Influence of Season, Population and Individual Characteristics on the Prevalence of Leptospira spp. in Bank Voles in North-West Germany
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. DNA Extraction
2.3. PCR Methods and Multilocus Sequence Typing
2.4. Statistical Analyses
3. Results
3.1. Collection of Rodents
3.2. Leptospira Prevalence, Genomospecies and ST Determination
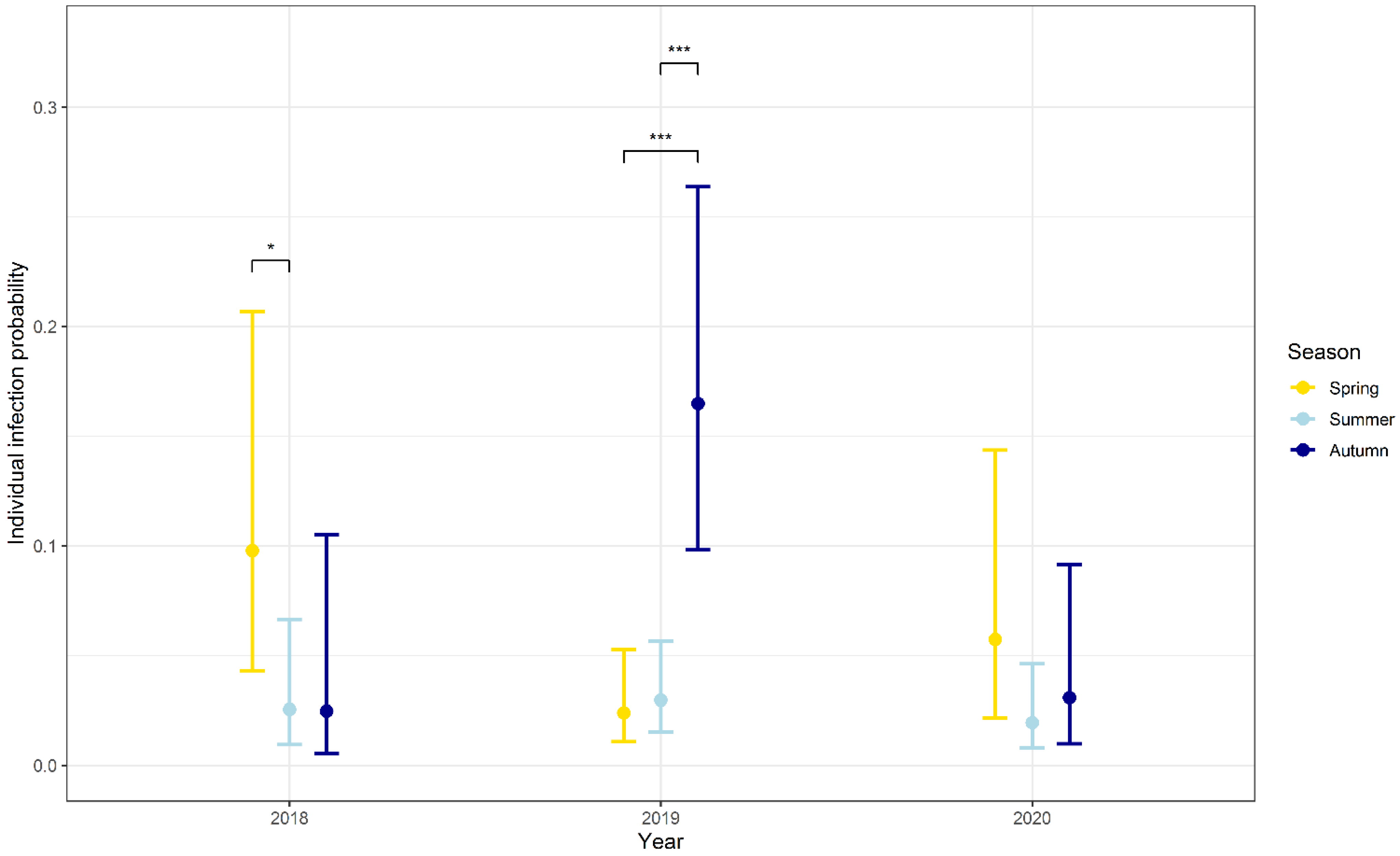
3.3. Influence of Seasonality, Body Weight, Sex and Abundance on the Probability of Infection
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Federal State | North Rhine-Westphalia | Lower Saxony | |
|---|---|---|---|
| Trapping location (Number of trapping sites per location) | NW1 (13 trapping sites) | LS3 (3 trapping sites) | |
| NW2 (3 trapping sites) | LS4 (6 trapping sites) | ||
| LS5 (2 trapping sites) | |||
| LS6 (2 trapping sites) | |||
| Trapping frequency | Three times yearly in spring (March to Mai), summer (July, August) and autumn (October to November) | Three times yearly in spring (April), summer (July) and autumn (October) | |
| Type of trap | Ugglan multiple capture live traps (Grahnab®, Gnosjö, Sweden) | Metal snap traps (Deufa, Neuburg, Germany) | Metal snap traps (Deufa, Neuburg, Germany) |
| Traps per night and trapping site/Number of nights/Control frequency | 49 traps/ 3 nights/ Checked twice a day | 49 traps/ 3 nights/ Checked once a day | 100 traps/ 3 nights/ Checked once a day |
| Bait | apple, rodent pellets, rolled oats, peanut curls | peanut butter with rolled oats | peanut butter with rolled oats |
| Additional information | Rodents captured alive were sampled and released, only rodents found dead were used in this study | ||
| Publication | Trapping according to [101] | Trapping according to [99] | Trapping according to [99] |
| Locus | Primer/BREAKProbe | Sequence (5′ to 3′) |
|---|---|---|
| lipl32 | lipl32-F | AAG CAT TAC CGC TTG TGG TG |
| lipl32-R | GAA CTC CCA TTT CAG CGA TT | |
| probe | 6FAM-AA AGC CAG GAC AAG CGC CG BHQ1 | |
| secY | secY-F | GAA TTT CTC TTT TGA TCT TCG |
| secY-R | GAG TTA GAG CTC AAA TCT AAG | |
| glmU | glmU-F | AGG ATA AGG TCG CTG TGG TA |
| glmU-R | AGT TTT TTT CCG GAG TTT CT | |
| pntA | pntA-F | TAG GAA ARA TGA AAC CRG GAA C |
| pntA-R | AAG AAG CAA GAT CCA CAA YTA C | |
| sucA | sucA-F | TCA TTC CAC TTY TAG ATA CGA T |
| sucA-R | TCT TTT TTG AAT TTT TGA CG | |
| tpiA | tpiA-F | TTG CAG GAA ACT GGA AAA TGA AT |
| tpiA-R | GTT TTA CRG AAC CHC CGT AGA GAA T | |
| pfkB | pfkB-F | CGG AGA GTT TTA TAA RAA GGA CAT |
| pfkB-R | AGA ACA CCC GCC GCA AAA CAA T | |
| mreA | mreA-F | GGC TCG CTC TYG ACG GAA A |
| mreA-R | TCC RTA ACT CAT AAA MGA CAA AGG | |
| caiB | caiB-F | CAA CTT GCG GAY ATA GGA GGA G |
| caiB-R | ATT ATG TTC CCC GTG AYT CG |
References
- Caimi, K.; Ruybal, P. Leptospira spp., a genus in the stage of diversity and genomic data expansion. Infect. Genet. Evol. 2020, 81, 104241. [Google Scholar] [CrossRef]
- Boonsilp, S.; Thaipadungpanit, J.; Amornchai, P.; Wuthiekanun, V.; Bailey, M.S.; Holden, M.T.G.; Zhang, C.; Jiang, X.; Koizumi, N.; Taylor, K.; et al. A single multilocus sequence typing (MLST) scheme for seven pathogenic Leptospira species. PLoS Negl. Trop. Dis. 2013, 7, e1954. [Google Scholar] [CrossRef] [PubMed]
- Picardeau, M. Leptospira and leptospirosis. Methods Mol. Biol. 2020, 2134, 271–275. [Google Scholar] [CrossRef] [PubMed]
- Costa, F.; Hagan, J.E.; Calcagno, J.; Kane, M.; Torgerson, P.; Martinez-Silveira, M.S.; Stein, C.; Abela-Ridder, B.; Ko, A.I. Global morbidity and mortality of leptospirosis: A systematic review. PLOS Negl. Trop. Dis. 2015, 9, e0003898. [Google Scholar] [CrossRef]
- Lau, C.L.; Smythe, L.D.; Craig, S.B.; Weinstein, P. Climate change, flooding, urbanisation and leptospirosis: Fuelling the fire? Trans. R. Soc. Trop. Med. Hyg. 2010, 104, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Munoz-Zanzi, C.; Groene, E.; Morawski, B.M.; Bonner, K.; Costa, F.; Bertherat, E.; Schneider, M.C. A systematic literature review of leptospirosis outbreaks worldwide, 1970–2012. Rev. Panam. Salud Públ. 2020, 44, e78. [Google Scholar] [CrossRef]
- Aviat, F.; Blanchard, B.; Michel, V.; Blanchet, B.; Branger, C.; Hars, J.; Mansotte, F.; Brasme, L.; de Champs, C.; Bolut, P.; et al. Leptospira exposure in the human environment in France: A survey in feral rodents and in fresh water. Comp. Immunol. Microbiol. Infect. Dis. 2009, 32, 463–476. [Google Scholar] [CrossRef]
- Desai, S.; Van Treeck, U.; Lierz, M.; Espelage, W.; Zota, L.; Sarbu, A.; Czerwinski, M.; Sadkowska-Todys, M.; Avdicová, M.; Reetz, J.; et al. Resurgence of field fever in a temperate country: An epidemic of leptospirosis among seasonal strawberry harvesters in Germany in 2007. Clin. Infect. Dis. 2009, 48, 691–697. [Google Scholar] [CrossRef]
- Habus, J.; Persic, Z.; Spicic, S.; Vince, S.; Stritof, Z.; Milas, Z.; Cvetnic, Z.; Perharic, M.; Turk, N. New trends in human and animal leptospirosis in Croatia, 2009–2014. Acta Trop. 2017, 168, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Vieira, M.L.; Gama-Simões, M.J.; Collares-Pereira, M. Human leptospirosis in Portugal: A retrospective study of eighteen years. Int. J. Infect. Dis. 2006, 10, 378–386. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Garvey, P.; Connell, J.; O’Flanagan, D.; McKeown, P. Leptospirosis in Ireland: Annual incidence and exposures associated with infection. Epidemiol. Infect. 2014, 142, 847–855. [Google Scholar] [CrossRef]
- Dupouey, J.; Faucher, B.; Edouard, S.; Richet, H.; Kodjo, A.; Drancourt, M.; Davoust, B. Human leptospirosis: An emerging risk in Europe? Comp. Immunol. Microbiol. Infect. Dis. 2014, 37, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Van Alphen, L.B.; Kunøe, A.L.; Ceper, T.; Kähler, J.; Kjelsø, C.; Ethelberg, S.; Krogfelt, K.A. Trends in human leptospirosis in Denmark, 1980 to 2012. Eurosurveillance 2015, 20, 21019. [Google Scholar] [CrossRef] [PubMed]
- European Centre for Disease Prevention and Control. Available online: https://www.ecdc.europa.eu/en (accessed on 11 March 2021).
- Robert Koch-Institut. SurvStat@RKI 2.0. Available online: https://survstat.rki.de (accessed on 1 March 2021).
- Ellis, W.A. Animal leptospirosis. In Leptospira and Leptospirosis, 1st ed.; Adler, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 99–137. ISBN 009783662523995. [Google Scholar]
- Collings, D.F. Leptospira interrogans infection in domestic and wild animals in Fiji. N. Z. Vet. J. 1984, 32, 21–24. [Google Scholar] [CrossRef] [PubMed]
- Sterling, C.R.; Thiermann, A.B. Urban rats as chronic carriers of leptospirosis: An ultrastructural investigation. Vet. Pathol. 1981, 18, 628–637. [Google Scholar] [CrossRef] [PubMed]
- Davis, S.; Calvet, E.; Leirs, H. Fluctuating rodent populations and risk to humans from rodent-borne zoonoses. Vector Borne Zoonotic Dis. 2005, 5, 305–314. [Google Scholar] [CrossRef]
- Perez, J.; Brescia, F.; Becam, J.; Mauron, C.; Goarant, C. Rodent abundance dynamics and leptospirosis carriage in an area of hyper-endemicity in New Caledonia. PLoS Negl. Trop. Dis. 2011, 5, e1361. [Google Scholar] [CrossRef]
- Bierque, E.; Thibeaux, R.; Girault, D.; Soupé-Gilbert, M.-E.; Goarant, C. A systematic review of Leptospira in water and soil environments. PLoS ONE 2020, 15, e0227055. [Google Scholar] [CrossRef] [PubMed]
- Haake, D.A.; Levett, P.N. Leptospirosis in humans. In Leptospira and Leptospirosis, 1st ed.; Adler, B., Ed.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 65–88. ISBN 009783662523995. [Google Scholar]
- Dadon, Y.; Haas, E.J.; Kaliner, E.; Anis, E.; Singer, S.R.; Atiya-Nasagi, Y.; Cohen-Dar, M.; Avramovich, E.; King, R.; Sued, O.; et al. Outbreak of human leptospirosis linked to contaminated water bodies in Northern Israel, June to August 2018. Eurosurveillance 2018, 23, 1800486. [Google Scholar] [CrossRef]
- Bharti, A.R.; Nally, J.E.; Ricaldi, J.N.; Matthias, M.A.; Diaz, M.M.; Lovett, M.A.; Levett, P.N.; Gilman, R.H.; Willig, M.R.; Gotuzzo, E.; et al. Leptospirosis: A zoonotic disease of global importance. Lancet Infect. Dis. 2003, 757–771. [Google Scholar] [CrossRef]
- Dechet, A.M.; Parsons, M.; Rambaran, M.; Mohamed-Rambaran, P.; Florendo-Cumbermack, A.; Persaud, S.; Baboolal, S.; Ari, M.D.; Shadomy, S.V.; Zaki, S.R.; et al. Leptospirosis outbreak following severe flooding: A rapid assessment and mass prophylaxis campaign; Guyana, January-February 2005. PLoS ONE 2012, 7, e39672. [Google Scholar] [CrossRef]
- Gaynor, K.; Katz, A.R.; Park, S.Y.; Nakata, M.; Clark, T.A.; Effler, P.V. Leptospirosis on Oahu: An outbreak associated with flooding of a university campus. Am. J. Trop. Med. Hyg. 2007, 76, 882–885. [Google Scholar] [CrossRef] [PubMed]
- Brockmann, S.; Piechotowski, I.; Bock-Hensley, O.; Winter, C.; Oehme, R.; Zimmermann, S.; Hartelt, K.; Luge, E.; Nöckler, K.; Schneider, T.; et al. Outbreak of leptospirosis among triathlon participants in Germany, 2006. BMC Infect. Dis. 2010, 10, 91. [Google Scholar] [CrossRef] [PubMed]
- Lau, C.L.; Townell, N.; Stephenson, E.; Van den Berg, D.; Craig, S.B. Leptospirosis: An important zoonosis acquired through work, play and travel. Aust. J. Gen. Pract. 2018, 47, 105–110. [Google Scholar] [CrossRef] [PubMed]
- Radl, C.; Müller, M.; Revilla-Fernandez, S.; Karner-Zuser, S.; De Martin, A.; Schauer, U.; Karner, F.; Stanek, G.; Balcke, P.; Hallas, A.; et al. Outbreak of leptospirosis among triathlon participants in Langau, Austria, 2010. Wien. Klin. Wochenschr. 2011, 123, 751–755. [Google Scholar] [CrossRef] [PubMed]
- Guillois, Y.; Bourhy, P.; Ayral, F.; Pivette, M.; Decors, A.; Grau, J.H.A.; Champenois, B.; Malhère, C.; Combes, B.; Richomme, C.; et al. An outbreak of leptospirosis among kayakers in Brittany, North-West France, 2016. Eurosurveillance 2018, 23. [Google Scholar] [CrossRef]
- Morgan, J.; Bornstein, S.L.; Karpati, A.M.; Bruce, M.; Bolin, C.A.; Austin, C.C.; Woods, C.W.; Lingappa, J.; Langkop, C.; Davis, B.; et al. Outbreak of leptospirosis among triathlon participants and community residents in Springfield, Illinois, 1998. Clin. Infect. Dis. 2002, 34, 1593–1599. [Google Scholar] [CrossRef] [PubMed]
- Jansen, A.; Schöneberg, I.; Frank, C.; Alpers, K.; Schneider, T.; Stark, K. Leptospirosis in Germany, 1962–2003. Emerg. Infect. Dis. 2005, 11, 1048–1054. [Google Scholar] [CrossRef]
- Narkkul, U.; Thaipadungpanit, J.; Srisawat, N.; Rudge, J.W.; Thongdee, M.; Pawarana, R.; Pan-Ngum, W. Human, animal, water source interactions and leptospirosis in Thailand. Sci. Rep. 2021, 11, 3215. [Google Scholar] [CrossRef]
- Meny, P.; Menéndez, C.; Ashfield, N.; Quintero, J.; Rios, C.; Iglesias, T.; Schelotto, F.; Varela, G. Seroprevalence of leptospirosis in human groups at risk due to environmental, labor or social conditions. Rev. Argent. Microbiol. 2019, 51, 324–333. [Google Scholar] [CrossRef]
- Mori, M.; Bourhy, P.; Le Guyader, M.; Van Esbroeck, M.; Djelouadji, Z.; Septfons, A.; Kodjo, A.; Picardeau, M. Pet rodents as possible risk for leptospirosis, Belgium and France, 2009 to 2016. Eurosurveillance 2017, 22, 30553. [Google Scholar] [CrossRef]
- Karpagam, K.B.; Ganesh, B. Leptospirosis: A neglected tropical zoonotic infection of public health importance—An updated review. Eur. J. Clin. Microbiol. Infect. Dis. 2020, 39, 835–846. [Google Scholar] [CrossRef]
- Yang, H.-Y.; Yen, T.-H.; Lin, C.-Y.; Chen, Y.-C.; Pan, M.-J.; Lee, C.-H.; Yu, C.-C.; Wu, M.-S.; Wu, S.-S.; Weng, C.-H.; et al. Early identification of leptospirosis as an ignored cause of multiple organ dysfunction syndrome. Shock 2012, 38, 24–29. [Google Scholar] [CrossRef]
- Maier, A.; Kaeser, R.; Thimme, R.; Boettler, T. Acute pancreatitis and vasoplegic shock associated with leptospirosis—A case report and review of the literature. BMC Infect. Dis. 2019, 19, 395. [Google Scholar] [CrossRef]
- Fernandez, P.S.; Kodjo, A.; Medkour, H.; Laidoudi, Y.; Dubourg, G.; Eldin, C.; Parola, P.; Davoust, B.; Lagier, J.-C. Autochthonous human and animal leptospirosis, Marseille, France. IDCases 2020, 21, e00899. [Google Scholar] [CrossRef] [PubMed]
- Ellis, T.; Imrie, A.; Katz, A.R.; Effler, P.V. Underrecognition of leptospirosis during a dengue fever outbreak in Hawaii, 2001–2002. Vector Borne Zoonotic Dis. 2008, 8, 541–547. [Google Scholar] [CrossRef]
- Briskin, E.A.; Casanovas-Massana, A.; Ryff, K.R.; Morales-Estrada, S.; Hamond, C.; Perez-Rodriguez, N.M.; Benavidez, K.M.; Weinberger, D.M.; Castro-Arellano, I.; Wunder, E.A.; et al. Seroprevalence, Risk Factors, and Rodent Reservoirs of Leptospirosis in an Urban Community of Puerto Rico, 2015. J. Infect. Dis. 2019, 220, 1489–1497. [Google Scholar] [CrossRef]
- Thiermann, A.B. The Norway rat as a selective chronic carrier of Leptospira icterohaemorrhagiae. J. Wildl. Dis. 1981, 17, 39–43. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, N.; Muto, M.; Tanikawa, T.; Mizutani, H.; Sohmura, Y.; Hayashi, E.; Akao, N.; Hoshino, M.; Kawabata, H.; Watanabe, H. Human leptospirosis cases and the prevalence of rats harbouring Leptospira interrogans in urban areas of Tokyo, Japan. J. Med Microbiol. 2009, 58, 1227–1230. [Google Scholar] [CrossRef] [PubMed]
- Holt, J.; Davis, S.; Leirs, H. A model of Leptospirosis infection in an African rodent to determine risk to humans: Seasonal fluctuations and the impact of rodent control. Acta Trop. 2006, 99, 218–225. [Google Scholar] [CrossRef] [PubMed]
- Krijger, I.M.; Ahmed, A.A.A.; Goris, M.G.A.; Cornelissen, J.B.W.J.; Koerkamp, P.W.G.G.; Meerburg, B.G. Wild rodents and insectivores as carriers of pathogenic Leptospira and Toxoplasma gondii in The Netherlands. Vet. Med. Sci. 2020, 6, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Essbauer, S.S.; Mayer-Scholl, A.; Poppert, S.; Schmidt-Chanasit, J.; Klempa, B.; Henning, K.; Schares, G.; Groschup, M.H.; Spitzenberger, F.; et al. Multiple infections of rodents with zoonotic pathogens in Austria. Vector Borne Zoonotic Dis. 2014, 14, 467–475. [Google Scholar] [CrossRef]
- Izquierdo-Rodríguez, E.; Fernández-Álvarez, Á.; Martín-Carrillo, N.; Marchand, B.; Feliu, C.; Miquel, J.; Foronda, P.; Quilichini, Y. Pathogenic Leptospira species in rodents from Corsica (France). PLoS ONE 2020, 15, e0233776. [Google Scholar] [CrossRef] [PubMed]
- Balážová, A.; Nosková, E.; Široký, P.; Durrant, C.; Baláž, V. Diversity and dynamics of zoonotic pathogens within a local community of small mammals. Biologia 2021, 76. [Google Scholar] [CrossRef]
- Millán, J.; Cevidanes, A.; Chirife, A.D.; Candela, M.G.; León-Vizcaíno, L. Risk factors of Leptospira infection in Mediterranean periurban micromammals. Zoonoses Public Health 2018, 65, e79–e85. [Google Scholar] [CrossRef]
- Jeske, K.; Emirhar, D.; García, J.T.; González-Barrio, D.; Olea, P.P.; Fons, F.R.; Schulz, J.; Mayer-Scholl, A.; Heckel, G.; Ulrich, R.G.; et al. Frequent Leptospira spp. detection, but absence of Tula orthohantavirus in Microtus voles, Northwestern Spain. J. Wildl. Dis. 2021, 57. [Google Scholar] [CrossRef]
- Tadin, A.; Tokarz, R.; Markotić, A.; Margaletić, J.; Turk, N.; Habuš, J.; Svoboda, P.; Vucelja, M.; Desai, A.; Jain, K.; et al. Molecular survey of zoonotic agents in rodents and other small mammals in Croatia. Am. J. Trop. Med. Hyg. 2016, 94, 466–473. [Google Scholar] [CrossRef]
- Jeske, K.; Jacob, J.; Drewes, S.; Pfeffer, M.; Heckel, G.; Ulrich, R.G.; Imholt, C. Hantavirus-Leptospira coinfections in small mammals from central Germany. Epidemiol. Infect. 2021, 149, 1–25. [Google Scholar] [CrossRef]
- Obiegala, A.; Woll, D.; Karnath, C.; Silaghi, C.; Schex, S.; Eßbauer, S.; Pfeffer, M. Prevalence and genotype allocation of pathogenic Leptospira species in small mammals from various habitat types in Germany. PLoS Negl. Trop. Dis. 2016, 10, e0004501. [Google Scholar] [CrossRef]
- Mayer-Scholl, A.; Hammerl, J.A.; Schmidt, S.; Ulrich, R.G.; Pfeffer, M.; Woll, D.; Scholz, H.C.; Thomas, A.; Nöckler, K. Leptospira spp. in rodents and shrews in Germany. Int. J. Environ. Res. Public Health 2014, 11, 7562–7574. [Google Scholar] [CrossRef]
- Heuser, E.; Fischer, S.; Ryll, R.; Mayer-Scholl, A.; Hoffmann, D.; Spahr, C.; Imholt, C.; Alfa, D.M.; Fröhlich, A.; Lüschow, D.; et al. Survey for zoonotic pathogens in Norway rat populations from Europe. Pest Manag. Sci. 2017, 73, 341–348. [Google Scholar] [CrossRef]
- Kocianová, E.; Kozuch, O.; Bakoss, P.; Rehácek, J.; Kovácová, E. The prevalence of small terrstrial mammals infected with tick-borne encephalitis virus and leptospirae in the foothills of the Bavarian forest, Germany. Appl. Parasitol. 1993, 34, 283–290. [Google Scholar]
- Obiegala, A.; Albrecht, C.; Dafalla, M.; Drewes, S.; Oltersdorf, C.; Turni, H.; Imholt, C.; Jacob, J.; Wagner-Wiening, C.; Ulrich, R.G.; et al. Leptospira spp. in small mammals from areas with low and high human hantavirus incidences in south-west Germany. Vector Borne Zoonotic Dis. 2017, 17, 312–318. [Google Scholar] [CrossRef]
- Fischer, S.; Mayer-Scholl, A.; Imholt, C.; Spierling, N.G.; Heuser, E.; Schmidt, S.; Reil, D.; Rosenfeld, U.M.; Jacob, J.; Nöckler, K.; et al. Leptospira genomospecies and sequence type prevalence in small mammal populations in Germany. Vector Borne Zoonotic Dis. 2018, 18, 188–199. [Google Scholar] [CrossRef]
- Boey, K.; Shiokawa, K.; Rajeev, S. Leptospira infection in rats: A literature review of global prevalence and distribution. PLoS Negl. Trop. Dis. 2019, 13, e0007499. [Google Scholar] [CrossRef] [PubMed]
- Piechotowski, I.; Brockmann, S.O.; Schwarz, C.; Winter, C.H.; Ranft, U.; Pfaff, G. Emergence of hantavirus in South Germany: Rodents, climate and human infections. Parasitol. Res. 2008, 103, 131–137. [Google Scholar] [CrossRef]
- Nationale Forschungsplattform für Zoonosen. RoBoPub. Available online: https://zoonosen.net/en/forschungsnetz/verbunde-nachwuchsgruppen/robopub (accessed on 3 May 2021).
- Stoddard, R.A.; Gee, J.E.; Wilkins, P.P.; McCaustland, K.; Hoffmaster, A.R. Detection of pathogenic Leptospira spp. through TaqMan polymerase chain reaction targeting the LipL32 gene. Diagn. Microbiol. Infect. Dis. 2009, 64, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Victoria, B.; Ahmed, A.; Zuerner, R.L.; Ahmed, N.; Bulach, D.M.; Quinteiro, J.; Hartskeerl, R.A. Conservation of the S10-spc-α locus within otherwise highly plastic genomes provides phylogenetic insight into the genus Leptospira. PLoS ONE 2008, 3, e2752. [Google Scholar] [CrossRef] [PubMed]
- Bates, D.; Mächler, M.; Bolker, B.; Walker, S. Fitting linear mixed-effects models using lme4. J. Stat. Softw. 2015, 67, 1–48. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2020. [Google Scholar]
- Lenth, R.V. Estimated marginal means, aka least-squares means. R Package Version 2018, 1, 3. [Google Scholar]
- University of Oxford. PubMLST. Available online: https://pubmlst.org/ (accessed on 27 April 2021).
- Helmholtz Zentrum für Umweltforschung. Dürremonitor Deutschland: Dürre 1952–2020 (Jährlich). Available online: https://www.ufz.de/index.php?de=47252 (accessed on 14 June 2021).
- Karaseva, E.V.; Chernukha, Y.G.; Piskunova, L.A. Results of studying the time of survival of pathogenic leptospira under natural conditions. J. Hyg. Epidemiol. Microbiol. Immunol 1973, 17, 339–345. [Google Scholar]
- Nau, L.H.; Obiegala, A.; Król, N.; Mayer-Scholl, A.; Pfeffer, M. Survival time of Leptospira kirschneri serovar Grippotyphosa under different environmental conditions. PLoS ONE 2020, 15, e0236007. [Google Scholar] [CrossRef]
- Woll, D.; Karnath, C.; Pfeffer, M.; Allgöwer, R. Genetic characterization of Leptospira spp. from beavers found dead in south-west Germany. Vet. Microbiol. 2012, 158, 232–234. [Google Scholar] [CrossRef]
- Ferreira, A.S.; Ahmed, A.; Rocha, T.; Vieira, M.L.; Paiva-Cardoso, M.D.N.; Mesquita, J.R.; Van der Linden, H.; Goris, M.; Thompson, G.; Hartskeerl, R.A.; et al. Genetic diversity of pathogenic leptospires from wild, domestic and captive host species in Portugal. Transbound. Emerg. Dis. 2020, 67, 852–864. [Google Scholar] [CrossRef]
- Piredda, I.; Ponti, M.N.; Palmas, B.; Noworol, M.; Pedditzi, A.; Rebechesu, L.; Chisu, V. Molecular typing of pathogenic Leptospira species isolated from wild mammal reservoirs in Sardinia. Animals 2021, 11, 1109. [Google Scholar] [CrossRef]
- Azhari, N.N.; Ramli, S.N.A.; Joseph, N.; Philip, N.; Mustapha, N.F.; Ishak, S.N.; Mohd-Taib, F.S.; Nor, S.M.; Yusof, M.A.; Sah, S.A.M.; et al. Molecular characterization of pathogenic Leptospira spp. in small mammals captured from the human leptospirosis suspected areas of Selangor state, Malaysia. Acta Trop. 2018, 188, 68–77. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, J.; Zhang, T.; Qiu, H.; Li, Z.; Zhang, E.; Li, S.; Chang, Y.-F.; Guo, X.; Jiang, X.; et al. Genetic characteristics of pathogenic Leptospira in wild small animals and livestock in Jiangxi Province, China, 2002–2015. PLoS Negl. Trop. Dis. 2019, 13, e0007513. [Google Scholar] [CrossRef] [PubMed]
- Balboni, A.; Zamagni, S.; Bertasio, C.; Boniotti, M.B.; Troìa, R.; Battilani, M.; Dondi, F. Identification of serogroups Australis and Icterohaemorrhagiae in two dogs with a severe form of acute leptospirosis in Italy. Pathogens 2020, 9, 351. [Google Scholar] [CrossRef] [PubMed]
- Moreno, L.Z.; Kremer, F.S.; Jaeger, L.H.; Loureiro, A.P.; Miraglia, F.; Eslabao, M.R.; Dellagostin, O.A.; Lilenbaum, W.; Moreno, A.M. Genomic characterization and comparative analysis of Leptospira interrogans serogroup Australis isolated from swine. Pathog. Dis. 2017, 75, ftx119. [Google Scholar] [CrossRef]
- Czopowicz, M.; Kaba, J.; Smith, L.; Szalus-Jordanow, O.; Nowicki, M.; Witkowski, L.; Frymus, T. Leptospiral antibodies in the breeding goat population of Poland. Vet. Rec. 2011, 169, 230. [Google Scholar] [CrossRef] [PubMed]
- Weis, S.; Rettinger, A.; Bergmann, M.; Llewellyn, J.R.; Pantchev, N.; Straubinger, R.K.; Hartmann, K. Detection of Leptospira DNA in urine and presence of specific antibodies in outdoor cats in Germany. J. Feline Med. Surg. 2017, 19, 470–476. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.H.; Moreno, L.Z.; Kremer, F.S.; Dellagostin, O.A.; Moreno, A.M.; Lilenbaum, W. Genomic characterization and comparative analysis of Leptospira kirschneri serogroup Grippotyphosa UC5/2011, a strain isolated after mare abortion: Implications for genital animal leptospirosis. Comp. Immunol. Microbiol. Infect. Dis. 2019, 64, 7–9. [Google Scholar] [CrossRef] [PubMed]
- Geisen, V.; Stengel, C.; Brem, S.; Müller, W.; Greene, C.; Hartmann, K. Canine leptospirosis infections—Clinical signs and outcome with different suspected Leptospira serogroups (42 cases). J. Small Anim. Pract. 2007, 48, 324–328. [Google Scholar] [CrossRef] [PubMed]
- Hamond, C.; Martins, G.; Bremont, S.; Medeiros, M.A.; Bourhy, P.; Lilenbaum, W. Molecular characterization and serology of Leptospira kirschneri (Serogroup Grippotyphosa) isolated from urine of a mare post-abortion in Brazil. Zoonoses Public Health 2016, 63, 191–195. [Google Scholar] [CrossRef]
- Cilia, G.; Bertelloni, F.; Piredda, I.; Ponti, M.N.; Turchi, B.; Cantile, C.; Parisi, F.; Pinzauti, P.; Armani, A.; Palmas, B.; et al. Presence of pathogenic Leptospira spp. in the reproductive system and fetuses of wild boars (Sus scrofa) in Italy. PLoS Negl. Trop. Dis. 2020, 14, e0008982. [Google Scholar] [CrossRef]
- Grippi, F.; Giudice, E.; Di Pietro, S.; Sciacca, C.; Santangelo, F.; Galluzzo, P.; Barreca, S.; Guercio, A. Leptospira interrogans serogroup Sejroe serovar Hardjo in aborting cows: Two herd cases in Sicily (Italy). J. Vet. Res. 2020, 64, 73–78. [Google Scholar] [CrossRef]
- Pinna, M.H.; Martins, G.; Loureiro, A.P.; Lilenbaum, W. Detection of bovine carriers of Leptospira by serological, bacteriological, and molecular tools. Trop. Anim. Health Prod. 2018, 50, 883–888. [Google Scholar] [CrossRef]
- Ivanova, S.; Herbreteau, V.; Blasdell, K.; Chaval, Y.; Buchy, P.; Guillard, B.; Morand, S. Leptospira and rodents in Cambodia: Environmental determinants of infection. Am. J. Trop. Med. Hyg. 2012, 86, 1032–1038. [Google Scholar] [CrossRef]
- Krøjgaard, L.H.; Villumsen, S.; Markussen, M.D.K.; Jensen, J.S.; Leirs, H.; Heiberg, A.-C. High prevalence of Leptospira spp. in sewer rats (Rattus norvegicus). Epidemiol. Infect. 2009, 137, 1586–1592. [Google Scholar] [CrossRef]
- Himsworth, C.G.; Bidulka, J.; Parsons, K.L.; Feng, A.Y.T.; Tang, P.; Jardine, C.M.; Kerr, T.; Mak, S.; Robinson, J.; Patrick, D.M. Ecology of Leptospira interrogans in Norway rats (Rattus norvegicus) in an inner-city neighborhood of Vancouver, Canada. PLoS Negl. Trop. Dis. 2013, 7, e2270. [Google Scholar] [CrossRef] [PubMed]
- Vanasco, N.B.; Sequeira, M.D.; Sequeira, G.; Tarabla, H.D. Associations between leptospiral infection and seropositivity in rodents and environmental characteristics in Argentina. Prev. Vet. Med. 2003, 60, 227–235. [Google Scholar] [CrossRef]
- Marchlewska-Koj, A.; Kolodziej, B.; Filimowska, A. Aggressive behavior of adult bank voles (Clethrionomys glareolus) towards conspecifics. Aggr. Behav. 1989, 15, 381–387. [Google Scholar] [CrossRef]
- Tamarin, R.H.; Ostfeld, R.S.; Pugh, S.R.; Bujalska, G. Social Systems and Population Cycles in Voles; Springer: Basel, Switzerland, 1990; ISBN 009783764324377. [Google Scholar]
- Gomes, C.K.; Guedes, M.; Potula, H.-H.; Dellagostin, O.A.; Gomes-Solecki, M. Sex matters: Male hamsters are more susceptible to lethal infection with lower doses of pathogenic Leptospira than female hamsters. Infect. Immun. 2018, 86. [Google Scholar] [CrossRef] [PubMed]
- Krijger, I.M.; Ahmed, A.A.A.; Goris, M.G.A.; Koerkamp, P.W.G.G.; Meerburg, B.G. Prevalence of Leptospira infection in rodents from Bangladesh. Int. J. Environ. Res. Public Health 2019, 16, 2113. [Google Scholar] [CrossRef] [PubMed]
- Morris, P. A review of mammalian age determination methods. Mammal Rev. 1972, 2, 69–104. [Google Scholar] [CrossRef]
- Mazurkiewicz, M. Density and weight structure of populations of the bank vole in open and enclosed areas. Acta Theriol. 1972, 17, 455–465. [Google Scholar] [CrossRef]
- Mills, J.N.; Ksiazek, T.G.; Peters, C.J.; Childs, J.E. Long-term studies of hantavirus reservoir populations in the southwestern United States: A synthesis. Emerg. Infect. Dis. 1999, 5, 135–142. [Google Scholar] [CrossRef] [PubMed]
- Birnbaum, S.; Shenberg, E.; Torten, M. The influence of maternal antibodies on the epidemiology of leptospiral carrier state in mice. Am. J. Epidemiol. 1972, 96, 313–317. [Google Scholar] [CrossRef] [PubMed]
- Korslund, L.; Steen, H. Small rodent winter survival: Snow conditions limit access to food resources. J. Anim. Ecol. 2006, 75, 156–166. [Google Scholar] [CrossRef] [PubMed]
- Binder, F.; Drewes, S.; Imholt, C.; Saathoff, M.; Below, D.A.; Bendl, E.; Conraths, F.J.; Tenhaken, P.; Mylius, M.; Brockmann, S.; et al. Heterogeneous Puumala orthohantavirus situation in endemic regions in Germany in summer 2019. Transbound. Emerg. Dis. 2020, 67, 502–509. [Google Scholar] [CrossRef]
- McMichael, A.J.; Woodruff, R.E.; Hales, S. Climate change and human health: Present and future risks. Lancet 2006, 367, 859–869. [Google Scholar] [CrossRef]
- Reil, D.; Rosenfeld, U.M.; Imholt, C.; Schmidt, S.; Ulrich, R.G.; Eccard, J.A.; Jacob, J. Puumala hantavirus infections in bank vole populations: Host and virus dynamics in Central Europe. BMC Ecol. 2017, 17, 9. [Google Scholar] [CrossRef] [PubMed]
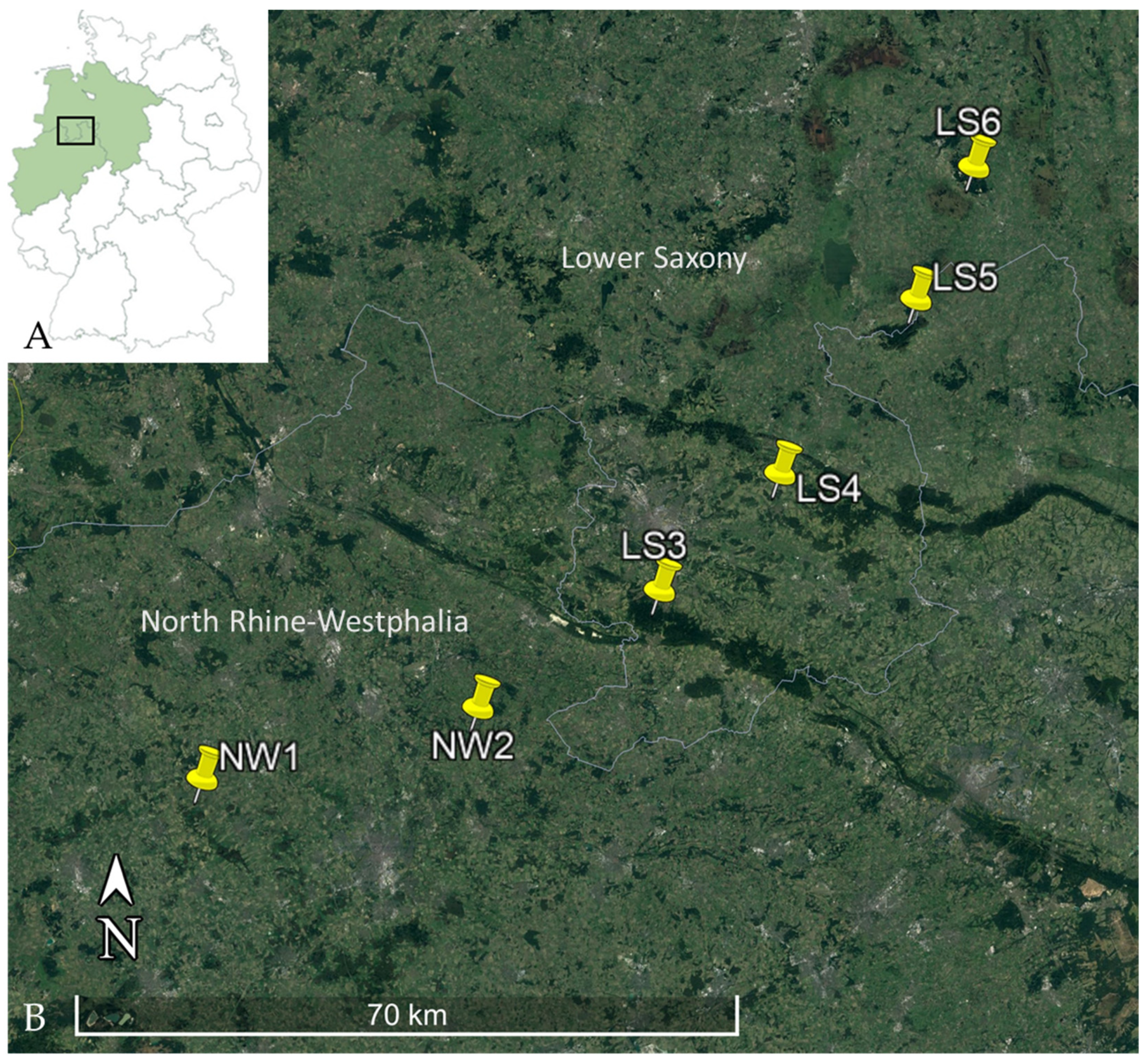


| Federal State | Total Number of Bank Voles | Trapping Location (see Figure 1) | Number of Leptospira DNA Positive/Total Number of Bank Voles Tested (lipl32-qPCR) (Percentage, 95% CI) | Number of secY-SLST/MLST Positive Bank Voles * | ||
|---|---|---|---|---|---|---|
| L. interrogans/ ST24 | L. kirschneri/ ST110 | L. borgpetersenii/ ST197 | ||||
| North Rhine-Westphalia | 782 | NW1 | 95/605 (15.7%, 12.9—18.9) | 55/15 | 8/3 | 2/1 |
| NW2 | 12/177 (6.8%, 3.6—11.5) | 7/5 | 1/0 | N/A | ||
| Lower Saxony | 1035 | LS3 | 1/160 (0.6, 0—3.4) | N/A | N/A | N/A |
| LS4 | 26/770 (4.3%, 2.2—4.9) | 17/9 | 2/1 | 3/2 | ||
| LS5 | 3/45 (6.7%, 1.4—18.3) | 1 | N/A | N/A | ||
| LS6 | 0/60 (0%, 0—6%) | N/A | N/A | N/A | ||
| total | 1817 | 137/1817 * (7.5%, 95% CI: 6.4—8.9) | 80/29 | 11/4 | 5/3 | |
| Factor | Estimate | Std. Error | z-Value | p-Value |
|---|---|---|---|---|
| Intercept | −3.676 | 0.783 | −4.696 | <0.001 |
| Season [spring] | 1.454 | 0.823 | 1.767 | 0.077 |
| Season [summer] | 0.032 | 0.857 | 0.037 | 0.971 |
| Year [2019] | 2.054 | 0.757 | 2.712 | 0.007 |
| Year [2020] | 0.225 | 0.909 | 0.248 | 0.804 |
| Year:season | Estimate | Std. Error | z-ratio | p-value |
| Year 2018 | ||||
| Autumn v. spring | −1.454 | 0.823 | −1.767 | 0.181 |
| Autumn v. summer | −0.032 | 0.857 | −0.037 | 0.999 |
| Spring v. summer | 1.423 | 0.567 | 2.511 | 0.032 |
| Year 2019 | ||||
| Autumn v. spring | 2.087 | 0.364 | 5.732 | <0.001 |
| Autumn v. summer | 1.868 | 0.285 | 6.554 | <0.001 |
| Spring v. summer | −0.219 | 0.400 | −0.546 | 0.848 |
| Year 2020 | ||||
| Autumn v. spring | −0.649 | 0.691 | −0.939 | 0.616 |
| Autumn v. summer | 0.473 | 0.643 | 0.735 | 0.743 |
| Spring v. summer | 1.122 | 0.582 | 1.928 | 0.131 |
| Random effects | Variance | Std.Dev. | ||
| Site | 1.460 | 1.208 | ||
| Year | 0.000 | 0.000 | ||
| Season:year | 0.000 | 0.000 | ||
| Factor | Estimate | Std. Error | z-Value | p-Value |
|---|---|---|---|---|
| Year 2018 | ||||
| Intercept | −4.746 | 1.277 | −3.717 | <0.001 |
| Sex [m] | 0.150 | 0.518 | 0.289 | 0.773 |
| Weight | 0.093 | 0.052 | 1.808 | 0.071 |
| Random effects | Variance | Std.Dev. | ||
| Site | 0.709 | 0.842 | ||
| Season | 0.137 | 0.370 | ||
| Year 2019 | ||||
| Intercept | −4.639 | 1.039 | −4.466 | <0.001 |
| Sex [m] | 0.637 | 0.262 | 2.433 | 0.015 |
| Weight | 0.069 | 0.035 | 2.001 | 0.045 |
| Random effects | Variance | Std.Dev. | ||
| Site | 1.614 | 1.271 | ||
| Season | 1.323 | 1.150 | ||
| Year 2020 | ||||
| Intercept | −8.284 | 1.680 | −4.931 | <0.001 |
| Sex [m] | 0.161 | 0.535 | 0.301 | 0.764 |
| Weight | 0.224 | 0.065 | 3.441 | <0.001 |
| Random effects | Variance | Std.Dev. | ||
| Site | 1.525 | 1.235 | ||
| Season | 0.000 | 0.000 | ||
| Factor | Estimate | Std. Error | z-Value | p-Value |
|---|---|---|---|---|
| Spring 2019 | ||||
| Intercept | −5.519 | 1.714 | −3.220 | 0.001 |
| Delayed abundance | −0.850 | 0.553 | −1.538 | 0.124 |
| Abundance | 0.352 | 0.219 | 1.605 | 0.109 |
| Random effects | Variance | Std.Dev. | ||
| Site | 2.802 | 1.674 | ||
| Summer 2019 | ||||
| Intercept | −2.845 | 0.602 | −4.727 | 0.000 |
| Delayed abundance | 0.102 | 0.049 | 2.078 | 0.038 |
| Abundance | −0.068 | 0.033 | −2.030 | 0.042 |
| Random effects | Variance | Std.Dev. | ||
| Site | 0.398 | 0.631 | ||
| Autumn 2019 | ||||
| Intercept | −1.284 | 0.576 | −2.230 | 0.026 |
| Delayed abundance | −0.097 | 0.034 | −2.865 | 0.004 |
| Abundance | 0.147 | 0.052 | 2.802 | 0.005 |
| Random effects | Variance | Std.Dev. | ||
| Site | 0.611 | 0.782 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Schmidt, E.; Obiegala, A.; Imholt, C.; Drewes, S.; Saathoff, M.; Freise, J.; Runge, M.; Jacob, J.; Mayer-Scholl, A.; Ulrich, R.G.; et al. Influence of Season, Population and Individual Characteristics on the Prevalence of Leptospira spp. in Bank Voles in North-West Germany. Biology 2021, 10, 933. https://doi.org/10.3390/biology10090933
Schmidt E, Obiegala A, Imholt C, Drewes S, Saathoff M, Freise J, Runge M, Jacob J, Mayer-Scholl A, Ulrich RG, et al. Influence of Season, Population and Individual Characteristics on the Prevalence of Leptospira spp. in Bank Voles in North-West Germany. Biology. 2021; 10(9):933. https://doi.org/10.3390/biology10090933
Chicago/Turabian StyleSchmidt, Elisabeth, Anna Obiegala, Christian Imholt, Stephan Drewes, Marion Saathoff, Jona Freise, Martin Runge, Jens Jacob, Anne Mayer-Scholl, Rainer G. Ulrich, and et al. 2021. "Influence of Season, Population and Individual Characteristics on the Prevalence of Leptospira spp. in Bank Voles in North-West Germany" Biology 10, no. 9: 933. https://doi.org/10.3390/biology10090933
APA StyleSchmidt, E., Obiegala, A., Imholt, C., Drewes, S., Saathoff, M., Freise, J., Runge, M., Jacob, J., Mayer-Scholl, A., Ulrich, R. G., & Pfeffer, M. (2021). Influence of Season, Population and Individual Characteristics on the Prevalence of Leptospira spp. in Bank Voles in North-West Germany. Biology, 10(9), 933. https://doi.org/10.3390/biology10090933







