Effect of Flowering Time-Related Genes on Biomass, Harvest Index, and Grain Yield in CIMMYT Elite Spring Bread Wheat
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Trait Measurements
2.3. Genotyping
2.4. Summary Statistics
2.5. Single Marker Regression and Stepwise Regression
3. Results
3.1. Phenotypic Variation of Trait Measurements
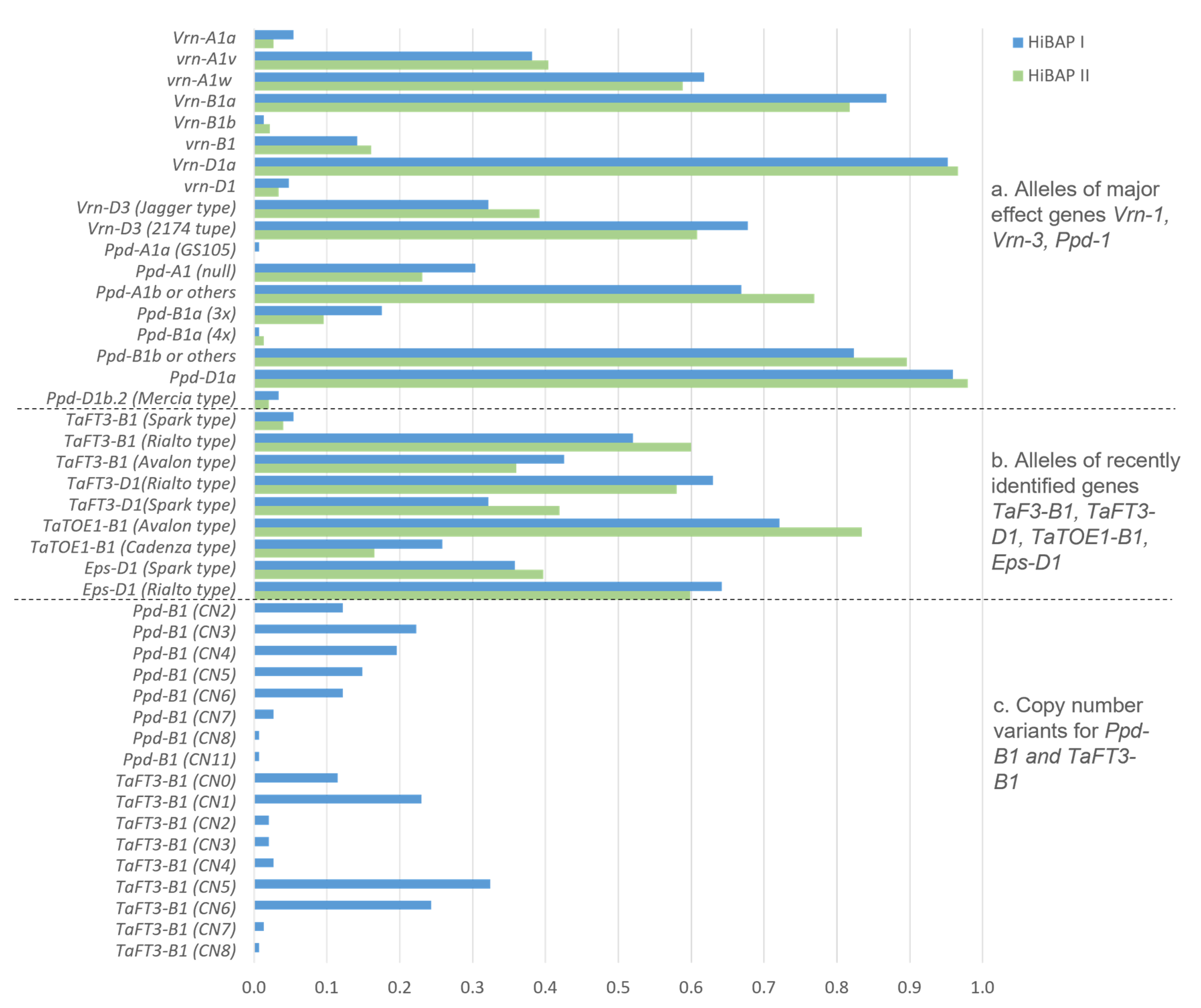
3.2. Frequencies of the Vrn, Ppd, and Eps Alleles
3.3. Allele Effects on Phenological Phases
3.4. Allele Effects on BM, HI and YLD
3.5. Allele Effects of TaTOE-B1 and TaFT3-B1 on Source- and Sink-Related Grain Yield Traits
4. Discussion
4.1. Estimations of the Allele Effects at Major and More Recently Identified Vrn, Ppd, and Eps Genes
4.2. Evaluation of the Possibility of Maximizing HI by Testing the Allele Effects on HI and Several of Its Physiological Components
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Salamini, F.; Özkan, H.; Brandolini, A.; Schäfer-Pregl, R.; Martin, W.F. Genetics and Geography of Wild Cereal Domestication in the Near East. Nat. Rev. Genet. 2002, 3, 429–441. [Google Scholar] [CrossRef] [PubMed]
- Shiferaw, B.; Smale, M.; Braun, H.-J.; Duveiller, E.; Reynolds, M.; Muricho, G. Crops that Feed the World 10. Past Successes and Future Challenges to the Role Played by Wheat in Global Food Security. Food Secur. 2013, 5, 291–317. [Google Scholar] [CrossRef] [Green Version]
- Van Bavel, J.; Reher, D.S. The Baby Boom and Its Causes: What We Know and What We Need to Know The Baby Boom and Its Causes: What We Know and What We Need to Know. Popul. Dev. Rev. 2013, 39, 257–288. [Google Scholar] [CrossRef]
- Asseng, S.; Ewert, F.; Martre, P.; Rötter, R.; Lobell, D.; Cammarano, D.; Kimball, B.A.; Ottman, M.J.; Wall, G.W.; White, J.W.; et al. Rising Temperatures Reduce Global Wheat Production. Nat. Clim. Chang. 2014, 5, 143–147. [Google Scholar] [CrossRef]
- Slafer, G.; Savin, R.; Sadras, V.O. Coarse and Fine Regulation of Wheat Yield Components in Response to Genotype and Environment. Field Crop. Res. 2014, 157, 71–83. [Google Scholar] [CrossRef]
- Trethowan, R.M. Defining a Genetic Ideotype for Crop Improvemento Title. In Crop Breeding Methods in Molecular Biology (Methods and Protocols); Fleury, D., Whitford, R., Eds.; Humana Press: New York, NY, USA, 2014; Volume 1145, pp. 1–20. [Google Scholar] [CrossRef]
- Chouard, P. Vernalization and Its Relations to Dormancy. Annu. Rev. Plant Physiol. 1960, 11, 191–238. [Google Scholar] [CrossRef]
- Kamran, A.; Iqbal, M.; Spaner, D. Flowering Time in Wheat (Triticum Aestivum L.): A Key Factor for Global Adaptability. Euphytica 2014, 197, 1–26. [Google Scholar] [CrossRef]
- Searle, I.; Coupland, G. Induction of Flowering by Seasonal Changes in Photoperiod. EMBO J. 2004, 23, 1217–1222. [Google Scholar] [CrossRef] [Green Version]
- Gomez, D.; Vanzetti, L.; Helguera, M.; Lombardo, L.; Fraschina, J.; Miralles, D. Effect of Vrn-1, Ppd-1 Genes and Earliness Per se on Heading Time in Argentinean Bread Wheat Cultivars. Field Crop. Res. 2014, 158, 73–81. [Google Scholar] [CrossRef]
- Langer, S.M.; Longin, C.F.H.; WuRschum, T. Flowering Time Control in European Winter Wheat. Front. Plant Sci. 2014, 5, 537. [Google Scholar] [CrossRef] [Green Version]
- Kiss, T.; Balla, K.; Veisz, O.; Láng, L.; Bedő, Z.; Griffiths, S.; Isaac, P.; Karsai, I. Allele Frequencies in the VRN-A1, VRN-B1 and VRN-D1 Vernalization Response and PPD-B1 and PPD-D1 Photoperiod Sensitivity Genes, and Their Effects on Heading in a Diverse Set of Wheat Cultivars (Triticum Aestivum L.). Mol. Breed. 2014, 34, 297–310. [Google Scholar] [CrossRef] [Green Version]
- Fu, D.; Szűcs, P.; Yan, L.; Helguera, M.; Skinner, J.S.; Von Zitzewitz, J.; Hayes, P.M.; Dubcovsky, J. Large Deletions within the First Intron in VRN-1 are Associated with Spring Growth Habit in Barley and Wheat. Mol. Genet. Genom. 2005, 273, 54–65. [Google Scholar] [CrossRef] [Green Version]
- Díaz, A.; Zikhali, M.; Turner, A.S.; Isaac, P.; Laurie, D.A. Copy Number Variation Affecting the Photoperiod-B1 and Vernalization-A1 Genes Is Associated with Altered Flowering Time in Wheat (Triticum Aestivum). PLoS ONE 2012, 7, e33234. [Google Scholar] [CrossRef] [Green Version]
- Beales, J.; Turner, A.; Griffiths, S.; Snape, J.; Laurie, D.A. A Pseudo-Response Regulator is Misexpressed in the Photoperiod Insensitive Ppd-D1a Mutant of Wheat (Triticum Aestivum L.). Theor. Appl. Genet. 2007, 115, 721–733. [Google Scholar] [CrossRef] [PubMed]
- Yan, L.; Helguera, M.; Kato, K.; Fukuyama, S.; Sherman, J.; Dubcovsky, J. Allelic Variation at the VRN-1 Promoter Region in Polyploid Wheat. Theor. Appl. Genet. 2004, 109, 1677–1686. [Google Scholar] [CrossRef] [Green Version]
- Dubcovsky, J.; Loukoianov, A.; Fu, D.; Valárik, M.; Sanchez, A.; Yan, L. Effect of Photoperiod on the Regulation of Wheat Vernalization Genes VRN1 and VRN2. Plant Mol. Biol. 2006, 60, 469–480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trevaskis, B.; Hemming, M.N.; Dennis, E.S.; Peacock, W.J. The Molecular Basis of Vernalization-Induced Flowering in Cereals. Trends Plant Sci. 2007, 12, 352–357. [Google Scholar] [CrossRef] [PubMed]
- Slafer, G.; Rawson, H. Sensitivity of Wheat Phasic Development to Major Environmental Factors: A Re-Examination of Some Assumptions Made by Physiologists and Modellers. Funct. Plant Biol. 1994, 21, 393–426. [Google Scholar] [CrossRef]
- Zikhali, M.; Griffiths, S. The Effect of Earliness per se (Eps) Genes on Flowering Time in Bread Wheat. Advances in Wheat Genetics: From Genome to Field. Proceedings of the 12th International Wheat Genetics Symposium; Ogihara, Y., Takumi, S., Handa, H., Eds.; Springer: Tokyo, Japan, 2015; pp. 339–345. [Google Scholar]
- Zikhali, M.; Leverington-Waite, M.; Fish, L.; Simmonds, J.; Orford, S.; Wingen, L.U.; Goram, R.; Gosman, N.; Bentley, A.; Griffiths, S. Validation of a 1DL Earliness Per se (eps) Flowering QTL in Bread Wheat (Triticum Aestivum). Mol. Breed. 2014, 34, 1023–1033. [Google Scholar] [CrossRef] [Green Version]
- Zikhali, M.; Wingen, L.U.; Griffiths, S. Delimitation of the Earliness per se D1(Eps-D1) Flowering Gene to a Subtelomeric Chromosomal Deletion in Bread Wheat (Triticum Aestivum). J. Exp. Bot. 2015, 67, 287–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zikhali, M.; Wingen, L.U.; Leverington-Waite, M.; Specel, S.; Griffiths, S. The Identification of New Candidate Genes Triticum Aestivum FLOWERING LOCUS T3-B1 (TaFT3-B1) and TARGET OF EAT1 (TaTOE1-B1) Controlling the Short-Day Photoperiod Response in Bread Wheat. Plant Cell Environ. 2017, 40, 2678–2690. [Google Scholar] [CrossRef] [Green Version]
- Sukumaran, S.; Lopes, M.S.; Dreisigacker, S.; Dixon, L.E.; Zikhali, M.; Griffiths, S.; Zheng, B.; Chapman, S.; Reynolds, M.P. Identification of Earliness Per Se Flowering Time Locus in Spring Wheat through a Genome-Wide Association Study. Crop Sci. 2016, 56, 2962-2672. [Google Scholar] [CrossRef]
- Shearman, V.J.; Scott, R.K.; Foulkes, M.J. Crop Physiology Metabolism Physiological Processes Associated with Wheat Yield Progress in the UK. Crop J. 2005, 45, 175–185. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P.; Manes, Y.; Singh, R.P.; Crossa, J.; Braun, H.J. Genetic Yield Gains and Changes in Associated Traits of CIMMYT Spring Bread Wheat in a “Historic” Set Representing 30 Years of Breeding. Crop Sci. 2012, 52, 1123–1131. [Google Scholar] [CrossRef]
- Aisawi, K.A.B.; Reynolds, M.P.; Singh, R.P.; Foulkes, J. The Physiological Basis of the Genetic Progress in Yield Potential of CIMMYT Spring Wheat Cultivars from 1966 to 2009. Crop Sci. 2015, 55, 1749–1764. [Google Scholar] [CrossRef]
- Reynolds, M.; Foulkes, J.; Slafer, G.; Berry, P.; Parry, M.; Snape, J.; Angus, W.J. Raising Yield Potential in Wheat. J. Exp. Bot. 2009, 60, 1899–1918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reynolds, M.; Foulkes, J.; Furbank, R.; Griffiths, S.; King, J.; Murchie, E.; Parry, M.; Slafer, G. Achieving Yield Gains in Wheat. Plant Cell Environ. 2012, 35, 1799–1823. [Google Scholar] [CrossRef]
- Molero, G.; Joynson, R.; Pinera-Chavez, F.J.; Gardiner, L.; Rivera-Amado, C.; Hall, A.; Reynolds, M.P. Elucidating the Genetic Basis of Biomass Accumulation and Radiation Use Efficiency in Spring Wheat and Its Role in Yield Potential. Plant Biotechnol. J. 2018, 17, 1276–1288. [Google Scholar] [CrossRef]
- Joynson, R.; Molero, G.; Coombes, B.; Gardiner, L.; Rivera-Amado, C.; Piñera-Chávez, F.J.; Evans, J.R.; Furbank, R.T.; Reynolds, M.P.; Hall, A. Uncovering Candidate Genes Involved in Photosynthetic Capacity Using Unexplored Genetic Variation in Spring Wheat. Plant Biotechnol. J. 2021. [Google Scholar] [CrossRef]
- Zadoks, J.; Chang, T.; Konzak, C.F. A Decimal Code for the Growth Stages of Cerals. Weed Res. 1974, 14, 415–421. [Google Scholar] [CrossRef]
- Angus, J.; Mackenzie, D.; Morton, R.; Schafer, C. Phasic Development in Field Crops II. Thermal and Photoperiodic Responses of Spring Wheat. Field Crop. Res. 1981, 4, 269–283. [Google Scholar] [CrossRef]
- Pask, A.; Pietragalla, J.; Mullan, D.; Reynolds, M. Physiological Breeding II: A Field Guide to Wheat Phenotyping. In Technical; CIMMYT: Texcoco, Mexico, 2012. [Google Scholar]
- Dreisigacker, S.; Sehgal, D.; Reyes Jaimez, A.; Luna Garrido, B.; Muñoz Savala, S.; Núñez Ríos, C. CIMMYT Wheat Molecular Genetics: Laboratory Protocols and Applications to Wheat Breeding; Mollins, J., Mall, S., Eds.; CIMMYT: Texcoco, Mexico, 2016. [Google Scholar]
- Würschum, T.; Rapp, M.; Miedaner, T.; Longin, C.F.H.; Leiser, W.L. Copy Number Variation of Ppd-B1 is the Major Determinant of Heading Time in Durum Wheat. BMC Genet. 2019, 20, 64–68. [Google Scholar] [CrossRef]
- Alvarado, G.; López, M.; Vargas, M.; Pacheco, Á.; Rodríguez, F.; Burgueño, J.; Crossa, J. META-R (Multi Environment Trail Analysis with R for Windows) Version 6.04. CIMMYT Research Data Software Repository Network, V23. 2015. Available online: https://data.cimmyt.org/dataset.xhtml?persistentId=hdl:11529/10201 (accessed on 23 January 2021).
- Alvarado, G.; Rodríguez, F.M.; Pacheco, A.; Burgueño, J.; Crossa, J.; Vargas, M.; Pérez-Rodríguez, P.; Lopez-Cruz, M.A. META-R: A Software to Analyze Data from Multi-Environment Plant Breeding Trials. Crop J. 2020, 8, 745–756. [Google Scholar] [CrossRef]
- Eagles, H.A.; Cane, K.; Trevaskis, B. Veery Wheats Carry an Allele of Vrn-A1 that has Implications for Freezing Tolerance in Winter Wheats. Plant Breed. 2011, 130, 413–418. [Google Scholar] [CrossRef]
- Hyles, J.; Bloomfield, M.T.; Hunt, J.R.; Trethowan, R.M.; Trevaskis, B. Phenology and related traits for wheat adaptation. Heredity 2020, 125, 417–430. [Google Scholar]
- Moorby, J.; Landsberg, J.J.; Cutting, C.V. Environmental Effects on Crop Physiology. J. Appl. Ecol. 1979, 16, 334. [Google Scholar] [CrossRef]
- Slafer, G.A. Wheat Development: Its Role in Phenotyping and Improving Crop Adaptation. In Wheat Physiological Breeding I: Interdisciplinary Approaches to Improve Crop Adaptation; Reynolds, M., Pask, A., Mullan, D., Eds.; CIMMYT: Texcoco, Mexico, 2012; pp. 107–121. [Google Scholar]
- Slafer, G.; Rawson, H. Responses to Photoperiod Change with Phenophase and Temperature during Wheat Development. Field Crop Res. 1996, 46, 1–13. [Google Scholar] [CrossRef]
- Appendino, M.L.; Slafer, G. Earliness Per se and Its Dependence upon Temperature in Diploid Wheat Lines Differing in the Major Gene Eps-Am1 Alleles. J. Agric. Sci. 2003, 141, 149–154. [Google Scholar] [CrossRef] [Green Version]
- Lantican, M.A.; Braun, H.-J.; Payne, T.S.; Singh, R.P.; Sonder, K.; Baum, M.; Braun, H.J.; Erenstein, O. Impacts of International Wheat Research 1994–2014; CIMMYT: Texcoco, Mexico, 2016. [Google Scholar]
- DeLacy, I.H.; Fox, P.N.; Corbett, J.D.; Crossa, J.; Rajaram, S.; Fischer, R.A.; Van Ginkel, M. Long-Term Association of Locations for Testing Spring Bread Wheat. Euphytica 1993, 72, 95–106. [Google Scholar] [CrossRef]
- Lillemo, M.; Van Ginkel, M.; Trethowan, R.M.; Hernandez, E.; Crossa, J. Differential Adaptation of CIMMYT Bread Wheat to Global High Temperature Environments. Crop Sci. 2005, 45, 2443–2453. [Google Scholar] [CrossRef]
- Crespo-Herrera, L.A.; Crossa, J.; Huerta-Espino, J.; Mondal, S.; Velu, G.; Juliana, P.; Vargas, M.; Pérez-Rodríguez, P.; Joshi, A.K.; Braun, H.J.; et al. Target Population of Environments for Wheat Breeding in India: Definition, Prediction and Genetic Gains. Front. Plant Sci. 2021, 12. [Google Scholar] [CrossRef]
- Goncharov, N. Response to Vernalization in Wheat: Its Quantitative or Qualitative Nature Nikolay. Cereal Res. Commun. 2004, 32, 55. [Google Scholar] [CrossRef]
- Pugsley, A. A Genetic Analysis of the Spring-Winter Habit of Growth in Wheat. Aust. J. Agric. Res. 1971, 22, 21–31. [Google Scholar] [CrossRef]
- Pugsley, A.T. Additional Genes Inhibiting Winter Habit in Wheat. Euphytica 1972, 21, 547–552. [Google Scholar] [CrossRef]
- Stelmakh, A. Genetic Effects of Vrn Genes on Heading Date and Agronomic Traits in Bread Wheat. Euphytica 1993, 65, 53–60. [Google Scholar] [CrossRef]
- Stelmakh, A. Genetic Systems Regulating Flowering Response in Wheat. Euphytica 1998, 100, 359–369. [Google Scholar] [CrossRef]
- Van Beem, J.; Mohler, V.; Lukman, R.; Van Ginkel, M.; William, M.; Crossa, J.; Worland, A.J. Analysis of Genetic Factors Influencing the Developmental Rate of Globally Important CIMMYT Wheat Cultivars. Crop Sci. 2005, 45, 2113–2119. [Google Scholar] [CrossRef]
- Stelmakh, A. Geographic Distribution of Vrn-Genes in Landraces and Improved Varieties of Spring Bread Wheat. Euphytica 1990, 45, 113–118. [Google Scholar] [CrossRef]
- Slafer, G.; Abeledo, L.; Miralles, D.; González, F.G.; Whitechurch, E. Photoperiod Sensitivity during Stem Elongation as an Avenue to Raise Potential Yield in Wheat. Euphytica 2001, 119, 191–197. [Google Scholar] [CrossRef]
- Miralles, D.J.; Slafer, G. Paper Presented at International Workshop On Increasing Wheat Yield Potential, Cimmyt, Obregon, Mexico, 20–24 March 2006 Sink Limitations to Yield in Wheat: How could it be reduced? J. Agric. Sci. 2007, 145, 139–149. [Google Scholar] [CrossRef] [Green Version]
- Gonzalez, F.G.; Slafer, G.A.; Miralles, D.J. Photoperiod during Stem Elongation in Wheat: Is its Impact on Fertile Floret and Grain Number Determination Similar to that of Radiation? Funct. Plant Biol. 2005, 32, 181–188. [Google Scholar] [CrossRef]
- González, F.G.; Slafer, G.; Miralles, D.J. Floret Development and Survival in Wheat Plants Exposed to Contrasting Photoperiod and Radiation Environments during Stem Elongation. Funct. Plant Biol. 2005, 32, 189–197. [Google Scholar] [CrossRef]
- Fischer, R.A. Number of Kernels in Wheat Crops and the Influence of Solar Radiation and Temperature. J. Agric. Sci. 1985, 105, 447–461. [Google Scholar] [CrossRef]
- Siddique, K.; Kirby, E.; Perry, M. Ear: Stem Ratio in Old and Modern Wheat Varieties; Relationship with Improvement in Number of Grains Per Ear and Yield. Field Crop. Res. 1989, 21, 59–78. [Google Scholar] [CrossRef]
- Sayre, K.D.; Rajaram, S.; Fischer, R.A. Yield Potential Progress in Short Bread Wheats in Northwest Mexico. Crop Sci. 1997, 37, 36–42. [Google Scholar] [CrossRef]
- Slafer, G.; Andrade, F. Genetic Improvement In Bread Wheat (Triticum Aestivum) Yield in Argentina. Field Crop. Res. 1989, 21, 289–296. [Google Scholar] [CrossRef]
- Calderini, D.F.; Dreccer, M.F.; Slafer, G.A. Genetic Improvement in Wheat Yield and Associated Traits. A Re-Examination of Previous Results and the Latest Trends. Plant Breed. 1995, 114, 108–112. [Google Scholar] [CrossRef]
- Slafer, G.A.; Elía, M.; Savin, R.; García, G.; Terrile, I.I.; Ferrante, A.; Miralles, D.J.; González, F.G. Fruiting Efficiency: An Alternative Trait to Further Rise Wheat Yield. Food Energy Secur. 2015, 4, 92–109. [Google Scholar] [CrossRef]
- González, F.G.; Miralles, D.J.; Slafer, G. Wheat Floret Survival as Related to Pre-Anthesis Spike Growth. J. Exp. Bot. 2011, 62, 4889–4901. [Google Scholar] [CrossRef]
- Lázaro, L.; Abbate, P.E. Cultivar Effects on Relationship between Grain Number and Photothermal Quotient or Spike Dry Weight in Wheat. J. Agric. Sci. 2011, 150, 442–459. [Google Scholar] [CrossRef]
- Bustos-Korts, D.; Hasan, A.K.; Reynolds, M.P.; Calderini, D. Combining High Grain Number and Weight through a DH-Population to Improve Grain Yield Potential of Wheat in High-Yielding Environments. Field Crop. Res. 2013, 145, 106–115. [Google Scholar] [CrossRef]
- Rivera-Amado, C.; Trujillo-Negrellos, E.; Molero, G.; Reynolds, M.P.; Sylvester-Bradley, R.; Foulkes, M.J. Optimizing Dry-Matter Partitioning for Increased Spike Growth, Grain Number and Harvest Index in Spring Wheat. Field Crop. Res. 2019, 240, 154–167. [Google Scholar] [CrossRef]
- Cossani, C.M.; Slafer, G.A.; Savin, R. Do Barley and Wheat (Bread and Durum) Differ in Grain Weight Stability through Seasons and Water–Nitrogen Treatments in a Mediterranean Location? Field Crop. Res. 2011, 121, 240–247. [Google Scholar] [CrossRef]
- Rharrabti, Y.; del Moral, L.G.; Villegas, D.; Royo, C. Durum Wheat Quality in Mediterranean Environments: III. Stability and Comparative Methods in Analysing G×E Interaction. Field Crop. Res. 2003, 80, 141–146. [Google Scholar] [CrossRef]
- Wu, X.; Tang, Y.; Li, C.; Wu, C. Characterization of the Rate and Duration of Grain Filling in Wheat in Southwestern China. Plant Prod. Sci. 2018, 21, 358–369. [Google Scholar] [CrossRef] [Green Version]






| HiBAP I | HiBAP II | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Trait | Mean | Range | H2 | G | G × Y | Y | Mean | Range | H2 | G | G × Y | Y |
| Developmental phases | ||||||||||||
| DTInB (days) | 60.8 | 53.7–66.8 | 0.83 | *** | *** | *** | 65.6 | 57.5–72.5 | 0.92 | *** | *** | *** |
| DTInB-DTA (days) | 15.7 | 14.3–17.2 | 0.57 | *** | *** | *** | 16.5 | 14.8–17.6 | 0.34 | ** | *** | *** |
| VGP/DTA (days) | 76.4 | 69.4–84.1 | 0.87 | *** | *** | *** | 82.1 | 73.7–90.6 | 0.93 | *** | *** | *** |
| GFD (days) | 38.5 | 34.4–45.3 | 0.63 | *** | *** | ** | 41.0 | 36.5–45.5 | 0.75 | *** | *** | ns |
| TTInB | 1007.4 | 886.0–1134.6 | 0.92 | *** | *** | *** | 1142.5 | 995.8–1248.2 | 0.91 | *** | *** | *** |
| TTInB-TTA | 279.9 | 248.8–319.7 | 0.55 | *** | *** | *** | 255.6 1 | 171.5–306.3 1 | - | - | - | - |
| TTVGP/TTA | 1287.3 | 1142.9–1443.1 | 0.94 | *** | *** | *** | 1398.1 | 1264.4–1535.6 | 0.91 | *** | *** | * |
| TTGFD | 716.6 | 678.0–796.5 | 0.64 | *** | *** | *** | 742.7 | 689.4–794.0 | 0.58 | *** | *** | *** |
| Agronomic and physiological traits | ||||||||||||
| BM_E40 (g/m2) | 146.6 | 136.0–156.9 | 0.32 | ns | ns | ns | 188.3 | 171.6–203.0 | 0.39 | *** | ns | *** |
| BM_InB (g/m2) | 424.9 | 387.2–453.7 | 0.24 | ** | *** | * | 569.4 | 466.6–652.6 | 0.61 | *** | *** | ns |
| BM_A7 (g/m2) | 857.7 | 778.9–945.7 | 0.62 | *** | ns | ns | 1036.4 | 870.6–1185.3 | 0.70 | *** | *** | ns |
| BM_PM (g/m2) | 1355.1 | 1254.5–1472.4 | 0.41 | ** | *** | * | 1341.4 | 1125.1–1471.9 | 0.73 | *** | ** | *** |
| HI | 0.47 | 0.42–0.51 | 0.73 | *** | * | ns | 0.47 | 0.41–0.52 | 0.83 | *** | *** | ** |
| YLD (kg/ha) | 5957 | 5303–6601 | 0.60 | *** | *** | ns | 6310 | 5111–7009 | 0.83 | *** | *** | ns |
| TaTOE-B1 | TaFT3-B1 | TaTOE-B1 + TaFT3-B1 | ||||||
|---|---|---|---|---|---|---|---|---|
| Panel | Avalon Type | Cadenza Type | Avalon Type | Rialto Type | Spark Type | Avalon + Avalon/Rialto | Cadenza + Avalon/Rialto | Cadenza + Spark Type |
| Allele frequency | ||||||||
| HIBAP I | 0.72 | 0.26 | 0.43 | 0.52 | 0.05 | 0.73 | 0.23 | 0.04 |
| HIBAP II | 0.83 | 0.17 | 0.36 | 0.60 | 0.04 | 0.83 | 0.14 | 0.03 |
| BM_PM | ||||||||
| HIBAP I | 1349 a | 1335 a | 1355 a | 1341 a | 1378 a | 1352 a | 1328 a | 1384 a |
| HIBAP II | 1342 a | 1327 a | 1337 a | 1346 a | 1302 a | 1344 a | 1332 a | 1302 a |
| HI | ||||||||
| HIBAP I | 0.474 a | 0.459 b | 0.473 a | 0.469 a | 0.455 a | 0.474 a | 0.459 b | 0.455 ab |
| HIBAP II | 0.486 a | 0.450 b | 0.493 a | 0.476 bc | 0.450 bc | 0.486 a | 0.455 b | 0.44 b |
| YLD (kg/ha) | ||||||||
| HIBAP I | 6010 a | 5740 b | 6010 a | 5910 a | 5840 a | 6020 a | 5740 b | 5800 ab |
| HIBAP II | 6350 a | 6050 b | 6350 a | 6310 a | 5990 a | 6360 a | 6110 b | 5910 b |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dreisigacker, S.; Burgueño, J.; Pacheco, A.; Molero, G.; Sukumaran, S.; Rivera-Amado, C.; Reynolds, M.; Griffiths, S. Effect of Flowering Time-Related Genes on Biomass, Harvest Index, and Grain Yield in CIMMYT Elite Spring Bread Wheat. Biology 2021, 10, 855. https://doi.org/10.3390/biology10090855
Dreisigacker S, Burgueño J, Pacheco A, Molero G, Sukumaran S, Rivera-Amado C, Reynolds M, Griffiths S. Effect of Flowering Time-Related Genes on Biomass, Harvest Index, and Grain Yield in CIMMYT Elite Spring Bread Wheat. Biology. 2021; 10(9):855. https://doi.org/10.3390/biology10090855
Chicago/Turabian StyleDreisigacker, Susanne, Juan Burgueño, Angela Pacheco, Gemma Molero, Sivakumar Sukumaran, Carolina Rivera-Amado, Matthew Reynolds, and Simon Griffiths. 2021. "Effect of Flowering Time-Related Genes on Biomass, Harvest Index, and Grain Yield in CIMMYT Elite Spring Bread Wheat" Biology 10, no. 9: 855. https://doi.org/10.3390/biology10090855
APA StyleDreisigacker, S., Burgueño, J., Pacheco, A., Molero, G., Sukumaran, S., Rivera-Amado, C., Reynolds, M., & Griffiths, S. (2021). Effect of Flowering Time-Related Genes on Biomass, Harvest Index, and Grain Yield in CIMMYT Elite Spring Bread Wheat. Biology, 10(9), 855. https://doi.org/10.3390/biology10090855







