Author Contributions
X.Y.: Conceptualization, Methodology, Software, Data Curation, Writing—Original Draft, Writing—Review & Editing; S.-H.W.: Validation, Investigation, Writing—Review & Editing, Visualization, Funding acquisition; J.M.G.: Methodology, Investigation, Writing—Review & Editing, Visualization; X.-W.J.: Conceptualization, Formal analysis, Resources, Writing—Review & Editing, Supervision, Project administration; D.S.G.: Software, Formal analysis, Resources, Data Curation, Writing—Original Draft, Project administration; Y.-D.Z.: Validation, Formal analysis, Investigation, Resources, Data Curation, Writing—Original Draft, Writing—Review & Editing, Supervision, Funding acquisition. All authors have read and agreed to the published version of the manuscript.
Figure 1.
Four example mammograms of one breast (a) Left MLO view images (b) Right MLO view images (c) Left CC view images (d) Right CC view images.
Figure 1.
Four example mammograms of one breast (a) Left MLO view images (b) Right MLO view images (c) Left CC view images (d) Right CC view images.
Figure 2.
Data flow in pre-processing module. Reference means the beast image is acquired from original mammograms by referring to the binarized mammograms.
Figure 2.
Data flow in pre-processing module. Reference means the beast image is acquired from original mammograms by referring to the binarized mammograms.
Figure 3.
Breast region extraction (a) Original mammogram (b) Binarized mammogram (c) Extracted breast region (d) Flipped breast region.
Figure 3.
Breast region extraction (a) Original mammogram (b) Binarized mammogram (c) Extracted breast region (d) Flipped breast region.
Figure 4.
Encoder-Decoder with dilated convolution in Deeplabv3+.
Figure 4.
Encoder-Decoder with dilated convolution in Deeplabv3+.
Figure 5.
CAM refinement module.
Figure 5.
CAM refinement module.
Figure 6.
Global channel attention module.
Figure 6.
Global channel attention module.
Figure 7.
The architecture encoder in PeMNet.
Figure 7.
The architecture encoder in PeMNet.
Figure 8.
The architecture of PeMNet with the backbone of InceptionResNetv2.
Figure 8.
The architecture of PeMNet with the backbone of InceptionResNetv2.
Figure 9.
Mammography image examples from OPTIMAM and INbreast datasets. (a) An example image from OPTIMAM. (b) An example image from INBreast.
Figure 9.
Mammography image examples from OPTIMAM and INbreast datasets. (a) An example image from OPTIMAM. (b) An example image from INBreast.
Figure 10.
A segmentation example from OPTIMAM by Deeplabv3+ with different backbones. (a) Pre-processed image. (b) : 98.29% of IoU, 99.73% of GPA, 97.70% of DSC, 95.50% of Jaccard, 99.89% of Sensitivity, and 99.72% of Specificity. (c) : 99.18% of IoU, 99.87% of GPA, 98.35% of DSC, 96.75% of Jaccard, 99.90% of Sensitivity, and 99.87% of Specificity. (d) : 99.35% of IoU, 99.90% of GPA, 98.64% of DSC, 97.32% of Jaccard, 100% of Sensitivity, and 99.89% of Specificity. (e) : 98.87% of IoU, 99.83% of GPA, 98.08% of DSC, 96.23% of Jaccard, 98.04% of Sensitivity, and 99.99% of Specificity. (f) : 98.99% of IoU, 99.85% of GPA, 98.30% of DSC, 96.65% of Jaccard, 98.15% of Sensitivity, and 100% of Specificity.
Figure 10.
A segmentation example from OPTIMAM by Deeplabv3+ with different backbones. (a) Pre-processed image. (b) : 98.29% of IoU, 99.73% of GPA, 97.70% of DSC, 95.50% of Jaccard, 99.89% of Sensitivity, and 99.72% of Specificity. (c) : 99.18% of IoU, 99.87% of GPA, 98.35% of DSC, 96.75% of Jaccard, 99.90% of Sensitivity, and 99.87% of Specificity. (d) : 99.35% of IoU, 99.90% of GPA, 98.64% of DSC, 97.32% of Jaccard, 100% of Sensitivity, and 99.89% of Specificity. (e) : 98.87% of IoU, 99.83% of GPA, 98.08% of DSC, 96.23% of Jaccard, 98.04% of Sensitivity, and 99.99% of Specificity. (f) : 98.99% of IoU, 99.85% of GPA, 98.30% of DSC, 96.65% of Jaccard, 98.15% of Sensitivity, and 100% of Specificity.
Figure 11.
A segmentation example from INbreast by Deeplabv3+ with different backbones. (a) Pre-processed image. (b) : 97.99% of IoU, 99.80% of GPA, 96.85% of DSC, 93.89% of Jaccard, 100% of Sensitivity, and 99.60% of Specificity. (c) : 98.60% of IoU, 99.86% of GPA, 97.39% of DSC, 94.92% of Jaccard, 100% of Sensitivity, and 99.72% of Specificity. (d) : 98.15% of IoU, 99.82% of GPA, 96.82% of DSC, 93.83% of Jaccard, 100% of Sensitivity, and 99.63% of Specificity. (e) : 98.87% of IoU, 99.83% of GPA, 98.08% of DSC, 96.23% of Jaccard, 98.04% of Sensitivity, and 99.99% of Specificity. (f) : 99.21% of IoU, 99.88% of GPA, 98.28% of DSC, 96.62% of Jaccard, 99.89% of Sensitivity, and 99.86% of Specificity.
Figure 11.
A segmentation example from INbreast by Deeplabv3+ with different backbones. (a) Pre-processed image. (b) : 97.99% of IoU, 99.80% of GPA, 96.85% of DSC, 93.89% of Jaccard, 100% of Sensitivity, and 99.60% of Specificity. (c) : 98.60% of IoU, 99.86% of GPA, 97.39% of DSC, 94.92% of Jaccard, 100% of Sensitivity, and 99.72% of Specificity. (d) : 98.15% of IoU, 99.82% of GPA, 96.82% of DSC, 93.83% of Jaccard, 100% of Sensitivity, and 99.63% of Specificity. (e) : 98.87% of IoU, 99.83% of GPA, 98.08% of DSC, 96.23% of Jaccard, 98.04% of Sensitivity, and 99.99% of Specificity. (f) : 99.21% of IoU, 99.88% of GPA, 98.28% of DSC, 96.62% of Jaccard, 99.89% of Sensitivity, and 99.86% of Specificity.
Figure 12.
A segmentation example from OPTIMAM by PeMNet with different backbones. (a) Pre-processed image. (b) : 99.00% of IoU, 99.70% of GPA, 98.41% of DSC, 96.87% of Jaccard, 99.53% of Sensitivity, and 99.88% of Specificity. (c) : 99.53% of IoU, 99.92% of GPA, 99.03% of DSC, 98.08% of Jaccard, 99.90% of Sensitivity, and 99.93% of Specificity. (d) : 99.57% of IoU, 99.78% of GPA, 99.20% of DSC, 98.41% of Jaccard, 99.59% of Sensitivity, and 99.97% of Specificity. (e) : 99.43% of IoU, 99.72% of GPA, 99.00% of DSC, 98.01% of Jaccard, 99.49% of Sensitivity, and 99.95% of Specificity. (f) : 99.49% of IoU, 99.76% of GPA, 99.03% of DSC, 98.08% of Jaccard, 99.56% of Sensitivity, and 99.95% of Specificity.
Figure 12.
A segmentation example from OPTIMAM by PeMNet with different backbones. (a) Pre-processed image. (b) : 99.00% of IoU, 99.70% of GPA, 98.41% of DSC, 96.87% of Jaccard, 99.53% of Sensitivity, and 99.88% of Specificity. (c) : 99.53% of IoU, 99.92% of GPA, 99.03% of DSC, 98.08% of Jaccard, 99.90% of Sensitivity, and 99.93% of Specificity. (d) : 99.57% of IoU, 99.78% of GPA, 99.20% of DSC, 98.41% of Jaccard, 99.59% of Sensitivity, and 99.97% of Specificity. (e) : 99.43% of IoU, 99.72% of GPA, 99.00% of DSC, 98.01% of Jaccard, 99.49% of Sensitivity, and 99.95% of Specificity. (f) : 99.49% of IoU, 99.76% of GPA, 99.03% of DSC, 98.08% of Jaccard, 99.56% of Sensitivity, and 99.95% of Specificity.
Figure 13.
A segmentation example from INbreast by PeMNet with different backbones. (a) Pre-processed image. (b) : 99.27% of IoU, 99.93% of GPA, 98.40% of DSC, 96.85% of Jaccard, 100% of Sensitivity, and 99.86% of Specificity. (c) : 99.42% of IoU, 99.94% of GPA, 98.55% of DSC, 97.14% of Jaccard, 100% of Sensitivity, and 99.89% of Specificity. (d) : 99.46% of IoU, 99.84% of GPA, 98.67% of DSC, 97.37% of Jaccard, 99.75% of Sensitivity, and 99.92% of Specificity. (e) : 99.64% of IoU, 99.96% of GPA, 98.85% of DSC, 97.72% of Jaccard, 100.00% of Sensitivity, and 99.93% of Specificity. (f) : 99.91% of IoU, 99.96% of GPA, 99.14% of DSC, 98.30% of Jaccard, 99.96% of Sensitivity, and 99.95% of Specificity.
Figure 13.
A segmentation example from INbreast by PeMNet with different backbones. (a) Pre-processed image. (b) : 99.27% of IoU, 99.93% of GPA, 98.40% of DSC, 96.85% of Jaccard, 100% of Sensitivity, and 99.86% of Specificity. (c) : 99.42% of IoU, 99.94% of GPA, 98.55% of DSC, 97.14% of Jaccard, 100% of Sensitivity, and 99.89% of Specificity. (d) : 99.46% of IoU, 99.84% of GPA, 98.67% of DSC, 97.37% of Jaccard, 99.75% of Sensitivity, and 99.92% of Specificity. (e) : 99.64% of IoU, 99.96% of GPA, 98.85% of DSC, 97.72% of Jaccard, 100.00% of Sensitivity, and 99.93% of Specificity. (f) : 99.91% of IoU, 99.96% of GPA, 99.14% of DSC, 98.30% of Jaccard, 99.96% of Sensitivity, and 99.95% of Specificity.
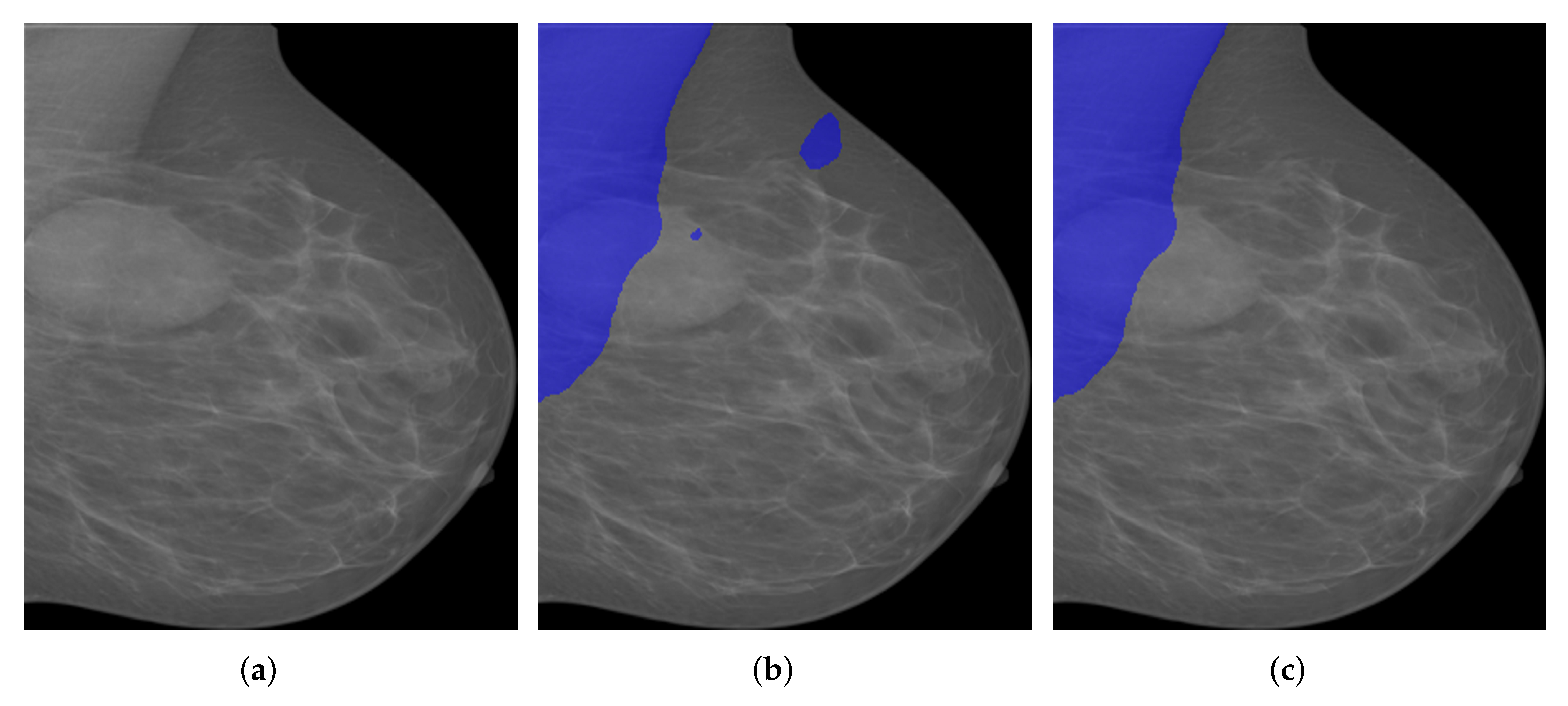
Figure 14.
A segmentation example by when tumor is adjoining to pectoral muscle. (a) Pre-processed image. (b) Original segmentation results by . (c) Post processed segmentation results.
Figure 14.
A segmentation example by when tumor is adjoining to pectoral muscle. (a) Pre-processed image. (b) Original segmentation results by . (c) Post processed segmentation results.
Figure 15.
A segmentation example by when image is of low contrast. (a) A low contrast mammography image. (b) Segmentation results by .
Figure 15.
A segmentation example by when image is of low contrast. (a) A low contrast mammography image. (b) Segmentation results by .
Figure 16.
Segmentation examples by with varied size of pectoral muscle. (a) Mammography images in the presence of the small pectoral muscle. (b) Segmentation results by . (c) Mammography images in the presence of the large pectoral muscle. (d) Segmentation results by .
Figure 16.
Segmentation examples by with varied size of pectoral muscle. (a) Mammography images in the presence of the small pectoral muscle. (b) Segmentation results by . (c) Mammography images in the presence of the large pectoral muscle. (d) Segmentation results by .
Table 1.
Training parameters for pectoral segmentation model.
Table 1.
Training parameters for pectoral segmentation model.
| Parameter | Value |
|---|
| Minibatch size | 32 |
| Initial learning rate | 1 × 10 |
| Maximum training epochs | 50 |
| Learning rate drop factor | 0.5 |
| Learning rate drop period | 20 |
| Shuffle per epoch | Yes |
| Loss function | Cross-entropy |
Table 2.
Dataset composition for pectoral muscle segmentation.
Table 2.
Dataset composition for pectoral muscle segmentation.
| Data | OPTIMAM | INbreast | Total |
|---|
| Training set | 545 | 160 | 705 |
| Testing set | 136 | 40 | 176 |
| Total | 682 | 704 | 881 |
Table 3.
Performance of the trained Deeplabv3+ based on different deep CNN models.
Table 3.
Performance of the trained Deeplabv3+ based on different deep CNN models.
| Model | IoU | GPA | DSC | Jaccard | Sensitivity | Specificity |
|---|
| 76.09 ± 10.08 | 92.13 ± 2.91 | 72.87 ± 10.38 | 60.77 ± 11.97 | 98.43 ± 0.43 | 91.37 ± 6.17 |
| 96.65 ± 0.57 | 99.30 ± 0.15 | 94.80 ± 0.82 | 90.47 ± 1.47 | 98.44 ± 0.42 | 99.41 ± 0.18 |
| 96.87 ± 0.75 | 99.35 ± 0.22 | 95.21 ± 1.28 | 91.19 ± 2.24 | 98.43 ± 0.66 | 99.47 ± 0.26 |
| 96.47 ± 0.34 | 99.27 ± 0.24 | 94.97 ± 0.57 | 90.68 ± 1.11 | 97.33 ± 0.62 | 99.52 ± 0.15 |
| 96.65 ± 0.55 | 99.30 ± 0.25 | 94.80 ± 0.92 | 90.52 ± 1.61 | 98.11 ± 0.62 | 99.46 ± 0.19 |
| 97.13 ± 0.28 | 99.42 ± 0.48 | 95.60 ± 0.40 | 92.29 ± 0.36 | 96.64 ± 1.96 | 99.77 ± 0.10 |
Table 4.
Number of training parameters of different models.
Table 4.
Number of training parameters of different models.
| Model | Number of Layers | Number of Parameters |
|---|
| 46 | 7,697,410 |
| 100 | 20,594,356 |
| 206 | 43,923,380 |
| 186 | 6,749,044 |
| 205 | 27,579,844 |
| 853 | 71,045,012 |
Table 5.
Performance of the trained PeMNets.
Table 5.
Performance of the trained PeMNets.
| Model | IoU | GPA | DSC | Jaccard | Sensitivity | Specificity |
|---|
| 96.98 ± 0.50 | 99.38 ± 0.20 | 95.33 ± 0.84 | 91.45 ± 1.53 | 98.28 ± 0.56 | 99.51 ± 0.18 |
| 96.78 ± 0.78 | 99.33 ± 0.21 | 95.06 ± 1.20 | 90.93 ± 2.11 | 98.41 ± 0.56 | 99.45 ± 0.24 |
| 96.45 ± 0.33 | 99.27 ± 0.24 | 94.78 ± 0.45 | 90.65 ± 0.87 | 97.43 ± 0.58 | 99.50 ± 0.13 |
| 96.70 ± 0.48 | 99.32 ± 0.49 | 94.93 ± 0.58 | 90.93 ± 1.12 | 97.61 ± 1.11 | 99.54 ± 0.18 |
| 97.46 ± 0.45 | 99.48 ± 0.29 | 96.30 ± 0.66 | 93.33 ± 1.04 | 97.12 ± 0.56 | 99.78 ± 0.07 |
Table 6.
Number of training parameters of PeMNets.
Table 6.
Number of training parameters of PeMNets.
| Model | Number of Layers | Number of Parameters | Parameters Increment |
|---|
| 116 | 20,760,500 | 166,144 |
| 220 | 44,909,876 | 986,496 |
| 200 | 6,927,964 | 178,920 |
| 226 | 28,787,168 | 1,207,324 |
| 874 | 80,670,948 | 9,625,936 |
Table 7.
Method comparison. The bold font indicates the best.
Table 7.
Method comparison. The bold font indicates the best.
| Method | Dataset | IoU | GPA | DSC | Jaccard | Sensitivity | Specificity |
|---|
| Shen et al. [18] | INbreast | - | - | 89.10 ± 16.54 | 84.61 ± 18.15 | - | - |
| Soleimani et al. [19] | INbreast | - | - | 95.60 ± 8.40 | 92.60 ± 10.60 | 95.20 ± 8.6 | 99.80 ± 1.80 |
| Ali et al. [21] | INbreast | 87.9 ± 4.5 | 95.00 ± 3.15 | 94.00 ± 3.72 | - | - | - |
| Rampun et al. [34] | INbreast | - | - | 89.60 ± 10.10 | 84.60 ± 15.60 | 89.60 ± 9.60 | 99.70 ± 0.80 |
| Guo et al. [20] | Private | - | - | 96.22 ± 0.05 | - | - | - |
| INbreast | 97.46 ± 0.45 | 99.48 ± 0.29 | 96.30 ± 0.66 | 93.33 ± 1.04 | 97.12 ± 0.56 | 99.78 ± 0.07 |
| (Our method) | OPTIMAM | | | | | | |