Perception of Biocontrol Potential of Bacillus inaquosorum KR2-7 against Tomato Fusarium Wilt through Merging Genome Mining with Chemical Analysis
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Strains and Culture Conditions
2.2. Dual Culture Assay
2.3. MALDI-TOF-MS Analysis of KR2-7 Secondary Metabolites
2.4. Genome Sequencing, Assembly, and Annotation
2.5. Genome Phylogeny
2.6. Pathway Analysis
2.7. Genome-Wide Identification of Secondary Metabolite Biosynthesis Gene Clusters
3. Results
3.1. General Genomic Features of Strain KR2-7
3.2. Genome Phylogeny
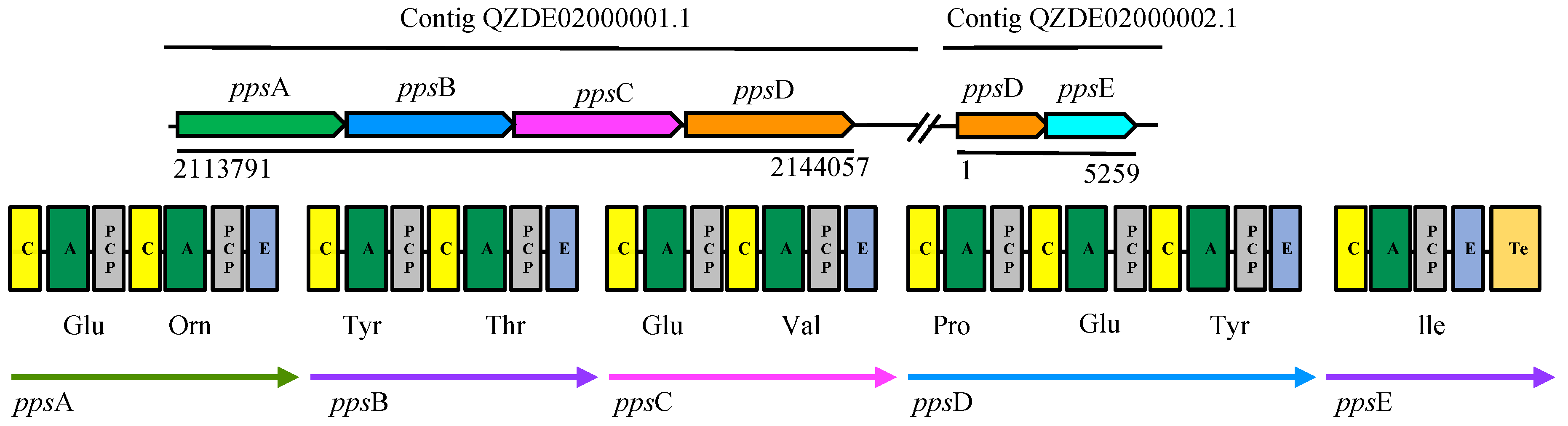
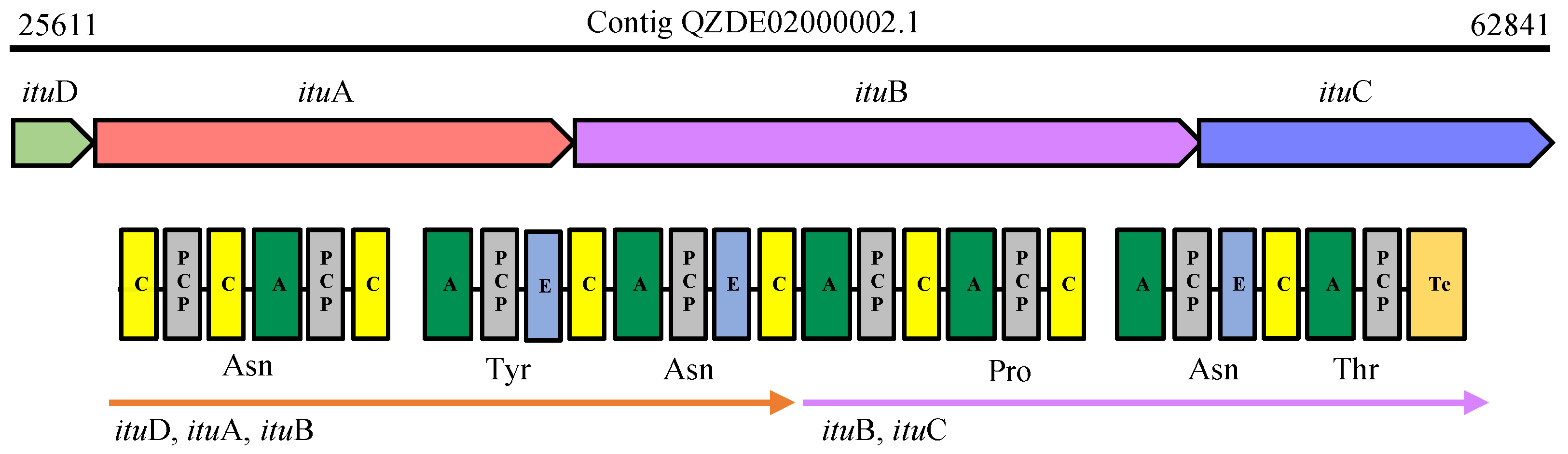
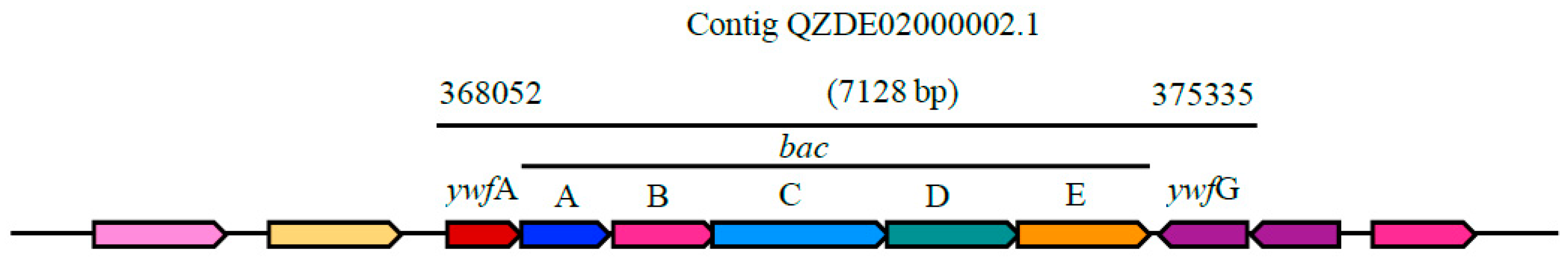
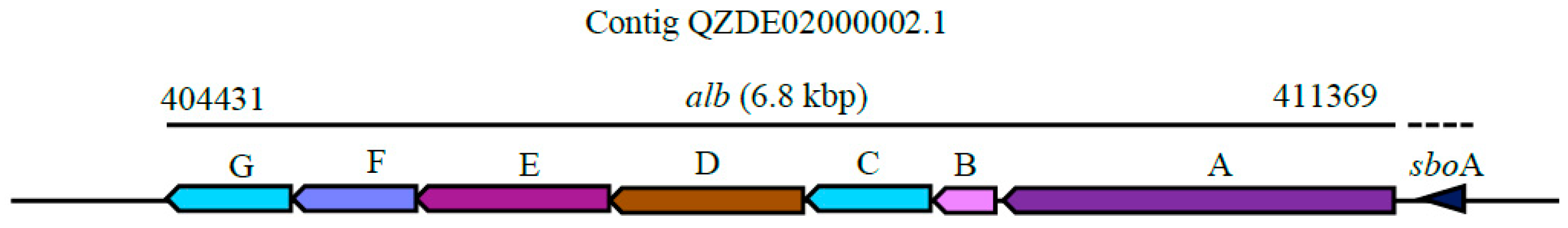
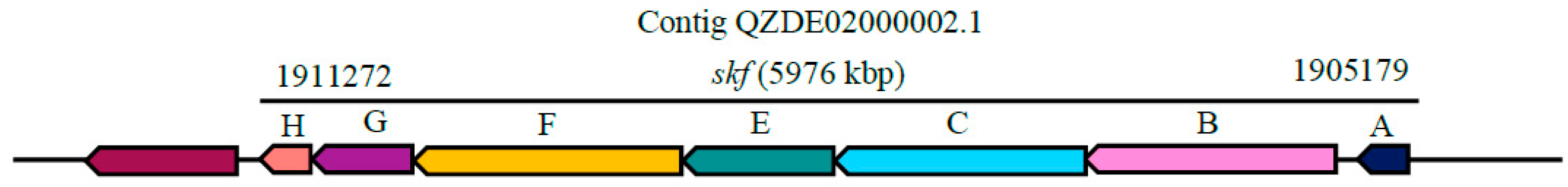
3.3. Secondary Metabolites Biosynthetic Gene Clusters in the KR2-7 Genome
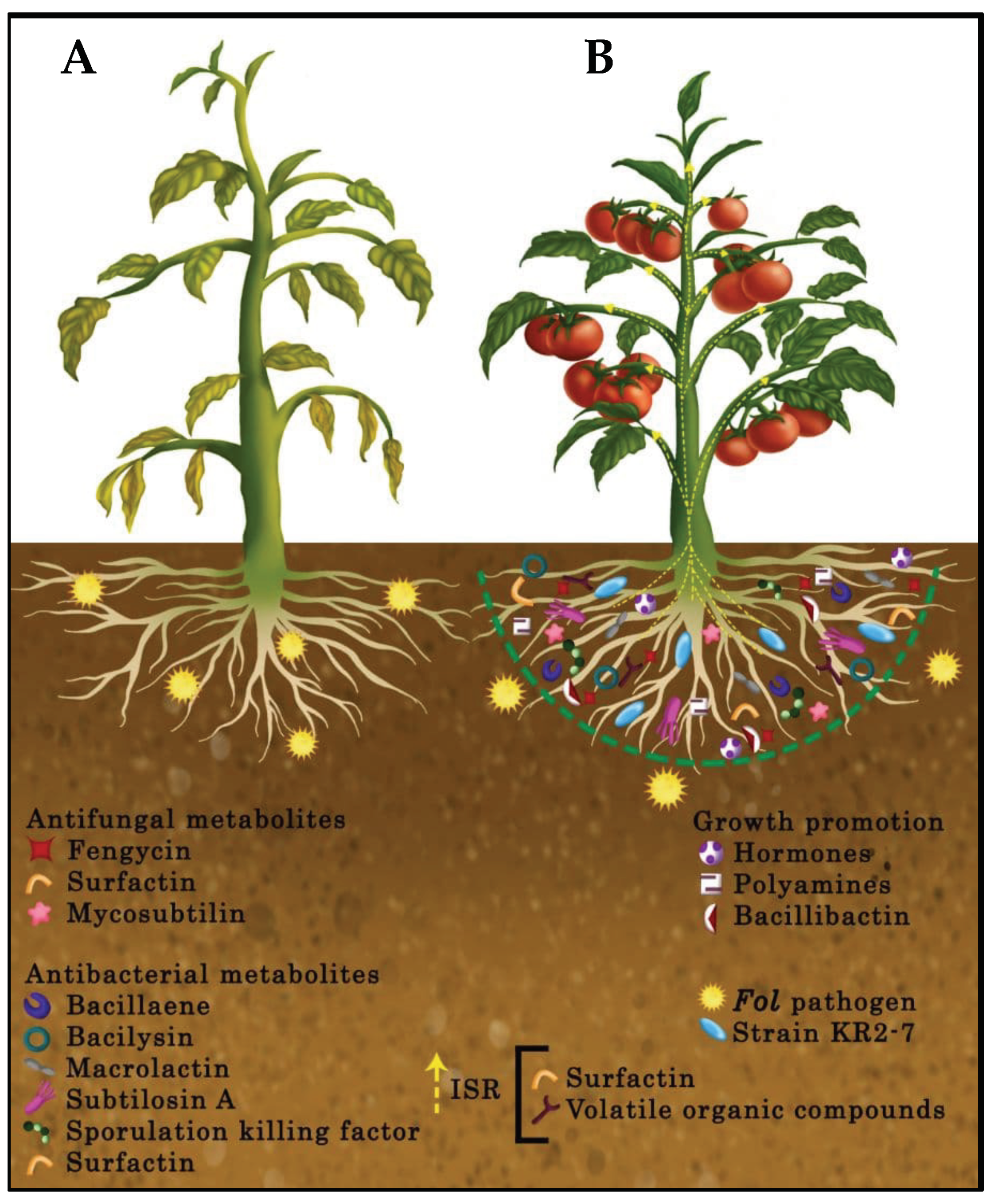
3.4. Antifungal Secondary Metabolites Production in Strain KR2-7
3.5. Antibacterial Secondary Metabolites Production in Strain KR2-7
3.6. Plant Colonization by Strain KR2-7
3.7. Genes Involved in Bacterium-Plant Interactions
4. Discussion
5. Conclusions
- (1)
- Inhibition of the pathogen growth through the diffusion of antifungal and antibacterial secondary metabolites and biofilm formation;
- (2)
- Stimulation of ISR in tomato via the production of surfactin and volatile organic compounds;
- (3)
- Promotion of plant health and growth by producing plant growth promotion hormones and polyamines, supplying iron for tomato, depriving the pathogen of iron, and relieving heavy metal stress in the soil as a result of siderophore bacillibactin activity;
- (4)
- Efficient colonization of plant roots.
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Agrios, G.N. Plant Pathology, 5th ed.; Elsevier Academic Press: London, UK, 2005; pp. 522–534. [Google Scholar]
- Borrero, C.; Ordovas, J.; Trillas, M.I.; Aviles, M. Tomato Fusarium wilt suppressiveness. The relationship between the organic plant growth media and their microbial communities as characterised by Biolog. Soil. Biol. Biochem. 2006, 38, 1631–1637. [Google Scholar] [CrossRef]
- Elmer, W.H. Effects of acibenzolar-S-methyl on the suppression of Fusarium wilt of cyclamen. Crop Prot. 2006, 25, 671–676. [Google Scholar] [CrossRef]
- L’Haridon, F.; Aimé, S.; Duplessis, S.; Alabouvette, C.; Steinberg, C.; Olivain, C. Isolation of differentially expressed genes during interactions between tomato cells and a protective or a non-protective strain of Fusarium oxysporum. Physiol. Mol. Plant Pathol. 2011, 76, 9–19. [Google Scholar] [CrossRef]
- Ajilogba, C.F.; Babalola, O.O. Integrated Management Strategies for Tomato Fusarium Wilt. Biocontrol. Sci. 2013, 18, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arie, T. Fusarium diseases of cultivated plants, control, diagnosis, and molecular and genetic studies. J. Pestic. Sci. 2019, 44, 275–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cawoy, H.; Bettiol, W.; Fickers, P.; Ongena, M. Bacillus-based biological control of plant diseases. In Pesticides in the Modern World-Pesticides Use and Management, 1st ed.; Stoytcheva, M., Ed.; IntechOpen Press: London, UK, 2011; pp. 273–302. Available online: https://www.intechopen.com/chapters/21989 (accessed on 21 May 2021).
- Raaijmakers, J.M.; Mazzola, M. Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu. Rev. Phytopathol. 2012, 50, 403–424. [Google Scholar] [CrossRef]
- Kohl, J.; Kolnaar, R.; Ravensberg, W.J. Mode of action of microbial biological control agents against plant diseases: Relevance beyond efficacy. Front. Plant Sci. 2019, 10, 845. Available online: https://www.frontiersin.org/article/10.3389/fpls.2019.00845 (accessed on 21 May 2021). [CrossRef] [Green Version]
- Conrath, U.; Beckers, G.J.M.; Langenbach, C.J.G.; Jaskiewicz, M.R. Priming for enhanced defense. Annu. Rev. Phytopathol. 2015, 53, 97–119. [Google Scholar] [CrossRef] [PubMed]
- Spadaro, D.; Droby, S. Development of biocontrol products for postharvest diseases of fruit: The importance of elucidating the mechanisms of action of yeast antagonists. Trends Food Sci. Technol. 2016, 47, 39–49. [Google Scholar] [CrossRef]
- Paterson, J.; Jahanshah, G.; Li, Y.; Wang, Q.; Mehnaz, S.; Gross, H. The contribution of genome mining strategies to the understanding of active principles of PGPR strains. FEMS Microbiol. Ecol. 2017, 93, fiw249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, X.; Koumoutsi, A.; Scholz, R.; Eisenreich, A.; Schneider, K.; Heinemeyer, I.; Morgenstern, B.; Voss, B.; Hess, W.R.; Reva, O.; et al. Comparative analysis of the complete genome sequence of the plant growth-promoting bacterium Bacillus amyloliquefaciens FZB42. Nat. Biotechnol. 2007, 25, 1007–1014. [Google Scholar] [CrossRef] [Green Version]
- Grady, E.N.; MacDonald, J.; Ho, M.T.; Weselowski, B.; McDowell, T.; Solomon, O.; Renaud, J.; Yuan, Z.C. Characterization and complete genome analysis of the surfactin-producing, plant-protecting bacterium Bacillus velezensis 9D-6. BMC Microbiol. 2019, 19, 5. [Google Scholar] [CrossRef]
- Knight, C.A.; Bowman, M.J.; Frederick, L.; Day, A.; Lee, C.; Dunlap, C.A. The first report of antifungal lipopeptide production by a Bacillus subtilis subsp. inaquosorum strain. Microbiol. Res. 2018, 216, 40–46. [Google Scholar] [CrossRef] [PubMed]
- Kamali, M.; Ahmadi, J.; Naeimi, S.; Guo, D. Characterization of Bacillus isolates from the rhizosphere of tomato suppressing Fusarium wilt disease. Acta Phytopathol. Entomol. Hung. 2019, 54, 53–68. [Google Scholar] [CrossRef]
- Skidmore, A.M.; Dickinson, C.H. Colony interactions and hyphal interference between Septoria Nodorumand phylloplane fungi. Trans. Brit. Mycol. Soc. 1976, 66, 57–64. [Google Scholar] [CrossRef]
- Vater, J.; Gao, X.; Hitzeroth, G.; Wilde, C.; Franke, P. “Whole cell”-matrix assisted laser desorption ionization-time of flight mass spectrometry, an emerging technique for efficient screening of biocombinatorial libraries of natural compounds- present state of research. Comb. Chem. High Throughput Screen. 2003, 6, 557–567. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Fedorova, N.D.; Jackson, J.D.; Jacobs, A.R.; Kiryutin, B.; Koonin, E.V.; Krylov, D.M.; Mazumder, R.; Mekhedov, S.L.; Nikolskaya, A.N.; et al. The COG database: An updated version includes eukaryotes. BMC Bioinform. 2003, 4, 41. [Google Scholar] [CrossRef] [Green Version]
- Bertels, F.; Silander, O.K.; Pachkov, M.; Rainey, P.B.; van Nimwegen, E. Automated reconstruction of whole-genome phylogenies from short- sequence reads. Mol. Biol. Evol. 2014, 31, 1077–1088. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 1981, 17, 368–376. [Google Scholar] [CrossRef]
- Tavare, S. Some probabilistic and statistical problems in the analysis of DNA sequences. Am. Math. Soc. 1986, 17, 57–86. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA and whole genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Goris, J.; Konstantinidis, K.T.; Klappenbach, J.A.; Coenye, T.; Vandamme, P.; Tiedje, J.M. DNA-DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 2007, 57, 81–91. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Richter, M.; Rosselló-Móra, R. Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. USA 2009, 106, 19126–19131. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meier-Kolthoff, J.P.; Auch, A.F.; Klenk, H.P.; Göker, M. Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinf. 2013, 14, 60. [Google Scholar] [CrossRef] [Green Version]
- Medema, M.H.; Blin, K.; Cimermancic, P.; de Jager, V.; Zakrzewski, P.; Fischbach, M.A.; Weber, T.; Takano, E.; Breitling, R. antiSMASH: Rapid identification, annotation, and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Res. 2011, 39, W339–W346. [Google Scholar] [CrossRef] [PubMed]
- Weber, T.; Blin, K.; Duddela, S.; Krug, D.; Kim, H.U.; Bruccoleri, R.; Lee, S.Y.; Fischbach, M.A.; Müller, R.; Wohlleben, W.; et al. antiSMASH 3.0- a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res. 2015, 43, W237–W243. [Google Scholar] [CrossRef] [Green Version]
- Dunlap, C.A.; Kim, S.J.; Kwon, S.W.; Rooney, A.P. Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp. plantarum and “Bacillus oryzicola” are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 2016, 66, 1212–1217. [Google Scholar] [CrossRef]
- Dunlap, C.A.; Bowman, M.J.; Rooney, A.P. Iturinic Lipopeptide diversity in the Bacillus subtilis species group–important antifungals for plant disease biocontrol applications. Front. Microbiol. 2019, 10, 1794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belbahri, L.; Chenari Bouket, A.; Rekik, I.; Alenezi, F.N.; Vallat, A.; Luptakova, L.; Petrovova, E.; Oszako, T.; Cherrad, S.; Vacher, S.; et al. Comparative genomics of Bacillus amyloliquefaciens strains reveals a core genome with traits for habitat adaptation and a secondary metabolites rich accessory genome. Front. Microbiol. 2017, 8, 1438. [Google Scholar] [CrossRef]
- Stein, T. Bacillus subtilis antibiotics: Structures, syntheses and specific functions. Mol. Microbiol. 2005, 56, 845–857. [Google Scholar] [CrossRef] [PubMed]
- Schneider, K.; Chen, X.; Vater, J.; Franke, P.; Nicholson, G.; Borriss, R.; Süssmuth, R.D. Macrolactin is the polyketide biosynthesis product of the pks2 cluster of Bacillus amyloliquefaciens FZB42. J. Nat. Prod. 2007, 70, 1417–1423. [Google Scholar] [CrossRef] [PubMed]
- Ongena, M.; Henry, G.; Thornart, P. The roles of cyclic lipopeptides in the biocontrol activity of Bacillus subtilis. In Recent Developments in Management of Plant Diseases, 1st ed.; Gisi, U., Chet, I., Gullino, M., Eds.; Springer Press: Dordrecht, The Netherlands, 2009; Volume 1, pp. 59–69. [Google Scholar] [CrossRef]
- Chen, L.; Heng, J.; Qin, S.; Bian, K. A comprehensive understanding of the biocontrol potential of Bacillus velezensis LM2303 against Fusarium head blight. PLoS ONE 2018, 13, e0198560. [Google Scholar] [CrossRef] [Green Version]
- Ongena, M.; Jacques, P. Bacillus lipopeptides: Versatile weapons for plant disease biocontrol. Trends Microbiol. 2008, 16, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Koumoutsi, A.; Chen, X.H.; Henne, A.; Liesegang, H.; Hitzeroth, G.; Franke, P.; Vater, J.; Borriss, R. Structural and functional characterization of gene clusters directing nonribosomal synthesis of bioactive cyclic lipopeptides in Bacillus amyloliquefaciens strain FZB42. J. Bacteriol. 2004, 186, 1084–1096. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimkic, I.; Stankovic, S.; Nišavic, M.; Petkovic, M.; Ristivojevic, P.; Fira, D.; Beric, T. The profile and antimicrobial activity of Bacillus lipopeptide extracts of five potential biocontrol strains. Front. Microbiol. 2017, 8, 925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mhammedi, A.; Peypoux, F.; Besson, F.; Michel, G. Bacillomycin f, a new antibiotic of iturin group: Isolation and characterization. J. Antibiot. 1982, 35, 306–311. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phae, C.G.; Shoda, M.; Kubota, H. Suppressive effect of Bacillus subtilis and its products on phytopathogenic microorganisms. J. Ferment. Bioeng. 1990, 69, 1–7. [Google Scholar] [CrossRef]
- Moyne, A.L.; Shelby, R.; Cleveland, T.E.; Tuzun, S. Bacillomycin D: An iturin with antifungal activity against Aspergillus flavus. J. Appl. Microbiol. 2001, 90, 622–629. [Google Scholar] [CrossRef]
- Yu, G.Y.; Sinclair, J.B.; Hartman, G.L.; Bertagnolli, B.L. Production of iturin A by Bacillus amyloliquefaciens suppressing Rhizoctonia solani. Soil Biol. Biochem. 2002, 34, 955–963. [Google Scholar] [CrossRef]
- Aranda, F.J.; Teruel, J.A.; Ortiz, A. Further aspects on the haemolytic activity of the antibiotic lipopeptide iturin A. Biochim. Biophys. Acta 2005, 17, 51–56. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Besson, F.; Michel, G. Action of mycosubtilin, an antifungal antibiotic of Bacillus subtilis, on the cell membrane of Saccharomyces cerevisiae. Microbios 1989, 59, 113–121. [Google Scholar]
- Garcia-Gutierrez, L.; Zeriouh, H.; Romero, D.; Cubero, J.; Vicente, A.; Perez-Garcia, A. The antagonistic strain Bacillus subtilis UMAF6639 also confers protection to melon plants against cucurbit powdery mildew by activation of jasmonate-and salicylic acid-dependent defence responses. Microb. Biotechnol. 2013, 6, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henry, G.; Deleu, M.; Jourdan, E.; Thonart, P.; Ongena, M. The bacterial lipopeptide surfactin targets the lipid fraction of the plant plasma membrane to trigger immune-related defence responses. Cell Microbiol. 2011, 13, 1824–1837. [Google Scholar] [CrossRef] [PubMed]
- Khong, N.G.; Randoux, B.; Tayeh, C.; Coutte, F.; Bourdon, N.; Tisserant, B.; Laruelle, F.; Jacques, P.; Reignault, P. Induction of resistance in wheat against powdery mildew by bacterial cyclic lipopeptides. Commun. Agric. Appl. Biol. Sci. 2012, 77, 39–51. [Google Scholar] [PubMed]
- Waewthongrak, W.; Leelasuphakul, W.; McCollum, G. Cyclic LIPopeptides from Bacillus subtilis ABS-S14 elicit defense-related gene expression in citrus fruit. PLoS ONE 2014, 9, e109386. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Hartmann, A.; Gao, X.; Borriss, R. Biocontrol mechanism by root-associated Bacillus amyloliquefaciens FZB42-a review. Front. Microbiol. 2015, 6, 780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Héloir, M.C.; Zhang, X.; Geissler, M.; Trouvelot, S.; Jacquens, L.; Henkel, M.; Su, X.; Fang, W.; Wang, Q.; et al. Surfactin and fengycin contribute to the protection of a Bacillus subtilis strain against grape downy mildew by both direct effect and defence stimulation. Mol. Plant Pathol. 2019, 20, 1037–1050. [Google Scholar] [CrossRef] [Green Version]
- Sarwar, A.; Hassan, M.N.; Imran, M.; Iqbal, M.; Majeed, S.; Brader, G.; Sessitsch, A.; Hafeez, F.Y. Biocontrol activity of surfactin A purified from Bacillus NH-100 and NH-217 against rice bakanae disease. Microbiol. Res. 2018, 209, 1–13. [Google Scholar] [CrossRef]
- Wu, S.; Liu, G.; Zhou, S.; Sha, Z.; Sun, C. Characterization of antifungal lipopeptide biosurfactants produced by marine bacterium Bacillus sp. CS30. Mar. Drugs 2019, 17, 199. [Google Scholar] [CrossRef] [Green Version]
- Krishnan, N.; Velramar, B.; Velu, R.K. Investigation of antifungal activity of surfactin against mycotoxigenic phytopathogenic fungus Fusarium moniliforme and its impact in seed germination and mycotoxicosis. Pestic. Biochem. Physiol. 2019, 155, 101–107. [Google Scholar] [CrossRef] [PubMed]
- Hazarika, D.J.; Goswami, G.; Gautom, T.; Parveen, A.; Das, P.; Barooah, M.; Boro, R.C. Lipopeptide mediated biocontrol activity of endophytic Bacillus subtilis against fungal phytopathogens. BMC Microbiol. 2019, 19, 71. [Google Scholar] [CrossRef]
- Fan, H.; Zhang, Z.; Li, Y.; Zhang, X.; Duan, Y.; Wang, Q. Biocontrol of bacterial fruit blotch by Bacillus subtilis 9407 via surfactin-mediated antibacterial activity and colonization. Front. Microbiol. 2017, 8, 1973. [Google Scholar] [CrossRef] [PubMed]
- Xiong, H.Q.; Li, Y.T.; Cai, Y.F.; Cao, Y.; Wang, Y. Isolation of Bacillus amyloliquefaciens JK6 and identification of its lipopeptides surfactin for suppressing tomato bacterial wilt. RSC Adv. 2015, 5, 82042–82049. [Google Scholar] [CrossRef]
- Baise, H.P.; Fall, R.; Vivanco, J.M. Biocontrol of Bacillus subtilis against infection of Arabidopsis roots by Pseudomonas syringae is facilitated by biofilm formation and surfactin production. Plant Physiol. 2004, 134, 307–319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernandes, P.; Arruda, I.R.; Santos, A.; Araújo, A.A.; Maior, A.M.; Ximenes, E. Antimicrobial activity of surfactants produced by Bacillus subtilis r14 against multidrug-resistant bacteria. Braz. J. Microbiol. 2007, 38, 704–709. [Google Scholar] [CrossRef] [Green Version]
- Patel, P.S.; Huang, S.; Fisher, S.; Pirnik, D.; Aklonis, C.; Dean, L.; Meyers, E.; Fernandes, P.; Mayerl, F. Bacillaene, a novel inhibitor of procaryotic protein synthesis produced by Bacillus subtilis: Production, taxonomy, isolation, physico-chemical characterization and biological activity. J. Antibiot. 1995, 48, 997–1003. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Um, S.; Fraimout, A.; Sapountzis, P.; Oh, D.C.; Poulsen, M. The fungus-growing termite Macrotermes natalensis harbors bacillaene-producing Bacillus sp. that inhibit potentially antagonistic fungi. Sci. Rep. 2013, 3, 3250. [Google Scholar] [CrossRef] [PubMed]
- Muller, S.; Strack, S.N.; Hoefler, B.C.; Straight, P.; Kearns, D.B.; Kirby, J.R. Bacillaene and sporulation protect Bacillus subtilis from predation by Myxococcus xanthus. Appl. Environ. Microbiol. 2014, 80, 5603–5610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Romero-Tabarez, M.; Jansen, R.; Sylla, M.; Lunsdorf, H.; Haussler, S.; Santosa, D.A.; Timmis, K.N.; Molinari, G. 7-O-malonyl macrolactin A, a new macrolactin antibiotic from Bacillus subtilis active against methicillin-resistant Staphylococcus aureus, vancomycin-resistant enterococci, and a small-colony variant of Burkholderia cepacia. Antimicrob. Agents Chemother. 2006, 50, 1701–1709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, J.; Li, B.; Zhang, N.; Waseem, R.; Shen, Q.; Huang, Q. Production of bacillomycin- and macrolactin-type antibiotics by Bacillus amyloliquefaciens NJN-6 for suppressing soilborne plant pathogens. J. Agric. Food Chem. 2012, 60, 2976–2981. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.H.; Scholz, R.; Borriss, M.; Junge, H.; Mogel, G.; Kunz, S.; Borriss, R. Difficidin and bacilysin produced by plant-associated Bacillus amyloliquefaciens are efficient in controlling fire blight disease. J. Biotechnol. 2009, 140, 38–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, L.; Wu, H.; Chen, L.; Yu, X.; Borriss, R.; Gao, X. Difficidin and bacilysin from Bacillus amyloliquefaciens FZB42 have antibacterial activity against Xanthomonas oryzae rice. Sci. Rep. 2015, 5, 12975. [Google Scholar] [CrossRef] [PubMed]
- Wu, L.; Wu, H.; Chen, L.; Lin, L.; Borriss, R.; Gao, X. Bacilysin overproduction in Bacillus amyloliquefaciens FZB42 markerless derivative strains FZBREP and FZBSPA enhances antibacterial activity. Appl. Microbiol. Biotechnol. 2015, 99, 4255–4263. [Google Scholar] [CrossRef] [PubMed]
- Steinborn, G.; Hajirezaei, M.R.; Hofemeister, J. bac genes for recombinant bacilysin and anticapsin production in Bacillus host strains. Arch. Microbiol. 2005, 183, 71–79. [Google Scholar] [CrossRef]
- Mahlstedt, S.A.; Walsh, C.T. Investigation of anticapsin biosynthesis reveals a four-enzyme pathway to tetrahydrotyrosine in Bacillus subtilis. Biochemistry 2010, 49, 912–923. [Google Scholar] [CrossRef] [Green Version]
- Babasaki, K.; Takao, T.; Shimonishi, Y.; Kurahashi, K. Subtilosin A, a new antibiotic peptide produced by Bacillus subtilis 168: Isolation, structural analysis, and biogenesis. J. Biochem. 1985, 98, 585–603. [Google Scholar] [CrossRef] [PubMed]
- Stein, T.; Düsterhus, S.; Stroh, A.; Entian, K.D. Subtilosin production by two Bacillus subtilis subspecies and variance of the sbo-alb cluster. Appl. Environ. Microbiol. 2004, 70, 2349–2353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sutyak, K.E.; Wirawan, R.E.; Aroutcheva, A.A.; Chikindas, M.L. Isolation of the Bacillus subtilis antimicrobial peptide subtilosin from the dairy product-derived Bacillus amyloliquefaciens. J. Appl. Microbiol. 2008, 104, 1067–1074. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shelburne, C.E.; An, F.Y.; Dholpe, V.; Ramamoorthy, A.; Lopatin, D.E.; Lantz, M.S. The spectrum of antimicrobial activity of the 18 bacteriocin subtilosin A. J. Antimicrob. Chemother. 2007, 59, 297–300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiedemann, I.; Breukink, E.; Kraaij, C.V.; Kuipers, O.S.; Bierbaum, G.; de Kruijff, B.; Sahl, H.G. Specific binding of nisin to the peptidoglycan precursor lipid II combines pore formation and inhibition of cell wall biosynthesis for potent antibiotic activity. J. Biol. Chem. 2001, 276, 1772–1779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thennarasu, S.; Lee, D.K.; Poon, A.; Kawulka, K.E.; Vederas, J.C.; Ramamoorthy, A. Membrane permeabilization, orientation, and antimicrobial mechanism of subtilosin A. Chem. Phys. Lipids 2005, 137, 38–51. [Google Scholar] [CrossRef] [PubMed]
- Zheng, G.; Hehn, R.; Zuber, P. Mutational analysis of sbo-alb locus of Bacillus subtilis: Identification of genes required for subtilosin production and immunity. J. Bacteriol. 2000, 182, 3266–3273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Pastor, J.E.; Hobbs, E.C.; Losick, R. Cannibalism by sporulating bacteria. Science 2003, 301, 510–513. [Google Scholar] [CrossRef] [PubMed]
- González-Pastor, J.E. Cannibalism: A social behavior in sporulating Bacillus subtilis. FEMS Microbiol. Rev. 2011, 35, 415–424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lin, D.; Qu, L.J.; Gu, H.; Chen, Z. A 3.1-kb genomic fragment of Bacillus subtilis encodes the protein inhibiting growth of Xanthomonas oryzae pv. oryzae. J. Appl. Microbiol. 2001, 91, 1044–1050. [Google Scholar] [CrossRef] [Green Version]
- Guo, S.; Li, X.; He, P.; Ho, H.; Wu, W.; He, Y. Whole-genome sequencing of Bacillus subtilis XF-1 reveals mechanisms for biological control and multiple beneficial properties in plants. J. Ind. Microbiol. Biotechnol. 2015, 42, 925–937. [Google Scholar] [CrossRef]
- Peypoux, F.; Bonmatin, J.M.; Wallach, J. Recent trends in the biochemistry of surfactin. Appl. Microbiol. Biotechnol. 1999, 51, 553–563. [Google Scholar] [CrossRef]
- Bonmatin, J.M.; Laprévote, O.; Peypoux, F. Diversity among microbial cyclic lipopeptides: Iturins and surfactins. Activity-structure relationships to design new bioactive agents. Comb. Chem. High Throughput Screen. 2003, 6, 541–556. [Google Scholar] [CrossRef]
- Zeriouh, H.; de Vicente, A.; Pérez-García, A.; Romero, D. Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 2013, 16, 2196–2211. [Google Scholar] [CrossRef]
- Wipat, A.; Harwood, C.R. The Bacillus subtilis genome sequence: The molecular blueprint of a soil bacterium. FEMS Microbiol. Ecol. 1999, 28, 1–9. [Google Scholar] [CrossRef]
- Bezzate, S.; Aymerich, S.; Chambert, R.; Czarnes, S.; Berge, O.; Heulin, T. Disruption of the Paenibacillus polymyxa levansucrase gene impairs its ability to aggregate soil in the wheat rhizosphere. Environ. Microbiol. 2000, 2, 333–342. [Google Scholar] [CrossRef] [PubMed]
- Kearns, D.B.; Chu, F.; Rudner, R.; Losick, R. Genes governing swarming in Bacillus subtilis and evidence for a phase variation mechanism controlling surface motility. Mol. Microbiol. 2004, 52, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Calvio, C.; Celandroni, F.; Ghelardi, E.; Amati, G.; Salvetti, S.; Ceciliani, F.; Galizzi, A.; Senesi, S. Swarming differentiation and swimming motility in Bacillus subtilis are controlled by swrA, a newly identified dicistronic operon. J. Bacteriol. 2005, 187, 5356–5366. [Google Scholar] [CrossRef] [Green Version]
- Tsuge, K.; Ohata, Y.; Shoda, M. Gene yerP, involved in surfactin self resistance in Bacillus subtilis. Antimicrob. Agents Chemother. 2001, 45, 3566–3573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, Y.; Yan, F.; Chen, Y.; Jin, C.; Guo, J.H.; Chai, Y. Poly-γ-glutamic acids contribute to biofilm formation and plant root colonization in selected environmental isolates of Bacillus subtilis. Front. Microbiol. 2016, 7, 1811. [Google Scholar] [CrossRef] [Green Version]
- Lazarevic, V.; Soldo, B.; Médico, N.; Pooley, H.; Bron, S.; Karamata, D. Bacillus subtilis alpha-Phosphoglucomutase is required for normal cell morphology and biofilm formation. Appl. Environ. Microbiol. 2005, 71, 39–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Branda, S.S.; González-Pastor, J.E.; Dervyn, E.; Ehrlich, S.D.; Losick, R.; Kolter, R. Genes involved in formation of structural multicellular communities by Bacillus subtilis. J. Bacteriol. 2004, 186, 3970–3979. [Google Scholar] [CrossRef] [Green Version]
- Kearns, D.B.; Chu, F.; Branda, S.S.; Kolter, R.; Losick, R. A master regulator for biofilm formation by Bacillus subtilis. Mol. Microbiol. 2005, 55, 739–749. [Google Scholar] [CrossRef]
- Branda, S.S.; González-Pastor, J.E.; Ben-Yehuda, S.; Losick, R.; Kolter, R. Fruiting body formation by Bacillus subtilis. Proc. Natl. Acad. Sci. USA 2001, 98, 11621–11626. [Google Scholar] [CrossRef] [Green Version]
- Chu, F.; Kearns, D.B.; McLoon, A.; Chai, Y.; Kolter, R.; Losick, R. A novel regulatory protein governing biofilm formation in Bacillus subtilis. Mol. Microbiol. 2008, 68, 1117–1127. [Google Scholar] [CrossRef] [Green Version]
- Chu, F.; Kearns, D.B.; Branda, S.S.; Kolter, R.; Losick, R. Targets of the master regulator of biofilm formation in Bacillus subtilis. Mol. Microbiol. 2006, 59, 1216–1228. [Google Scholar] [CrossRef]
- Harman, G.E. Multifunctional fungal plant symbionts: New tools to enhance plant growth and productivity. New Phytol. 2011, 189, 647–649. [Google Scholar] [CrossRef]
- Shafi, J.; Tian, H.; Ji, M. Bacillus species as versatile weapons for plant pathogens: A review. Biotechnol. Biotechnol. Equip. 2017, 31, 446–459. [Google Scholar] [CrossRef] [Green Version]
- Holtmann, G.; Bakker, E.P.; Uozumi, N.; Bremer, E. KtrAB and KtrCD: Two K+ uptake systems in Bacillus subtilis and their role in adaptation to hypertonicity. J. Bacteriol. 2003, 185, 1289–1298. [Google Scholar] [CrossRef] [Green Version]
- Miethke, M.; Klotz, O.; Linne, U.; May, J.J.; Beckering, C.L.; Marahiel, M.A. Ferri-bacillibactin uptake and hydrolysis in Bacillus subtilis. Mol. Microbiol. 2006, 61, 1413–1427. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.; Tian, Z.; Luo, Y.; Cheng, Y.; Long, C. Antagonistic Activity and the Mechanism of Bacillus amyloliquefaciens DH-4 Against Citrus Green Mold. Phytopathology 2018, 108, 1253–1262. [Google Scholar] [CrossRef] [Green Version]
- Rajkumar, M.; Ae, N.; Prasad, M.N.; Freitas, H. Potential of siderophore-producing bacteria for improving heavy metal phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Radzki, W.; Gutierrez Manero, F.J.; Algar, E.; Lucas Garcıa, J.A.; Garcıa-Villaraco, A.; Ramos Solano, B. Bacterial siderophores efficiently provide iron to iron-starved tomato plants in hydroponics culture. Antonie Leeuwenhoek 2013, 104, 321–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lurthy, T.; Cantat, C.; Jeudy, C.; Declerck, P.; Gallardo, K.; Barraud, C.; Leroy, F.; Ourry, A.; Lemanceau, P.; Salon, C.; et al. Impact of bacterial siderophores on iron status and ionome in pea. Front. Plant Sci. 2020, 11, 730. [Google Scholar] [CrossRef]
- Arguelles-Arias, A.; Ongena, M.; Halimi, B.; Lara, Y.; Brans, A.; Joris, B.; Fickers, P. Bacillus amyloliquefaciens GA1 as a source of potent antibiotics and other secondary metabolites for biocontrol of plant pathogens. Microb. Cell Fact. 2009, 8, 63. [Google Scholar] [CrossRef] [Green Version]
- Yu, X.; Ai, C.; Xin, L.; Zhou, G. The siderophore-producing bacterium, Bacillus subtilis CAS15, has a biocontrol effect on Fusarium wilt and promotes the growth of pepper. Eur. J. Soil Biol. 2011, 47, 138–145. [Google Scholar] [CrossRef]
- Woo, S.M.; Kim, S.D. Structural identification of siderophore (AH18) from Bacillus subtilis AH18, a biocontrol agent of phytophthora blight disease in red-pepper. Kor. J. Microbiol. Biotechnol. 2008, 36, 326–335. [Google Scholar] [CrossRef]
- Tahir, H.A.; Gu, Q.; Wu, H.; Raza, W.; Hanif, A.; Wu, L.; Colman, M.V.; Gao, X. Plant Growth Promotion by Volatile Organic Compounds Produced by Bacillus subtilis SYST2. Front. Microbiol. 2017, 8, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.; Farag, M.A.; Park, H.B.; Kloepper, W.J.; Lee, S.H.; Ryu, C.M. Induced resistance by a long-chain bacterial volatile: Elicitation of plant systemic defense by a C13 volatile produced by Paenibacillus polymyxa. PLoS ONE 2012, 7, e48744. [Google Scholar] [CrossRef]
- Park, Y.S.; Dutta, S.; Ann, M.; Raaijmakers, J.M.; Park, K. Promotion of plant growth by Pseudomonas fluorescens strain SS101 via novel volatile organic compounds. Biochem. Biophys. Res. Commun. 2015, 461, 361–365. [Google Scholar] [CrossRef] [PubMed]
- Tahir, H.A.; Gu, Q.; Wu, H.; Niu, Y.; Huo, R.; Gao, X. Bacillus volatiles adversely affect the physiology and ultra-structure of Ralstonia solanacearum and induce systemic resistance in tobacco against bacterial wilt. Sci. Rep. 2017, 7, 40481. [Google Scholar] [CrossRef]
- Zhang, X.; Zhang, R.; Bao, T.; Yang, T.; Xu, M.; Li, H.; Xu, Z.; Rao, Z. Moderate expression of the transcriptional regulator ALsR enhances acetoin production by Bacillus subtilis. J. Ind. Microbiol. Biotechnol. 2013, 40, 1067–1076. [Google Scholar] [CrossRef]
- Goupil-feuillerat, N.; Cocaign-Bousquet, M.; Godon, J.J.; Ehrlich, S.D.; Renault, P. Dual Role of α—Acetolactate Decarboxylase in Lactococcus lactis subsp. Lactis. J. Bacteriol. 1997, 179, 6285–6293. [Google Scholar] [CrossRef] [Green Version]
- Zhou, C.; Ma, Z.; Zhu, L.; Xiao, X.; Xie, Y.; Zhu, J.; Wang, J. Rhizobacterial strain Bacillus megaterium BOFC15 induces cellular polyamine changes that improve plant growth and drought resistance. Int. J. Mol. Sci. 2016, 17, 976. [Google Scholar] [CrossRef] [PubMed]
- Rooney, A.P.; Price, N.P.J.; Ehrhardt, C.; Sewzey, J.L.; Bannan, J.D. Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int. J. Syst. Evol. Microbiol. 2009, 59, 2429–2436. [Google Scholar] [CrossRef]
- Yi, H.; Chun, J.; Cha, C.J. Genomic insights into the taxonomic status of the three subspecies of Bacillus subtilis. Syst. Appl. Microbiol. 2014, 37, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, L.K.; Roberts, M.S.; Cohan, F.M. Relationship of Bacillus subtilis clades associated with strains 168 and W23: A proposal for Bacillus subtilis subsp. subtilis subsp. nov. and Bacillus subtilis subsp. spizizenii subsp. nov. Int. J. Syst. Bacteriol. 1999, 49, 1211–1215. [Google Scholar] [CrossRef]
- Brito, P.H.; Chevreux, B.; Serra, C.R.; Schyns, G.; Henriques, A.O.; Pereira-Leal, J.B. Genetic competence drives genome diversity in Bacillus subtilis. Genome Biol. Evol. 2018, 10, 108–124. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dunlap, C.A.; Bowman, M.J.; Zeigler, D.I. Promotion of Bacillus subtilis subsp. inaquosorum, Bacillus subtilis subsp. spizizenii and Bacillus subtilis subsp. stercoris to species status. Antonie Leeuwenhoek 2020, 113, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Wang, Y.; Li, J.; Shen, Q.; Zhang, R. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 2012, 97, 8823–8830. [Google Scholar] [CrossRef] [PubMed]
- Chowdhury, S.P.; Dietel, K.; Rändler, M.; Schmid, M.; Junge, H.; Borriss, R.; Hartmann, A.; Grosch, R. Effects of Bacillus amyloliquefaciens FZB42 on lettuce growth and health under pathogen pressure and its impact on the rhizosphere bacterial community. PLoS ONE 2013, 8, e68818. [Google Scholar] [CrossRef] [Green Version]
- Zhu, M.L.; Wu, X.Q.; Wang, Y.H.; Dai, Y. Role of biofilm formation by Bacillus pumilus HR10 in biocontrol against pine seedling damping-off disease caused by Rhizoctonia solani. Forests 2020, 11, 652. [Google Scholar] [CrossRef]
- Steller, S.; Vater, J. Purification of the fengycin synthetase multienzyme system from Bacillus subtilis b213. J. Chromatogr. B Biomed. Sci. Appl. 2000, 737, 267–275. [Google Scholar] [CrossRef]
- Deleu, M.; Paquot, M.; Nylander, T. Fengycin interaction with lipid monolayers at the air-aqueous interface-implications for the effect of fengycin on biological membranes. J. Colloid Interface Sci. 2005, 283, 358–365. [Google Scholar] [CrossRef]
- Wang, J.; Liu, J.; Chen, H.; Yao, J. Characterization of Fusarium graminearum inhibitory lipopeptide from Bacillus subtilis IB. Appl. Microbiol. Biotechnol. 2007, 76, 889–894. [Google Scholar] [CrossRef] [PubMed]
- Hanif, A.; Zhang, F.; Li, P.; Li, C.; Xu, Y.; Zubair, M.; Zhang, M.; Jia, D.; Zhao, X.; Liang, J.; et al. Fengycin produced by Bacillus amyloliquefaciens FZB42 inhibits Fusarium graminearum growth and mycotoxins biosynthesis. Toxins 2019, 11, 295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ongena, M.; Jourdan, E.; Adam, A.; Paquot, M.; Brans, A.; Joris, B.; Arpigny, J.L.; Thonart, P. Surfactin and fengycin lipopeptides of Bacillus subtilis as elicitors of induced systemic resistance in plants. Environ. Microbiol. 2007, 9, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Xu, Z.; Ling, N.; Yuan, Y.; Yang, X.; Chen, L.; Shen, B.; Shen, Q. Isolation and identification of lipopeptides produced by B. subtilis SQR 9 for suppressing Fusarium wilt of cucumber. Sci. Hortic. 2012, 135, 32–39. [Google Scholar] [CrossRef]
- Yuan, J.; Raza, W.; Huang, Q.; Shen, Q. The ultrasound-assisted extraction and identification of antifungal substances from B. amyloliquefaciensstrain NJN-6 suppressing Fusarium oxysporum. J. Basic Microbiol. 2012, 52, 721–730. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Sun, C. Fengycins, cyclic lipopeptides from marine Bacillus subtilis strains, kill the plant-pathogenic fungus Magnaporthe grisea by inducing reactive oxygen species production and chromatin condensation. Appl. Environ. Microbiol. 2018, 84, e00445-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cao, Y.; Pi, H.; Chandrangsu, P.; Li, Y.; Wang, Y.; Zhou, H.; Xiong, H.; Helman, J.D.; Cai, Y. Antagonism of two plant-growth promoting Bacillus velezensis isolates against Ralstonia solanacearum and Fusarium oxysporum. Sci Rep. 2018, 8, 4360. [Google Scholar] [CrossRef]
- Singh, B.; Satyanarayana, T. Microbial phytases in phosphorus acquisition and plant growth promotion. Physiol. Mol. Biol. Plants 2011, 17, 93–103. [Google Scholar] [CrossRef] [Green Version]
- Idriss, E.E.; Makarewicz, O.; Farouk, A.; Rosner, K.; Greiner, R.; Bochow, H.; Richter, T.; Borriss, R. Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 2002, 148, 2097–2109. [Google Scholar] [CrossRef] [Green Version]
- Kumar, V.; Singh, P.; Jorquera, M.A.; Sangwan, P.; Kumar, P.; Verma, A.K.; Agrawal, S. Isolation of phytase-producing bacteria from Himalayan soils and their effect on growth and phosphorus uptake of Indian mustard (Brassica juncea). World J. Microbiol. Biotechnol. 2013, 29, 1361–1369. [Google Scholar] [CrossRef]
- Liu, L.; Li, A.; Chen, J.; Su, Y.; Li, Y.; Ma, S. Isolation of a phytase-producing bacterial strain from agricultural soil and its characterization and application as an effective eco-friendly phosphate solubilizing bioinoculant. Commun. Soil Sci. Plant 2018, 49, 984–994. [Google Scholar] [CrossRef]
- Orozco-Mosqueda, M.C.; Duan, J.; DiBernardo, M.; Zetter, E.; Campos-García, J.; Glick, B.R.; Santoyo, G. The production of ACC deaminase and trehalose by the plant growth promoting bacterium Pseudomonas sp. UW4 synergistically protect tomato plants against salt stress. Front. Microbiol. 2019, 10, 1392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duca, D.R.; Rose, D.R.; Glick, B.R. Indole acetic acid overproduction transformants of the rhizobacterium Pseudomonas sp. UW4. Antonie Leeuwenhoek 2018, 111, 1645–1660. [Google Scholar] [CrossRef] [PubMed]
- Moon, J.-H.; Won, S.-J.; Maung, C.E.H.; Choi, J.-H.; Choi, S.-I.; Ajuna, H.B.; Ahn, Y.S. Bacillus velezensis CE 100 Inhibits Root Rot Diseases (Phytophthora spp.) and Promotes Growth of Japanese Cypress (Chamaecyparis obtusa Endlicher) Seedlings. Microorganisms 2021, 9, 821. [Google Scholar] [CrossRef]





| Species | Strain | Accession Number | ANI (%) | GGDC |
|---|---|---|---|---|
| Bacillus altitudinis | P-10 | NZ_CP024204.1 | 71.39 | 0.2368 |
| Bacillus amyloliquefaciens | B15 | NZ_CP014783.1 | 77.31 | 0.2087 |
| CC178 | NC_022653.1 | 77.35 | 0.2082 | |
| FZB42 | NC_009725.1 | 77.65 | 0.2081 | |
| L-H15 | NZ_CP010556.1 | 77.38 | 0.2104 | |
| L-S60 | NZ_CP011278.1 | 77.37 | 0.2101 | |
| S499 | NZ_CP014700.1 | 77.36 | 0.2097 | |
| Y2 | NC_017912.1 | 77.43 | 0.2086 | |
| Bacillus atrophaeus | GQJK17 | NZ_CP022653.1 | 80.4 | 0.1895 |
| UCMB-5137 | NZ_CP011802.1 | 80.3 | 0.192 | |
| Bacillus cellulasensis | GLB197 | NZ_CP018574.1 | 71.45 | 0.2368 |
| Bacillus flexus | KLBMP 4941 | NZ_CP016790.1 | 68.97 | 0.1585 |
| Bacillus megaterium | YC4-R4 | NZ_CP026736.1 | 68.87 | 0.1619 |
| Bacillus muralis | G25-68 | NZ_CP017080.1 | 68.82 | 0.1484 |
| Bacillus mycoides | Gnyt1 | NZ_CP020743.1 | 68.46 | 0.1317 |
| Bacillus oceanisediminis | 2691 | NZ_CP015506.1 | 69.26 | 0.1506 |
| Bacillus paralicheniformis | MDJK30 | NZ_CP020352.1 | 73.09 | 0.2242 |
| Bacillus pumilus | SAFR-032 | NC_009848.4 | 71.3 | 0.2341 |
| Bacillus pumilus | TUAT1 | NZ_AP014928.1 | 71.2 | 0.2366 |
| Bacillus sp. | B25 (2016b) | CP016285.1 | 68.41 | 0.177 |
| WP8 | NZ_CP010075.1 | 71.11 | 0.2347 | |
| Bacillus subtilis | BSn5 | NC_014976.1 | 93.06 | 0.0706 |
| HJ5 | NZ_CP007173.1 | 93.1 | 0.0703 | |
| XF-1 | NC_020244.1 | 93.01 | 0.0707 | |
| Bacillus subtilis subsp. inaquosorum | KCTC 13429 | NZ_CP029465.1 | 99.26 | 0.0075 |
| Bacillus subtilis subsp. inaquosorum | DE111 | NZ_CP013984.1 | 98.8 | 0.01222 |
| Bacillus subtilis subsp. spizizenii | W23 | NC_014479.1 | 94.18 | 0.0585 |
| Bacillus subtilis subsp. subtilis | 168 | NC_000964.3 | 93.03 | 0.0707 |
| Bacillus thuringiensis subsp. kurstaki | HD-1 | NZ_CP004870.1 | 68.83 | 0.1575 |
| Bacillus vallismortis | NBIF-001 | NZ_CP020893.1 | 77.57 | 0.2081 |
| Bacillus velezensis | SQR9 | NZ_CP006890.1 | 77.38 | 0.2095 |
| Bacillus weihenstephanensis | KBAB4 | NC_010184.1 | 68.46 | 0.1386 |
| Metabolite | Synthetase Type | Gene Cluster | Function | Gene Similarity with Strain KR2-7 | |||
|---|---|---|---|---|---|---|---|
| B. inaquosorum KR2-7 | B. subtilis 168 | B. velezensis FZB42 | 168 | FZB42 | |||
| Bacillaene | PKS-NRPS | pksABCDE, acpK, pksFGHIJLMNRS | pksABCDE, acpk, pksFGHIJLMNRS | baeBCDE, acpK, baeGHIJLMNRS | Antibacterial | 89.63% | 75.25% |
| Bacillibactin | NRPS | dhbABCEF | dhbABCEF | dhbABCEF | Nutrient uptake | 92.25% | 73.07% |
| Bacilysin | NRPS | bacABCDE, ywfAG | bacABCDEFG, ywfA | bacABCDE, ywfAG | Antibacterial | 93.50% | 80.67% |
| Fengycin | NRPS | ppsABCDE | ppsABCDE | fenEABCD | Antifungal | 92.01% | 72.05% |
| Macrolactin | PKS | pksJL, baeE | - | pks2ABCDEFGHI | Antibacterial | - | 74.12% |
| Bacillomycin F | NRPS | ituD, ituABC | - | - | Antifungal, ISR | - | - |
| Sporulation killing factor | Head-to-tail cyclised peptide | skfABCEFGH | skfABCEFGH | - | Antibacterial | 96.08% | - |
| Subtilosin A | Thiopeptide | sboA, albABCDEFG | sboA, albABCDEFG | - | Antibacterial | 91.86% | - |
| Surfactin | NRPS | srfAA, AB, AC, AD | srfAA, AB, AC, AD | srfAA, AB, AC, AD | Antifungal, Antibacterial, Colonization, ISR | 92.20% | 74.65% |
| Mass Peak (m/z) | Assignment | Reference | |
|---|---|---|---|
| On PDA control | 1501.9 | Ala-6-C16 fengycin [M + H, Na, K]+ | [38] |
| 1515.9 | Ala-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1529.9 | Val-6-C16 fengycin [M + H, Na, K]+ | [38] | |
| 1543.8 | Val-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| On PDA dual culture | 1471.9 | Ala-6-C15 fengycin [M + H, Na, K]+ | [38] |
| 1485.7 | C16-Fengycin A [M + Na] | [39] | |
| 1487.9 | Ala-6-C15 fengycin [M + H, Na, K]+ | [38] | |
| 1499.9 | Ala-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1501.9 | Ala-6-C16 fengycin [M + H, Na, K]+ | [38] | |
| 1513.8 | C18-fengycin A [M + Na] | [39] | |
| 1515.9 | Ala-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1527.8 | Val-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1529.9 | Val-6-C16 fengycin [M + H, Na, K]+ | [38] | |
| 1543.8 | Val-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1501.9 | Ala-6-C16 fengycin [M + H, Na, K]+ | [38] | |
| 1515.9 | Ala-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1527.8 | Val-6-C16 fengycin [M + H, Na, K]+ | [38] | |
| 1529.9 | Val-6-C17 fengycin [M + H, Na, K]+ | [38] | |
| 1543.8 | Ala-6-C15 fengycin [M + H, Na, K]+ | [38] |
| Mass Peak (m/z) | Assignment | Reference | |
|---|---|---|---|
| On PDA control | 1106.6 | C17-iturin [M + Na]+ | [18] |
| 1122.6 | C17-iturin [M + K]+ | [18] | |
| 1134.6 | C19-iturin [M + Na]+ | [18] | |
| 1136.6 | C18-iturin [M + K]+ | [18] | |
| On PDA dual culture | 1092.6 | C16-iturin [M + Na]+ | [18] |
| 1098.6 | C16-iturin [M + H]+ | [18] | |
| 1106.6 | C17-iturin [M + Na]+ | [18] | |
| 1112.6 | C19-iturin [M + H]+ | [18] | |
| 1120.6 | C18-iturin [M + Na]+ | [18] | |
| 1122.6 | C17-iturin [M + K]+ | [18] | |
| 1134.6 | C19-iturin [M + Na]+ | [18] | |
| 1136.6 | C18-iturin [M + K]+ | [18] | |
| 1150.6 | C19-iturin [M + K]+ | [18] |
| Mass Peak (m/z) | Assignment | Reference | |
|---|---|---|---|
| On PDA control | 1044.6 | C14-surfactin [M + Na, K]+ | [18] |
| 1046.6 | C13-surfactin [M + K]+ | [18] | |
| 1058.6 | C15-surfactin [M + Na]+ | [18] | |
| 1060.5 | C14-surfactin [M + Na, K]+ | [18] | |
| 1074.6 | C15-surfactin [M + Na, K]+ | [18] | |
| On PDA dual culture | 1030.6 | C13-surfactin [M + Na]+ | [18] |
| 1032.7 | C13-surfactin [M + K]+ | [18] | |
| 1044.6 | C14-surfactin [M + Na, K]+ | [18] | |
| 1046.6 | C13-surfactin [M + K]+ | [18] | |
| 1058.6 | C15-surfactin [M + Na]+ | [18] | |
| 1060.5 | C14-surfactin [M + Na, K]+ | [18] | |
| 1074.6 | C15-surfactin [M + Na, K]+ | [18] |
| Bioactivity | Gene/Gene Cluster | From | To | Product | Remark |
|---|---|---|---|---|---|
| Root colonization | yfiQ | 1181545 | 1180457 | Putative membrane-bound acyltransferase YfiQ | Involved in surface adhesion [13,84] |
| sacB | 2852367 | 2850946 | Levan sucrase | Levan contributed to the aggregation of wheat root-adhering soil [85] | |
| Swarming motility | swrB | 319026 | 318514 | Swarming motility protein swrB | Essential for swarming motility [86] |
| swrC | 1348113 | 1344916 | Swarming motility protein SwrC | Self-resistance to surfactin [86] | |
| sfp | 1712320 | 1712994 | 4’-phosphopantetheinyl transferase sfp | Necessary for lipopeptide and polyketide synthesis which is essential for surface motility and biofilm formation [13,86] | |
| swrAA | 2770348 | 2770776 | Swarming motility protein swrAA | Essential for swarming motility [87] | |
| swrAB | 2770855 | 2772051 | Swarming motility protein swrAB | Essential for swarming motility [87] | |
| efp | 3829945 | 3830526 | Elongation factor P | Essential for swarming motility [88] | |
| flhA | 328577 | 326544 | Flagellar biosynthesis protein flhA | Flagellar assembly | |
| flhB | 329692 | 328610 | Flagellar biosynthetic protein flhB | ||
| fliR | 330489 | 329692 | Flagellar biosynthetic protein fliR | ||
| fliQ | 330757 | 330479 | Flagellar biosynthetic protein FliQ | ||
| fliP | 331428 | 330763 | Flagellar biosynthetic protein fliP | ||
| fliY | 333625 | 332483 | Flagellar motor switch phosphatase FliY | ||
| fliM | 334613 | 333606 | Flagellar motor switch protein FliM | ||
| fliL | 335060 | 334638 | Flagellar protein FliL | ||
| flgG | 336163 | 335312 | Flagellar basal-body rod protein flgG | ||
| fliK | 338009 | 336546 | Probable flagellar hook-length control protein | ||
| fliJ | 339085 | 338642 | Flagellar FliJ protein | ||
| fliI | 340404 | 339088 | Flagellum-specific ATP synthase | ||
| fliH | 341153 | 340401 | Probable flagellar assembly protein fliH | ||
| fliG | 342162 | 341146 | Flagellar motor switch protein FliG | ||
| fliF | 343785 | 342175 | Flagellar M-ring protein | ||
| fliE | 344151 | 343831 | Flagellar hook-basal body complex protein FliE | ||
| flgC | 344618 | 344163 | Flagellar basal-body rod protein flgC | ||
| flgB | 345010 | 344615 | Flagellar basal-body rod protein flgB | ||
| motA | 654985 | 655887 | Motility protein A | ||
| motB | 655835 | 656650 | Motility protein B | ||
| Swarming motility | flhO | 2629891 | 2630742 | Flagellar hook-basal body complex protein flhO | Flagellar assembly |
| flhP | 2630949 | 2631584 | Flagellar hook-basal body complex protein flhP | ||
| flgM | 2751851 | 2752117 | Negative regulator of flagellin synthesis | ||
| flgK | 2752634 | 2754151 | Flagellar hook-associated protein 1 | ||
| flgL | 2754161 | 2755057 | Flagellar hook-associated protein 3 | ||
| hag | 2756494 | 2757405 | Flagellin | ||
| fliD | 2757987 | 2759483 | Flagellar hook-associated protein 2 | ||
| fliS | 2759505 | 2759906 | Flagellar protein fliS | ||
| fliT | 2759903 | 2760244 | Flagellar protein FliT | ||
| cheD | 320333 | 319833 | Chemoreceptor glutamine deamidase CheD | Bacterial chemotaxi | |
| cheC | 320959 | 320330 | CheY-P phosphatase CheC | ||
| cheW | 321457 | 320978 | Chemotaxis protein CheW | ||
| cheA | 323488 | 321470 | Chemotaxis protein CheA | ||
| cheB | 324555 | 323485 | Chemotaxis response regulator protein-glutamate methylesterase | ||
| cheY | 332463 | 332095 | Chemotaxis protein CheY | ||
| cheV | 616969 | 616058 | Chemotaxis protein CheV | ||
| cheR | 3987238 | 3988155 | Chemotaxis protein methyltransferase | ||
| Biofilm formation | pgsA | 273607 | 273026 | CDP-diacylglycerol-glycerol-3-phosphate3-Phosphatidyl transferase | Member of pgsB-pgsC-pgsA-pgsE gene cluster encoding PGA which is contributed to robustness and complex morphology of the colony biofilms [89] |
| pgcA | 1083817 | 1082072 | Phosphoglucomutase | Phosphoglucomutase plays an important role in biofilm formation [90] | |
| ybdK | 1903365 | 1902379 | Sensor histidine kinase ybdK | Transcriptional regulation of biofilm formation [91,92] | |
| sigW | 1928876 | 1928313 | RNA polymerase sigma factor sigW | Transcriptional regulation of biofilm formation [91,92] | |
| sigH | 2011463 | 2010807 | RNA polymerase sigma-H factor | Involves in the initial stage of biofilm formation [93] | |
| abrB | 2082858 | 2083148 | Transition state regulatory protein AbrB | Transcriptional regulation of biofilm formation [94] | |
| epsC-O | 2731363 | 2874940 | Gene cluster for capsular poly-saccharide biosynthesis | Encoding exopolysaccharide which is essential for biofilm formation [91] | |
| lytS | 3426013 | 3427803 | Sensor protein lytS | Transcriptional regulation of biofilm formation [91,92] | |
| yqxM | 3813390 | 3814151 | Protein yqxM | Belongs to yqxM-sipW-tasA gene cluster that is necessary for biofilm formation [95] | |
| tasA | 3814771 | 3815556 | Spore coat-associated protein N | Required for development of complex colony architecture [94] | |
| sinR | 3816019 | 3815651 | HTH-type transcriptional regulator sinR | Transcriptional regulation of biofilm formation [91,92] | |
| sinI | 3816289 | 3816020 | Protein sinI | Transcriptional regulation of biofilm formation [91,92] | |
| Biofilm formation | spo0A | 3849786 | 3850625 | Stage 0 sporulation protein A | Involved in the initial stage of biofilm formation [93] |
| resE | 3951273 | 3953042 | Sensor histidine kinase resE | Transcriptional regulation of biofilm formation [91,92] | |
| ymcA | 261856 | 261425 | Protein ymcA | These genes are involved in the development of multicellular communities [91] | |
| ylbF | 467892 | 467434 | Regulatory protein ylbF | ||
| yqeK | 3725618 | 3726187 | Protein yqeK | ||
| sipW | 3814123 | 3814707 | Signal peptidase I W | ||
| Mineral assimilation | moaD | 594413 | 594180 | Molybdopterin synthase sulfur carrier subunit | Nitrogen assimilation |
| moaE | 594879 | 594406 | Molybdopterin synthase catalytic subunit | ||
| moaC | 1469542 | 1469000 | Molybdenum cofactor biosynthesis protein C | ||
| moaA | 2592667 | 2593692 | Molybdenum cofactor biosynthesis protein A | ||
| moaB | 3369200 | 3369751 | Molybdenum cofactor biosynthesis protein B | ||
| nasA-F | 1758586 | 1768809 | Gene cluster for Nitrate transport and reduction | Nitrogen assimilation | |
| narK | 2532129 | 2533373 | Nitrite extrusion protein | ||
| fnr | 2533450 | 2534190 | Anaerobic regulatory protein | ||
| arfM | 2534994 | 2535563 | Probable transcription regulator arfM | ||
| narG-J | 2535783 | 2542171 | Gene cluster for Nitrate reductase | ||
| nrgB | 2619035 | 2618670 | Nitrogen regulatory PII-like protein | ||
| nrgA | 2620246 | 2619032 | Ammonium transporter nrgA | ||
| mgtE | 694316 | 692961 | Magnesium transporter mgtE | Magnesium assimilation | |
| corA | 3806910 | 3807869 | Magnesium transport protein CorA | ||
| mntH | 1626982 | 1628259 | Manganese transport protein mntH | Manganese assimilation | |
| mntA-D | 3237914 | 3241819 | Gene cluster for Manganese binding/transport protein | ||
| mntR | 3824735 | 3825202 | Transcriptional regulator mntR | ||
| ktrC | 573312 | 572647 | Ktr system potassium uptake protein C | Potassium assimilation | |
| ykqA | 574212 | 573469 | Putative gamma-glutamylcyclo transferase ykqA | ||
| ktrD | 674275 | 672926 | Ktr system potassium uptake protein D | ||
| yugO | 3174241 | 3173258 | Putative potassium channel protein yugO | ||
| ktrB | 3203187 | 3201850 | Ktr system potassium uptake protein B | ||
| ktrA | 3203862 | 3203194 | Ktr system potassium uptake protein A | ||
| Mineral assimilation | yclQ | 1683673 | 1682711 | Ferrichrome ABC transporter | Iron assimilation |
| yvrC | 2985220 | 2986197 | Putative iron binding lipoprotein yvrC | ||
| yusV | 3009672 | 3010583 | Putative iron (III) ABC transport ATPase component | ||
| dhbABCEF | 3101063 | 3112861 | Gene cluster encoding Bacillibactin | ||
| tuaA-H | 2733487 | 2742546 | Gene cluster for teichuronic acid biosynthesis | Bivalent cations assimilation | |
| yvdK | 2839830 | 2842166 | Glycosyl hydrolase yvdK | Ferrochrome assimilation | |
| Plant growth promotion/ISR | alsR | 2672398 | 2671511 | HTH-type transcriptional regulator alsR | These genes encode enzymes of the biosynthetic pathway from pyruvate to 3-hydroxy-2-butanone |
| alsS | 2672549 | 2674270 | Acetolactate synthase | ||
| alsD | 2674320 | 2675099 | Alpha-acetolactate decarboxylase | ||
| bdhA | 1448068 | 1449108 | (R, R)-butanediol dehydrogenase | This gene encodes enzyme to acatlyse 3-hydroxy-2-butanone to 2,3-butanediol | |
| Plant growth promotion | yhcX | 1092936 | 1091395 | Carbon-nitrogen hydrolase | These genes are involved in indole acetic acid biosynthesis |
| ysnE | 3489604 | 3489101 | N-acetyltransferase | ||
| dhaS | 4136609 | 4135263 | Putative aldehyde dehydrogenase dhaS | ||
| phy | 4084788 | 4085936 | 3-phytase | Phytase hormones biosynthesis gene | |
| treR | 1237083 | 1236367 | Trehalose gene cluster transcriptional repressor | These genes are involved in trehalose biosynthesis | |
| treA | 1238792 | 1237104 | Trehalose-6-phosphate hydrolase | ||
| treP | 1240275 | 1238863 | PTS system trehalose-specific EIIBC component | ||
| ilvH | 3492606 | 3493124 | Acetolactate synthase small subunit | These genes are parts of leucine, valine, and isoleucine biosynthesis pathway | |
| ilvB | 3490858 | 3492609 | Acetolactate synthase large subunit | ||
| ilvC | 3493148 | 3494176 | Ketol-acid reductoisomerase | ||
| speA | 501879 | 503375 | Arginine decarboxylase | These genes may transform amino acids to plant growth-promoting substances [80] | |
| speE | 2512298 | 2513128 | Spermidine synthase | ||
| speB | 2513189 | 2514061 | Agmatinase |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamali, M.; Guo, D.; Naeimi, S.; Ahmadi, J. Perception of Biocontrol Potential of Bacillus inaquosorum KR2-7 against Tomato Fusarium Wilt through Merging Genome Mining with Chemical Analysis. Biology 2022, 11, 137. https://doi.org/10.3390/biology11010137
Kamali M, Guo D, Naeimi S, Ahmadi J. Perception of Biocontrol Potential of Bacillus inaquosorum KR2-7 against Tomato Fusarium Wilt through Merging Genome Mining with Chemical Analysis. Biology. 2022; 11(1):137. https://doi.org/10.3390/biology11010137
Chicago/Turabian StyleKamali, Maedeh, Dianjing Guo, Shahram Naeimi, and Jafar Ahmadi. 2022. "Perception of Biocontrol Potential of Bacillus inaquosorum KR2-7 against Tomato Fusarium Wilt through Merging Genome Mining with Chemical Analysis" Biology 11, no. 1: 137. https://doi.org/10.3390/biology11010137
APA StyleKamali, M., Guo, D., Naeimi, S., & Ahmadi, J. (2022). Perception of Biocontrol Potential of Bacillus inaquosorum KR2-7 against Tomato Fusarium Wilt through Merging Genome Mining with Chemical Analysis. Biology, 11(1), 137. https://doi.org/10.3390/biology11010137







