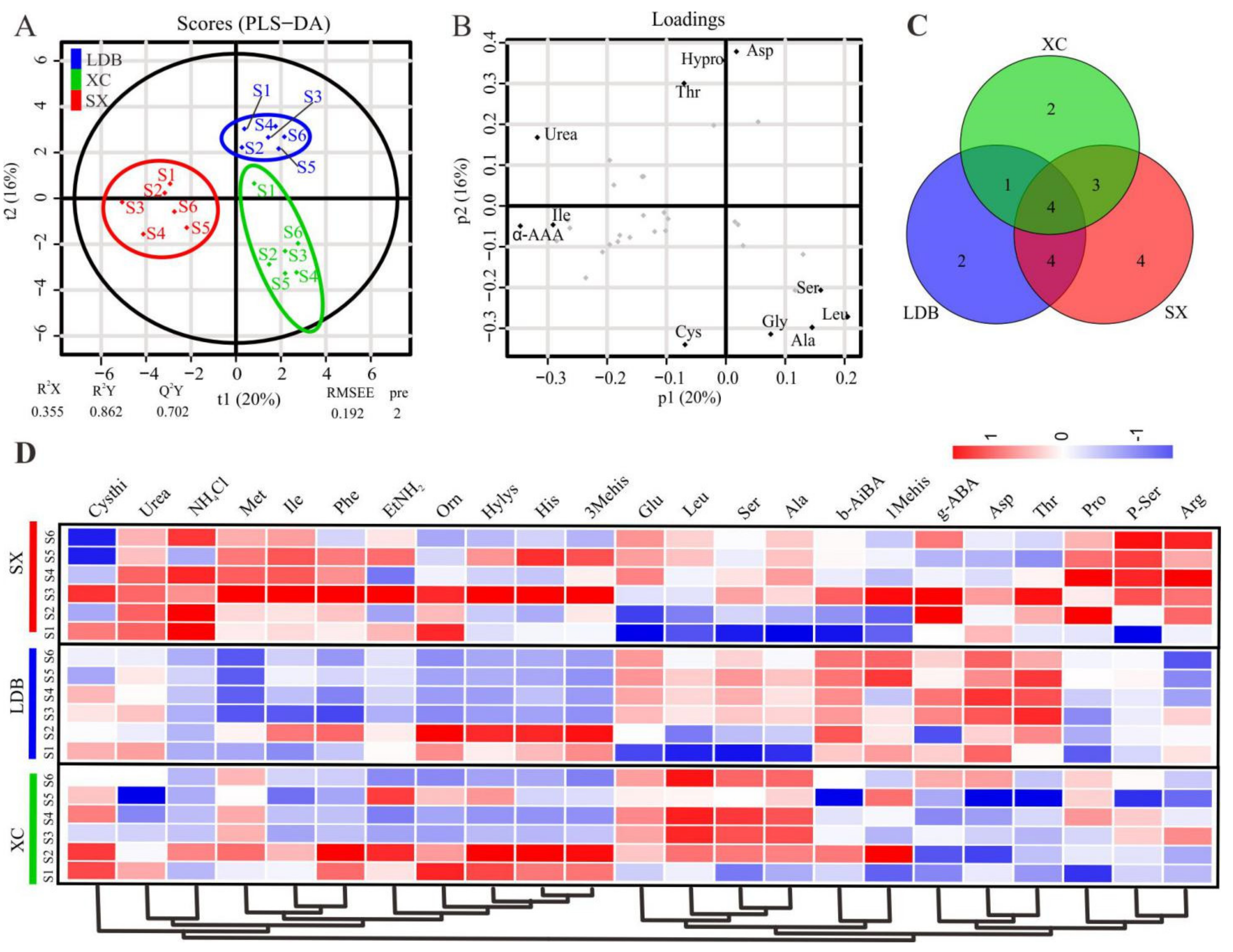
3.2. Multivariate Analysis of Identified FAAs
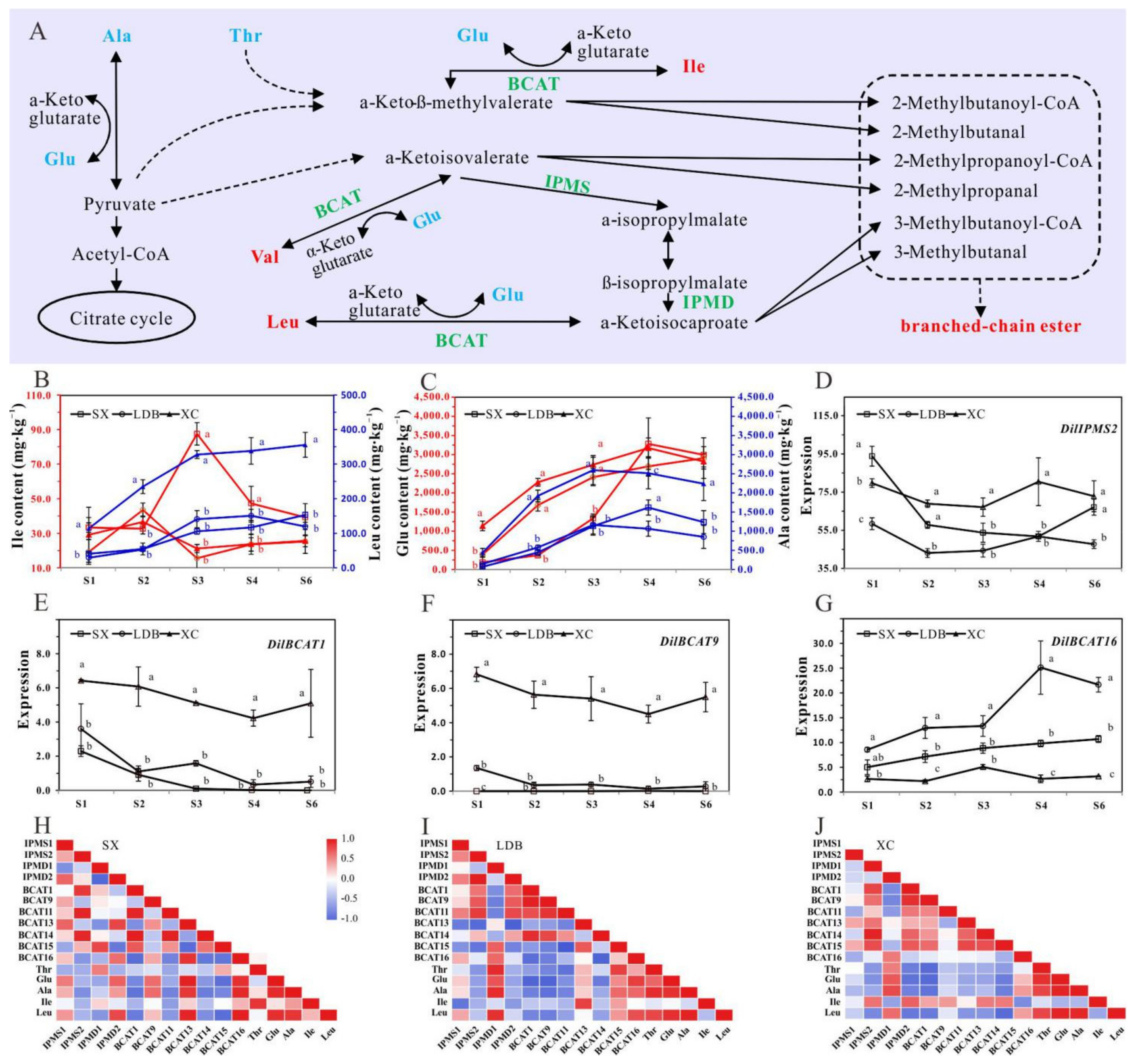
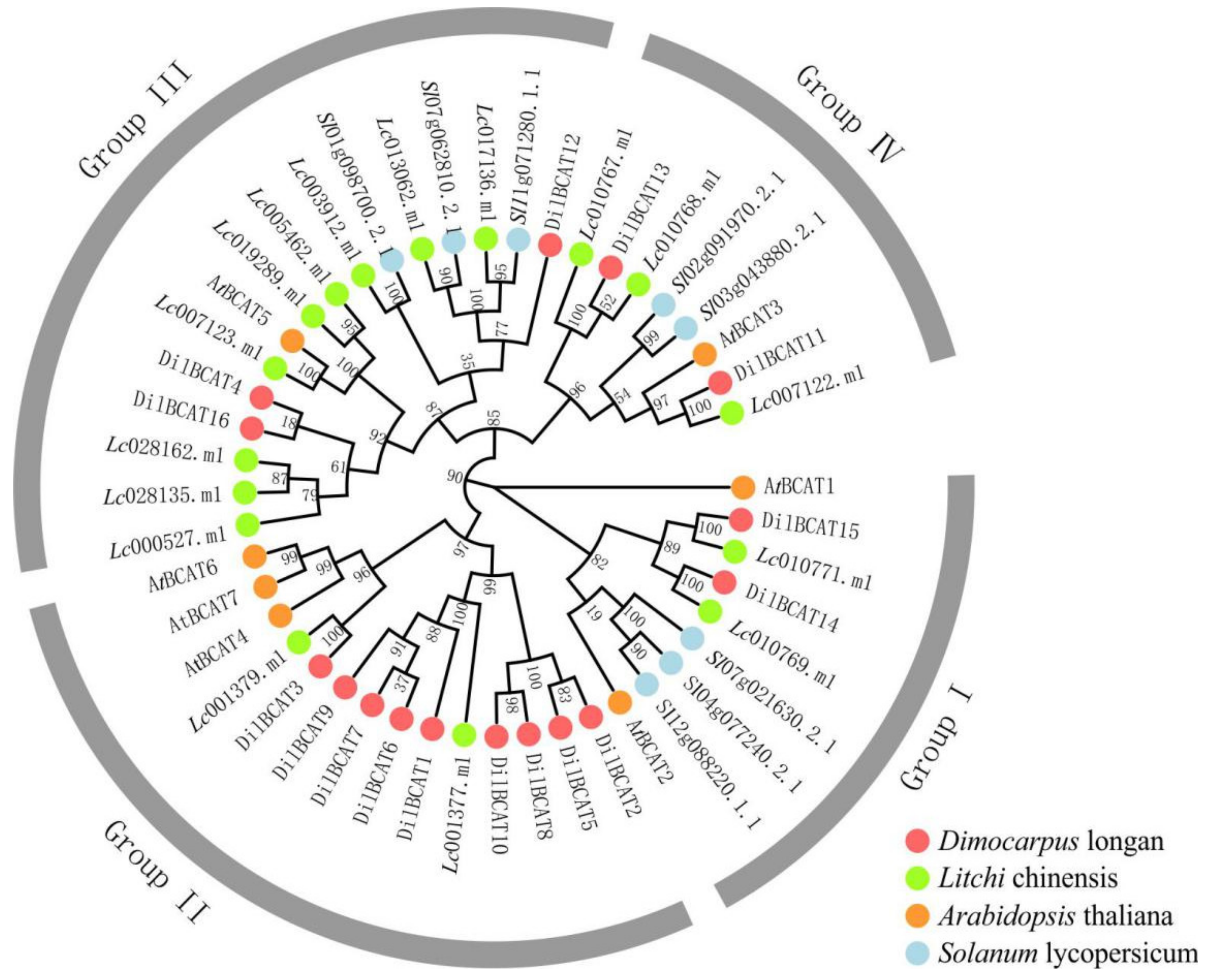
3.3. Identification and Expression Analysis of Gene Family Related to BCAAs Synthesis and Degradation during Fruit Development
4. Discussion
4.1. FAAs Profile of Longan Cultivars with Different Aroma Types during Fruit Development
4.2. Genes Involved in the Differential Accumulation of BCAAs in Pulp of Longan Cultivars with Different Aroma Types
4.3. BCAAs-Derived Volatile Compounds in Longan Fruit
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Huang, F.; Liu, H.J.; Zhang, R.F.; Dong, L.H.; Liu, L.; Ma, Y.X.; Jia, X.C.; Wang, G.J.; Zhang, M.W. Physicochemical properties and prebiotic activities of polysaccharides from longan pulp based on different extraction techniques. Carbohydr. Polym. 2019, 206, 344–351. [Google Scholar] [CrossRef]
- Yvon, M.; Rijnen, L. Cheese flavour formation by amino acid catabolism. Int. Dairy J. 2001, 11, 185–201. [Google Scholar] [CrossRef]
- Thage, B.V.; Broe, M.L.; Petersen, M.H.; Bennedsen, M.; Ardo, Y. Aroma development in semi-hard reduced-fat cheese added Lactobacillus paracasei strains with different aminotransferase profiles. Int. Dairy J. 2005, 15, 795–805. [Google Scholar] [CrossRef]
- Zhang, R.F.; Khan, S.A.; Lin, Y.S.; Guo, D.L.; Pan, X.W.; Liu, L.; Wei, Z.C.; Zhang, Y.; Deng, Y.Y.; Zhang, M.W. Phenolic profiles and cellular antioxidant activity of longan pulp of 24 representative Chinese cultivars. Int. J. Food Prop. 2018, 21, 746–759. [Google Scholar] [CrossRef] [Green Version]
- Tang, Y.Y.; He, X.M.; Sun, J.; Li, C.B.; Li, L.; Sheng, J.F.; Xin, M.; Li, Z.C.; Zheng, F.J.; Liu, G.M.; et al. Polyphenols and alkaloids in byproducts of longan fruits (Dimocarpus longan Lour.) and their bioactivities. Molecules 2019, 24, 1186. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Guo, D.L.; Han, D.M.; Pan, X.W.; Li, J.G. A comprehensive insight into the metabolic landscape of fruit pulp, peel, and seed in two longan (Dimocarpus longan Lour.) varieties. Int. J. Remote Sens. 2020, 23, 1527–1539. [Google Scholar] [CrossRef]
- Yang, W.H.; Zhu, X.C.; Bu, J.H.; Hu, G.B.; Wang, H.C.; Huang, X.M. Effects of bagging on fruit development and quality in cross-winter off-season longan. Sci. Hortic. 2009, 120, 194–200. [Google Scholar] [CrossRef]
- Wall, M.M. Ascorbic acid and mineral composition of longan (Dimocarpus longan), lychee (Litchi chinensis) and rambutan (Nephelium lappaceum) cultivars grown in Hawaii. J. Food Compost. Anal. 2006, 19, 655–663. [Google Scholar] [CrossRef]
- Hu, W.S.; Jiang, J.M.; Huang, A.P.; Zheng, S.Q. GC-MS analysis on aromatics of Wanxiang longan using different extraction fibers. Fujian J. Agric. Sci. 2015, 30, 1149–1154. [Google Scholar] [CrossRef]
- Ardo, Y. Flavour formation by amino acid catabolism. Biotechnol. Adv. 2006, 24, 238–242. [Google Scholar] [CrossRef]
- Kustyawati, M.M. Flavor formation in fruits and vegetables. J. Teknol. Ind. Hasil Pertan. 2008, 13, 31–36. [Google Scholar]
- Zhang, S.L.; Chen, K.S. Molecular Physiology of Fruit Quality Development and Regulation; China Agriculture Press: Beijing, China, 2007; pp. 190–191. [Google Scholar]
- Sugimoto, N.; Daniel, J.A.; Beaudry, R. Changes in free amino acid content in “Jonagold” apple fruit as related to branched-chain ester production, ripening, and senescence. J. Am. Soc. Hortic. Sci. 2011, 136, 429–440. [Google Scholar] [CrossRef] [Green Version]
- Alsmairat, N.; Engelgau, P.; Beaudry, R. Changes in free amino acid content in the flesh and peel of ‘cavendish’ banana fruit as related to branched-chain ester production, ripening, and senescence. J. Am. Soc. Hortic. Sci. 2018, 143, 370–380. [Google Scholar] [CrossRef]
- Wyllie, S.G.; Fellman, J.K. Formation of volatile branched chain esters in bananas (Musa sapientum L.). J. Agric. Food Chem. 2000, 48, 3493–3496. [Google Scholar] [CrossRef]
- Gonda, I.; Bar, E.; Portnoy, V.; Lev, S.; Burger, J.; Schaffer, A.A.; Tadmor, Y.; Gepstein, S.; Giovannoni, J.J.; Katzir, N.; et al. Branched-chain and aromatic amino acid catabolism into aroma volatiles in Cucumis melo L. fruit. J. Exp. Bot. 2010, 61, 1111–1123. [Google Scholar] [CrossRef] [Green Version]
- Sikdar, M.S.I.; Kim, J.S. Isolation of a gene encoding 3-isopropylmalate dehydrogenase from rice. Russ. J. Plant. Physiol. 2011, 58, 190–196. [Google Scholar] [CrossRef]
- He, Y.Q.; Cheng, J.P.; He, Y.; Yang, B.; Cheng, Y.H.; Yang, C.; Zhang, H.S.; Wang, Z.F. Influence of isopropylmalate synthase OsIPMS1 on seed vigour associated with amino acid and energy metabolism in rice. Plant. Biotechnol. J. 2019, 17, 322–337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.Q.; Ke, L.J.; Xiang, L.W.; Gao, G.Z.; Rao, Q.F.; Zhou, J.W. Analysis of amino acids in arillus longan during heating process. Amino Acids Biotic Resour. 2009, 31, 14–16. [Google Scholar] [CrossRef]
- Peng, Y.; Zhou, R.J. Determination and analysis of aroma and nutrient components in fresh juices of two kinds of longan. Sci. Technol. Food Ind. 2017, 38, 263–266. [Google Scholar] [CrossRef]
- Huang, A.P.; Zheng, S.Q. Changes of flesh flavonoids and amino acids concentration during the fruit development of longan (Dimocarpus longan Lour.) cultivar. Chin. Trop. Crops 2010, 31, 1519–1523. [Google Scholar] [CrossRef]
- Zheng, S.Q.; Zheng, J.G. Analysis on component and content amino acid in longan (Dimocarpus longan Lour.) germplasm. In Proceedings of the Second Symposium of Tropical and Subtropical Fruit Tree Branch of Chinese Horticultural Society and National Longan Industry Development Forum, Fuzhou, China, 19–21 September 2008; pp. 580–585. [Google Scholar]
- Hu, W.S.; Huang, A.P.; Jiang, F.; Jiang, J.M.; Chen, X.P.; Zheng, S.Q. Identification and genetic diversity of reciprocal hybrids in longan (Dimocarpus longan) by SSR. Acta Hortic. Sin. 2015, 42, 1899–1908. [Google Scholar] [CrossRef]
- Chen, Z.H.; Hu, Z.Y.; Lv, E.L.; Zhao, L.; Li, Y.P.; Xu, B.Q. Changes in amino acid contents of shuangjianyuhebao litchi during different storage conditions. Mod. Food Sci. Technol. 2013, 29, 1955–1960. [Google Scholar] [CrossRef]
- Maloney, G.S.; Kochevenko, A.; Tieman, D.M.; Tohge, T.; Krieger, U.; Zamir, D.; Taylor, M.G.; Fernie, A.R.; Klee, H.J. Characterization of the branched-chain amino acid aminotransferase enzyme family in tomato. Plant. Physiol. 2010, 153, 925–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Diebold, R.; Schuster, J.; Däschner, K.; Binder, S. The branched-chain amino acid transaminase gene family in arabidopsis encodes plastid and mitochondrial proteins. Plant. Physiol. 2002, 129, 540–550. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Thevenot, E.A.; Roux, A.; Xu, Y.; Ezan, E.; Junot, C. Analysis of the human adult urinary metabolome variations with age, body mass index and gender by implementing a comprehensive workflow for univariate and OPLS statistical analysis. J. Proteome Res. 2015, 14, 3322–3335. [Google Scholar] [CrossRef]
- Schulze-Siebert, D.; Heineke, D.; Scharf, H.; Schultz, G. Pyruvatederived amino acids in spinach chloroplasts: Synthesis and regulation during photosynthetic carbon metabolism. Plant. Physiol. 1984, 76, 465–471. [Google Scholar] [CrossRef] [Green Version]
- Hagelstein, P.; Sieve, B.; Klein, M.; Jans, H.; Schultz, G. Leucine synthesis in chloroplasts: Leucine/isoleucine aminotransferase and valine aminotransferase are different enzymes in spinach chloroplasts. J. Plant. Physiol. 1997, 150, 23–30. [Google Scholar] [CrossRef]
- Wang, J.Y.; Zhu, S.G.; Xu, C.F. Biochemistry: Volume II; Higher Education Press: Beijing, China, 2012; pp. 351–353. [Google Scholar]
- Efeyan, A.; Comb, W.C.; Sabatini, D.M. Nutrient-sensing mechanisms and pathways. Nature 2015, 517, 302–310. [Google Scholar] [CrossRef] [Green Version]
- Kocada, T.; Oezdemir, K.S.; Goekmen, V. Effects of infusion conditions and decaffeination on free amino acid profiles of green and black tea. Food Res. Intl. 2013, 53, 720–725. [Google Scholar] [CrossRef]
- Choi, S.H.; Ahn, J.B.; Kim, H.J.; Im, N.K.; Kozukue, N.; Levin, C.E.; Friedman, M. Changes in free amino acid, protein, and flavonoid content in Jujube (Ziziphus jujube) fruit during eight stages of growth and antioxidative and cancer cell inhibitory effects by extracts. J. Agric. Food Chem. 2012, 60, 10245–10255. [Google Scholar] [CrossRef] [PubMed]
- Khan, S.A.; Liu, L.; Lai, T.; Zhang, R.F.; Wei, Z.C.; Xiao, J.; Deng, Y.Y.; Zhang, M.W. Phenolic profile, free amino acids composition and antioxidant potential of dried longan fermented by lactic acid bacteria. J. Food Sci. Technol. 2018, 55, 4782–4791. [Google Scholar] [CrossRef] [PubMed]
- Xing, A.Q.; Last, R.L. A regulatory hierarchy of the Arabidopsis branched-chain amino acid metabolic network. Plant. Cell 2017, 29, 1480–1499. [Google Scholar] [CrossRef] [PubMed]
- Nozawa, A.; Takano, J.; Miwa, K.; Nakagawa, Y.; Fujiwara, T. Cloning of cDNAs encoding isopropylmalate dehydrogenase from arabidopsis thaliana and accumulation patterns of their transcripts. Biosci. Biotechnol. Biochem. 2005, 69, 806–810. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Galant, A.; Pang, Q.; Strul, J.M.; Balogun, S.F.; Jez, J.M.; Chen, S. Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana. J. Biol. Chem. 2011, 286, 28794–28801. [Google Scholar] [CrossRef] [Green Version]
- Martin, G.; Baurens, F.C.; Droc, G.; Rouard, M.; Cenci, A.; Kilian, A.; Hastie, A.; Doležel, J.; Aury, J.M.; Alberti, A.; et al. Improvement of the banana “Musa acuminata” reference sequence using NGS data and semi-automated bioinformatics methods. BMC Genom. 2016, 17, 243. [Google Scholar] [CrossRef] [Green Version]
- Ning, J.; Moghe, G.D.; Leong, B.; Kim, J.; Ofner, I.; Wang, Z.; Adams, C.; Jones, A.D.; Zamir, D.; Last, R.L. A feedback-iInsensitive isopropylmalate synthase affects acylsugar composition in cultivated and wild tomato. Plant. Physiol. 2015, 169, 1821–1835. [Google Scholar] [CrossRef] [Green Version]
- Gerbling, H.; Gerhardt, B. Peroxisomal degradation of branched-chain 2-oxo acids. Plant. Physiol. 1989, 91, 1387–1392. [Google Scholar] [CrossRef] [Green Version]
- Trinh, T.T.T.; Woon, W.Y.; Yu, B.; Curran, P.; Liu, S.Q. Effect of L-isoleucine and L-phenylalanine addition on aroma compound formation during longan juice fermentation by a co-culture of saccharomyces cerevisiae and williopsis saturnus. S. Afr. J. Enol. Vitic. 2010, 31, 116–124. [Google Scholar] [CrossRef]
- Trinh, T.T.T.; Bin, Y.; Curran, P.; Liu, S.Q. Formation of aroma compounds during longan juice fermentation by williopsis saturnus var. saturnus with the addition of selected amino acids. J. Food Process. Preserv. 2012, 36, 198–206. [Google Scholar] [CrossRef]
- Zhang, Y. Aroma Compounds in Longan (Dimocarpus longan Lour.) Juice and Aroma Variation during Processing and Storage. Ph.D. Thesis, Huazhong Agricultural University, Wuhan, China, 2010; pp. 28–31. [CrossRef]
- An, K.J.; Liu, H.C.; Fu, M.Q.; Qian, M.C.; Yu, Y.S.; Wu, J.J.; Xiao, G.S.; Xu, Y.J. Identification of the cooked off-flavor in heat-sterilized lychee (Litchi chinensis Sonn.) juice by means of molecular sensory science. Food Chem. 2019, 301, 125282. [Google Scholar] [CrossRef] [PubMed]
- Tressl, R.; Drawert, F. Biogenesis of banana volatiles. J. Agric. Food Chem. 1973, 21, 560–565. [Google Scholar] [CrossRef]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hu, W.; Wang, B.; Ali, M.M.; Chen, X.; Zhang, J.; Zheng, S.; Chen, F. Free Amino Acids Profile and Expression Analysis of Core Genes Involved in Branched-Chain Amino Acids Metabolism during Fruit Development of Longan (Dimocarpus longan Lour.) Cultivars with Different Aroma Types. Biology 2021, 10, 807. https://doi.org/10.3390/biology10080807
Hu W, Wang B, Ali MM, Chen X, Zhang J, Zheng S, Chen F. Free Amino Acids Profile and Expression Analysis of Core Genes Involved in Branched-Chain Amino Acids Metabolism during Fruit Development of Longan (Dimocarpus longan Lour.) Cultivars with Different Aroma Types. Biology. 2021; 10(8):807. https://doi.org/10.3390/biology10080807
Chicago/Turabian StyleHu, Wenshun, Baiyu Wang, Muhammad Moaaz Ali, Xiuping Chen, Jisen Zhang, Shaoquan Zheng, and Faxing Chen. 2021. "Free Amino Acids Profile and Expression Analysis of Core Genes Involved in Branched-Chain Amino Acids Metabolism during Fruit Development of Longan (Dimocarpus longan Lour.) Cultivars with Different Aroma Types" Biology 10, no. 8: 807. https://doi.org/10.3390/biology10080807
APA StyleHu, W., Wang, B., Ali, M. M., Chen, X., Zhang, J., Zheng, S., & Chen, F. (2021). Free Amino Acids Profile and Expression Analysis of Core Genes Involved in Branched-Chain Amino Acids Metabolism during Fruit Development of Longan (Dimocarpus longan Lour.) Cultivars with Different Aroma Types. Biology, 10(8), 807. https://doi.org/10.3390/biology10080807






