Architectural Distortion-Based Digital Mammograms Classification Using Depth Wise Convolutional Neural Network
Abstract
:Simple Summary
Abstract
1. Introduction
- Proposed an automated computer-aided diagnostic system based on architectural distortion using computer vision and depth-wise deep learning techniques to predict breast cancer from digital mammograms. Applied the image pixel-wise segmentation using a computer vision algorithm to extract architectural distortion ROIs from the digital mammogram image in the first phase.
- In the second phase, employed a depth-wise V-Net 64 convolutional neural network to extract automatic features from ADs ROIs and classify them into malignant and benign ROIs. Moreover, use machine learning and deep learning algorithms, such as shuffelnet, mobilenet, support vector machine, k-nearest neighbor, and random forest, to classify these ROIs.
- Proposed method obtained higher accuracy than machine learning and with the previous studies. Furthermore, evaluated proposed model with other metrics to enhance the diagnostic ability of the model.
- Evaluated the proposed method on three datasets, the local private PINUM and publicly available CBIS-DDSM and DDSM dataset that makes a fair comparison of the proposed model with others.
2. Related Works and Techniques
2.1. Conventional Deep Learning Mammogram Classification
2.2. Conventional Machine Learning Mammogram Classification
3. Materials
Databases
4. Methods
4.1. Proposed Method
4.2. Image Preprocessing
| Algorithm 1 Image preprocessing algorithm 1. |
|
4.3. Image Augmentation
4.4. Pixel Wise Segmentation
- The image is to be segmented as a targeted image , where P representing a pixel array vector having N elements that has belongs to the specific category as:
- The pixel and represents the vertical w and horizontal h pixels, where and are the elements of pixel vector. The dot product has performed as:
- represents each object in a pixel array belonging to classes 0 and 1. The pixel-wise prediction can be improved on which we can generate the segmentation results.
4.5. Architectural Distortion ROI’s Detection
4.6. Depth-Wise-CNN Architecture
4.7. Depth-Wise-V-Net64 Training
4.8. Standard Classifiers
4.9. Evaluation Metrics
5. Results Analysis
5.1. Experimental Configuration
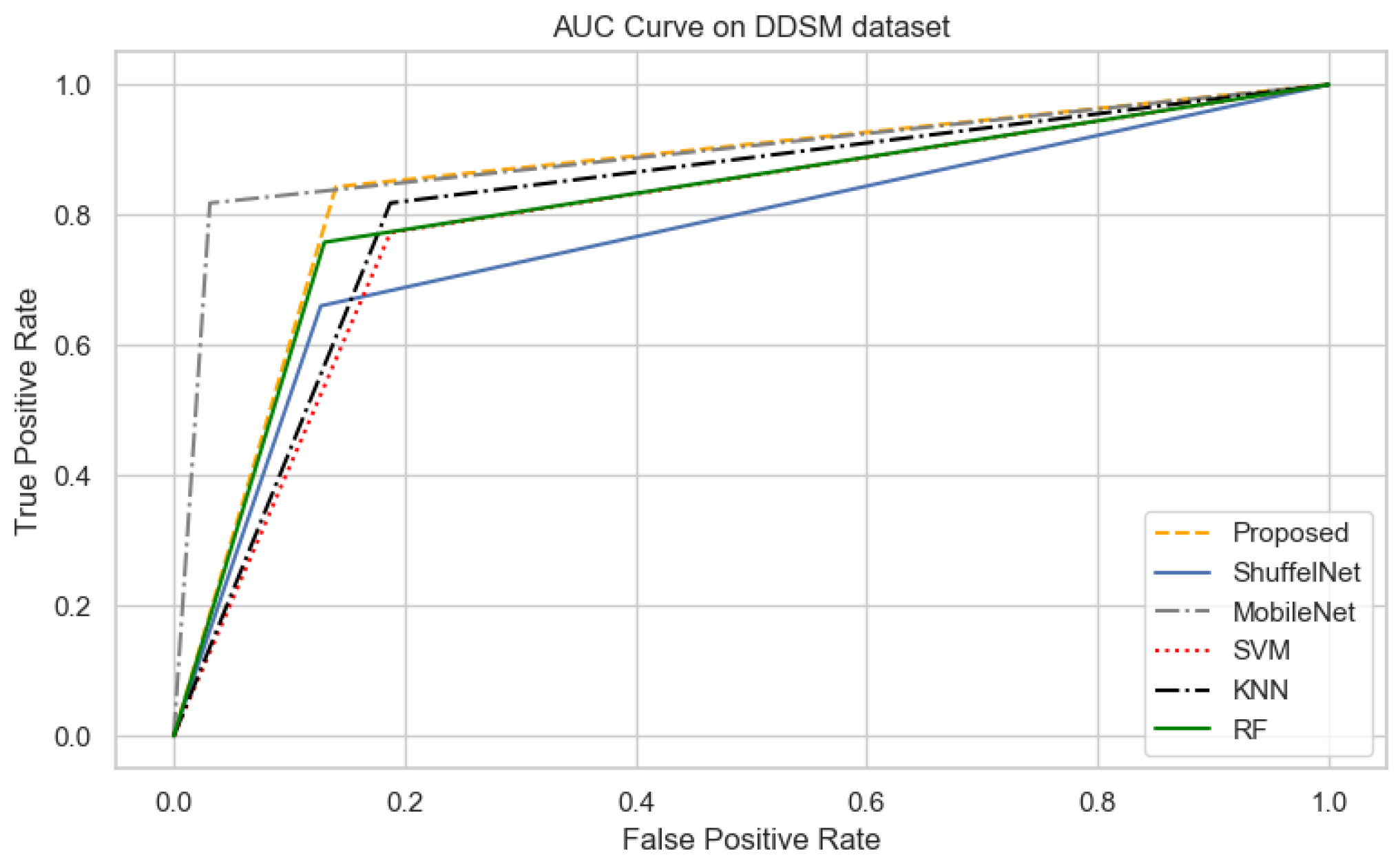
5.2. Comparison between Proposed Method, ShuffelNet, MobileNet and SVM, KNN, RF
5.3. Results Comparison between Proposed Method and Previous Studies
6. Discussion
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- WHO. Fact Sheet World Health Organization; WHO. 2019. Available online: https://www.who.int/news-room/fact-sheets/detail/cancer (accessed on 16 December 2021).
- Ribli, D.; Horváth, A.; Unger, Z.; Pollner, P.; Csabai, I. Detecting and classifying lesions in mammograms with deep learning. Sci. Rep. 2018, 8, 1–7. [Google Scholar]
- Radiology, A.C. Mammography and Breast Imaging Resoruces. Available online: https://www.acr.org/Clinical-Resources/Breast-Imaging-Resources (accessed on 16 December 2021).
- Gaur, S.; Dialani, V.; Slanetz, P.J.; Eisenberg, R.L. Architectural distortion of the breast. Am. J. Roentgenol. 2013, 201, 662–670. [Google Scholar] [CrossRef]
- Heidari, M.; Mirniaharikandehei, S.; Liu, W.; Hollingsworth, A.B.; Liu, H.; Zheng, B. Development and assessment of a new global mammographic image feature analysis scheme to predict likelihood of malignant cases. IEEE Trans. Med. Imaging 2019, 39, 1235–1244. [Google Scholar] [CrossRef]
- Murali, S.; Dinesh, M. Model based approach for detection of architectural distortions and spiculated masses in mammograms. Int. J. Comput. Sci. Eng. 2011, 3, 3534. [Google Scholar]
- Banik, S.; Rangayyan, R.M.; Desautels, J.L. Detection of architectural distortion in prior mammograms. IEEE Trans. Med. Imaging 2010, 30, 279–294. [Google Scholar] [CrossRef] [PubMed]
- Jasionowska, M.; Przelaskowski, A.; Rutczynska, A.; Wroblewska, A. A two-step method for detection of architectural distortions in mammograms. In Information Technologies in Biomedicine; Springer: Berlin, Germany, 2010; pp. 73–84. [Google Scholar]
- Kamra, A.; Jain, V.; Singh, S.; Mittal, S. Characterization of architectural distortion in mammograms based on texture analysis using support vector machine classifier with clinical evaluation. J. Digit. Imaging 2016, 29, 104–114. [Google Scholar] [CrossRef]
- Liu, X.; Zhai, L.; Zhu, T.; Yang, Z. Architectural distortion recognition based on a subclass technique and the sparse representation classifier. In Proceedings of the 2016 9th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Datong, China, 15–17 October 2016; pp. 422–426. [Google Scholar]
- Ciurea, A.I.; Ciortea, C.A.; Ștefan, P.A.; Lisencu, L.A.; Dudea, S.M. Differentiating Breast Tumors from Background Parenchymal Enhancement at Contrast-Enhanced Mammography: The Role of Radiomics A Pilot Reader Study. Diagnostics 2021, 11, 1248–1265. [Google Scholar]
- Massafra, R.; Bove, S.; Lorusso, V.; Biafora, A.; Comes, M.C.; Didonna, V.; Diotaiuti, S.; Fanizzi, A.; Nardone, A.; Nolasco, A.; et al. Radiomic Feature Reduction Approach to Predict Breast Cancer by Contrast-Enhanced Spectral Mammography Images. Diagnostics 2021, 11, 684. [Google Scholar] [CrossRef]
- De Oliveira, H.C.; Moraes, D.R.; Reche, G.A.; Borges, L.R.; Catani, J.H.; de Barros, N.; Melo, C.F.; Gonzaga, A.; Vieira, M.A. A new texture descriptor based on local micro-pattern for detection of architectural distortion in mammographic images. In Proceedings of the Medical Imaging 2017: Computer-Aided Diagnosis. International Society for Optics and Photonics, SPIE Medical Imaging, Orlando, FL, USA, 3 March 2017; Volume 10134, pp. 101342–101357. [Google Scholar]
- De Oliveira, H.C.; Mencattini, A.; Casti, P.; Catani, J.H.; de Barros, N.; Gonzaga, A.; Martinelli, E.; da Costa Vieira, M.A. A cross-cutting approach for tracking architectural distortion locii on digital breast tomosynthesis slices. Biomed. Signal Process. Control 2019, 50, 92–102. [Google Scholar] [CrossRef]
- Palma, G.; Bloch, I.; Muller, S. Detection of masses and architectural distortions in digital breast tomosynthesis images using fuzzy and a contrario approaches. Pattern Recognit. 2014, 47, 2467–2480. [Google Scholar] [CrossRef]
- Cai, Q.; Liu, X.; Guo, Z. identifying architectural distortion in mammogram images via a SE-DenseNet model and twice transfer learning. In Proceedings of the 2018 11th International Congress on Image and Signal Processing, BioMedical Engineering and Informatics (CISP-BMEI), Beijing, China, 13–15 October 2018; pp. 1–6. [Google Scholar]
- Bahl, M.; Baker, J.A.; Kinsey, E.N.; Ghate, S.V. Architectural distortion on mammography: Correlation with pathologic outcomes and predictors of malignancy. Am. J. Roentgenol. 2015, 205, 1339–1345. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Zhang, L.; Wang, Z.; Lv, Q.; Yi, Z. Deep neural networks with region-based pooling structures for mammographic image classification. IEEE Trans. Med. Imaging 2020, 39, 2246–2255. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Li, M.; Wang, H.; Jiang, H.; Yao, Y.; Zhang, H.; Xin, J. Breast cancer detection using extreme learning machine based on feature fusion with CNN deep features. IEEE Access 2019, 7, 105146–105158. [Google Scholar] [CrossRef]
- Wu, N.; Phang, J.; Park, J.; Shen, Y.; Huang, Z.; Zorin, M.; Jastrzebski, S.; Fevry, T.; Katsnelson, J.; Kim, E.; et al. Deep neural networks improve radiologists performance in breast cancer screening. IEEE Trans. Med. Imaging 2019, 39, 1184–1194. [Google Scholar] [CrossRef] [Green Version]
- Khan, H.N.; Shahid, A.R.; Raza, B.; Dar, A.H.; Alquhayz, H. Multi-view feature fusion based four views model for mammogram classification using convolutional neural network. IEEE Access 2019, 7, 165724–165733. [Google Scholar] [CrossRef]
- Soleimani, H.; Michailovich, O.V. On Segmentation of Pectoral Muscle in Digital Mammograms by Means of Deep Learning. IEEE Access 2020, 8, 204173–204182. [Google Scholar] [CrossRef]
- Hao, D.; Zhang, L.; Sumkin, J.; Mohamed, A.; Wu, S. Inaccurate Labels in Weakly-Supervised Deep Learning: Automatic Identification and Correction and Their Impact on Classification Performance. IEEE J. Biomed. Health Inform. 2020, 24, 2701–2710. [Google Scholar] [CrossRef]
- Sun, D.; Wang, M.; Li, A. A multimodal deep neural network for human breast cancer prognosis prediction by integrating multi-dimensional data. IEEE/ACM Trans. Comput. Biol. Bioinform. 2018, 16, 841–850. [Google Scholar] [CrossRef]
- Guan, Y.; Wang, X.; Li, H.; Zhang, Z.; Chen, X.; Siddiqui, O.; Nehring, S.; Huang, X. Detecting Asymmetric Patterns and Localizing Cancers on Mammograms. Patterns 2020, 1, 100106–100120. [Google Scholar] [CrossRef]
- Singh, V.K.; Rashwan, H.A.; Romani, S.; Akram, F.; Pandey, N.; Sarker, M.M.K.; Saleh, A.; Arenas, M.; Arquez, M.; Puig, D.; et al. Breast tumor segmentation and shape classification in mammograms using generative adversarial and convolutional neural network. Expert Syst. Appl. 2020, 139, 112855–112870. [Google Scholar] [CrossRef]
- Song, R.; Li, T.; Wang, Y. Mammographic Classification Based on XGBoost and DCNN With Multi Features. IEEE Access 2020, 8, 75011–75021. [Google Scholar] [CrossRef]
- Shen, T.; Wang, J.; Gou, C.; Wang, F.Y. Hierarchical Fused Model With Deep Learning and Type-2 Fuzzy Learning for Breast Cancer Diagnosis. IEEE Trans. Fuzzy Syst. 2020, 28, 3204–3218. [Google Scholar] [CrossRef]
- Guan, S.; Loew, M. Breast cancer detection using synthetic mammograms from generative adversarial networks in convolutional neural networks. J. Med. Imaging 2019, 6, 031411–031432. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Zhuang, S.; Li, D.A.; Zhao, J.; Ma, Y. Benign and malignant classification of mammogram images based on deep learning. Biomed. Signal Process. Control 2019, 51, 347–354. [Google Scholar] [CrossRef]
- Ionescu, G.V.; Fergie, M.; Berks, M.; Harkness, E.F.; Hulleman, J.; Brentnall, A.R.; Cuzick, J.; Evans, D.G.; Astley, S.M. Prediction of reader estimates of mammographic density using convolutional neural networks. J. Med. Imaging 2019, 6, 031405–031425. [Google Scholar] [CrossRef] [Green Version]
- Falconí, L.G.; Pérez, M.; Aguilar, W.G. Transfer learning in breast mammogram abnormalities classification with mobilenet and nasnet. In Proceedings of the 2019 International Conference on Systems, Signals and Image Processing (IWSSIP), Osijek, Croatia, 5–7 June 2019; pp. 109–114. [Google Scholar]
- Gnanasekaran, V.S.; Joypaul, S.; Sundaram, P.M.; Chairman, D.D. Deep learning algorithm for breast masses classification in mammograms. IET Image Process. 2020, 14, 2860–2868. [Google Scholar] [CrossRef]
- Shen, R.; Yan, K.; Tian, K.; Jiang, C.; Zhou, K. Breast mass detection from the digitized X-ray mammograms based on the combination of deep active learning and self-paced learning. Future Gener. Comput. Syst. 2019, 101, 668–679. [Google Scholar] [CrossRef]
- Shen, T.; Gou, C.; Wang, J.; Wang, F.Y. Simultaneous segmentation and classification of mass region from mammograms using a mixed-supervision guided deep model. IEEE Signal Process. Lett. 2019, 27, 196–200. [Google Scholar] [CrossRef]
- Shaymaa, A.H.; Sayed, M.S.; Abdalla, M.I.; Rashwan, M.A. Detection of breast cancer mass using MSER detector and features matching. Multimed. Tools Appl. 2019, 78, 20239–20262. [Google Scholar]
- Wang, H.; Feng, J.; Zhang, Z.; Su, H.; Cui, L.; He, H.; Liu, L. Breast mass classification via deeply integrating the contextual information from multi-view data. Pattern Recognit. 2018, 80, 42–52. [Google Scholar] [CrossRef]
- Wang, R.; Ma, Y.; Sun, W.; Guo, Y.; Wang, W.; Qi, Y.; Gong, X. Multi-level nested pyramid network for mass segmentation in mammograms. Neurocomputing 2019, 363, 313–320. [Google Scholar] [CrossRef]
- Birhanu, M.A.; Karssemeijer, N.; Gubern-Merida, A.; Kallenberg, M. A deep learning method for volumetric breast density estimation from processed full field digital mammograms. In Proceedings of the Medical Imaging 2019: Computer-Aided Diagnosis. International Society for Optics and Photonics, SPIE Medical Imaging, San Diego, CA, USA, 13 March 2019; Volume 10950, pp. 109500–109525. [Google Scholar]
- Fan, J.; Wu, Y.; Yuan, M.; Page, D.; Liu, J.; Ong, I.M.; Peissig, P.; Burnside, E. Structure-leveraged methods in breast cancer risk prediction. J. Mach. Learn. Res. 2016, 17, 2956–2970. [Google Scholar]
- Loizidou, K.; Skouroumouni, G.; Nikolaou, C.; Pitris, C. An automated breast micro-calcification detection and classification technique using temporal subtraction of mammograms. IEEE Access 2020, 8, 52785–52795. [Google Scholar] [CrossRef]
- Zebari, D.A.; Zeebaree, D.Q.; Abdulazeez, A.M.; Haron, H.; Hamed, H.N.A. Improved Threshold Based and Trainable Fully Automated Segmentation for Breast Cancer Boundary and Pectoral Muscle in Mammogram Images. IEEE Access 2020, 8, 203097–203116. [Google Scholar] [CrossRef]
- Chakraborty, J.; Midya, A.; Mukhopadhyay, S.; Rangayyan, R.M.; Sadhu, A.; Singla, V.; Khandelwal, N. Computer-aided detection of mammographic masses using hybrid region growing controlled by multilevel thresholding. J. Med Biol. Eng. 2019, 39, 352–366. [Google Scholar] [CrossRef]
- Beham, M.P.; Tamilselvi, R.; Roomi, S.M.; Nagaraj, A. Accurate Classification of Cancer in Mammogram Images. In Innovations in Electronics and Communication Engineering; Springer: Berlin, Germany, 2019; pp. 71–77. [Google Scholar]
- Liu, N.; Qi, E.S.; Xu, M.; Gao, B.; Liu, G.Q. A novel intelligent classification model for breast cancer diagnosis. Inf. Process. Manag. 2019, 56, 609–623. [Google Scholar] [CrossRef]
- Yang, L.; Xu, Z. Feature extraction by PCA and diagnosis of breast tumors using SVM with DE-based parameter tuning. Int. J. Mach. Learn. Cybern. 2019, 10, 591–601. [Google Scholar] [CrossRef]
- Obaidullah, S.M.; Ahmed, S.; Gonçalves, T.; Rato, L. RMID: A novel and efficient image descriptor for mammogram mass classification. In Proceedings of the Conference on Information Technology, Systems Research and Computational Physics, Cham, Switzerland, 11–13 January 2020; pp. 229–240. [Google Scholar]
- PAEC. Pakistan Atomic Energy Commission Punjab Institue of Nuclear Medicine (PINUM Faisalabad). 2020. Available online: http://www.paec.gov.pk/Medical/Centres/ (accessed on 16 December 2021).
- Archive, C.I. Curated Breast Imaging Digital Database for Screening Mammography(DDSM). 2021. Available online: https://wiki.cancerimagingarchive.net/display/Public/CBIS-DDSM (accessed on 16 December 2021).
- University of South Florida. Digital Database for Screening Mammography (DDSM). 2021. Available online: http://www.eng.usf.edu/cvprg/Mammography/Database.html (accessed on 16 December 2021).
- Ting, F.F.; Tan, Y.J.; Sim, K.S. Convolutional neural network improvement for breast cancer classification. Expert Syst. Appl. 2019, 120, 103–115. [Google Scholar] [CrossRef]
- Wang, M.; Li, P. Label fusion method combining pixel greyscale probability for brain MR segmentation. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef]
- Samreen, N.; Moy, L.; Lee, C.S. Architectural Distortion on Digital Breast Tomosynthesis: Management Algorithm and Pathological Outcome. J. Breast Imaging 2020, 2, 424–435. [Google Scholar] [CrossRef]
- Guo, Q.; Shao, J.; Ruiz, V. Investigation of Support Vector Machine for the Detection of Architectural Distortion in Mammographic Images; Journal of Physics: Conference Series; Institute of Physics and Engineering in Medicine (IPEM) and IOP: London, UK, 2005; Volume 15, pp. 15–35. [Google Scholar]
- Vedalankar, A.V.; Gupta, S.S.; Manthalkar, R.R. Addressing architectural distortion in mammogram using AlexNet and support vector machine. Inform. Med. Unlocked 2021, 23, 100551. [Google Scholar] [CrossRef]
- Pete, W.B. How Many Images Do You Need to Train A Neural Net-Work? 2017. Available online: https://petewarden.com/2017/12/14/how-many-images-do-you-need-to-train-a-neural-network (accessed on 16 December 2021).























| Mammogram Label | Category | Images | Dataset |
|---|---|---|---|
| Benign (0) | Original | 425 | PINUM |
| Malignant (1) | Original | 152 | PINUM |
| Benign (0) | Augmented | 2550 | PINUM |
| Malignant (1) | Augmented | 912 | PINUM |
| Benign (0) | AD ROIs | 75 | PINUM |
| Malignant (1) | AD ROIs | 75 | PINUM |
| Benign (0) | Original | 1740 | CBIS-DDSM |
| Malignant (1) | Original | 1828 | CBIS-DDSM |
| Benign (0) | AD ROIs | 100 | CBIS-DDSM |
| Malignant (1) | AD ROIs | 100 | CBIS-DDSM |
| Benign (0) | Original | 2500 | DDSM |
| Malignant (1) | Original | 3000 | DDSM |
| Benign (0) | AD ROIs | 100 | DDSM |
| Malignant (1) | AD ROIs | 100 | DDSM |
| Sr | Augmentation Techniques | Performance Values |
|---|---|---|
| 1 | Rotation | 45, 90, 135, 180, 360 |
| 2 | Flipping | Left, Right, Top, Bottom |
| 3 | Sharpen (lightness value) | 0.5–1.5 |
| 4 | D-skew (angle) | 15, 40 |
| 5 | Contrast (intensity value) | 20–60% |
| 6 | Brightness (darkness values) | 15–55% |
| Network Layers | Filters | Filter Size | Padding | Stride | Output Shape |
|---|---|---|---|---|---|
| Input Image | - | - | - | - | |
| DW_Conv2D | 64 | same | |||
| Activataion_Relu | - | - | - | - | |
| Max_Pooling | 1 | - | 0 | ||
| DW_Conv2D | 64 | same | |||
| Activataion_Relu | - | - | - | - | |
| Max_Pooling | 1 | - | 0 | ||
| DW_Conv2D | 64 | same | |||
| Activataion_Relu | - | - | - | - | |
| Max_Pooling | 1 | - | 0 | ||
| Dropout (0.5) | - | - | - | - | |
| FC1_Flatten_4 | - | - | - | - | (6400) |
| FC2_Dense_5 | 64 | - | - | - | (6400) |
| Sigmoid | - | - | - | - | [0/1] |
| Configuration | Values |
|---|---|
| Batch Size | 16 |
| Learning Rate | 0.001 |
| Epochs | 20 |
| Optimization function | Adam |
| Loss Function | binary_crossentropy |
| Target Size | [320, 240] |
| histogram_freq | 1 |
| Tarin Split | 0.6 |
| Validation Split | 0.2 |
| Algorithms | Accuracy | F1-Score | Precision | Recall | Sensitivity | AUC |
|---|---|---|---|---|---|---|
| Proposed | 0.95 | 0.87 | 0.90 | 0.89 | 0.99 | 0.91 |
| ShuffelNet | 0.91 | 0.81 | 0.86 | 0.76 | 0.95 | 0.79 |
| MobileNet | 0.89 | 0.77 | 0.61 | 0.73 | 0.85 | 0.79 |
| SVM | 0.87 | 0.72 | 0.88 | 0.61 | 0.97 | 0.69 |
| KNN | 0.83 | 0.63 | 0.84 | 0.51 | 0.96 | 0.59 |
| RF | 0.90 | 0.81 | 0.89 | 0.75 | 0.96 | 0.75 |
| Algorithms | Accuracy | F1-Score | Precision | Recall | Sensitivity | AUC |
|---|---|---|---|---|---|---|
| Proposed | 0.97 | 0.96 | 0.94 | 0.98 | 0.95 | 0.98 |
| ShuffelNet | 0.93 | 0.69 | 0.75 | 0.73 | 0.84 | 0.69 |
| MobileNet | 0.90 | 0.93 | 0.82 | 0.83 | 0.87 | 0.61 |
| SVM | 0.73 | 0.69 | 0.73 | 0.66 | 0.79 | 0.67 |
| KNN | 0.80 | 0.78 | 0.79 | 0.78 | 0.82 | 0.81 |
| RF | 0.95 | 0.95 | 0.93 | 0.97 | 0.94 | 0.89 |
| Algorithms | Accuracy | F1-Score | Precision | Recall | Sensitivity | AUC |
|---|---|---|---|---|---|---|
| Proposed | 0.98 | 0.90 | 0.96 | 0.86 | 0.96 | 0.85 |
| ShuffelNet | 0.87 | 0.74 | 0.83 | 0.76 | 0.89 | 0.69 |
| MobileNet | 0.90 | 0.84 | 0.85 | 0.84 | 0.88 | 0.81 |
| SVM | 0.80 | 0.76 | 0.74 | 0.77 | 0.81 | 0.79 |
| KNN | 0.81 | 0.78 | 0.75 | 0.82 | 0.81 | 0.81 |
| RF | 0.91 | 0.88 | 0.95 | 0.82 | 0.90 | 0.78 |
| Authors | Problem | Method | Database | Images | Accuracy |
|---|---|---|---|---|---|
| [6] | Architectural Distortion Detection | SVM, MLP | DDSM | 190 | 0.89 |
| [7] | Architectural Distortion Detection | Bayesian, SELF ANN | Private | 1745 | N/A |
| [8] | Architectural Distortion Detection | Differential direction method | DDSM | 33 | 0.83 |
| [9] | Architectural Distortion Detection | SVM | DDSM | 147 | 0.92 |
| [10] | Architectural Distortion Detection | Sparse classifier | DDSM | 69 | 0.91 |
| [13] | Architectural Distortion Detection | MLP | FFDM | 300 | 0.83 |
| [14] | Architectural Distortion tracking | LDA | FFDM | 37 | N/A |
| [55] | Architectural Distortion tracking | CNN | CBIS-DDSM | 334 | 0.92 |
| Proposed | Architectural Distortion Detection | Depth-wise 2DCNN | Private (PINUM) | 3462 | 0.95 |
| Proposed | Architectural Distortion Detection | Depth-wise 2DCNN | CBIS-DDSM | 3568 | 0.97 |
| Proposed | Architectural Distortion Detection | Depth-wise 2DCNN | DDSM | 5500 | 0.98 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rehman, K.u.; Li, J.; Pei, Y.; Yasin, A.; Ali, S.; Saeed, Y. Architectural Distortion-Based Digital Mammograms Classification Using Depth Wise Convolutional Neural Network. Biology 2022, 11, 15. https://doi.org/10.3390/biology11010015
Rehman Ku, Li J, Pei Y, Yasin A, Ali S, Saeed Y. Architectural Distortion-Based Digital Mammograms Classification Using Depth Wise Convolutional Neural Network. Biology. 2022; 11(1):15. https://doi.org/10.3390/biology11010015
Chicago/Turabian StyleRehman, Khalil ur, Jianqiang Li, Yan Pei, Anaa Yasin, Saqib Ali, and Yousaf Saeed. 2022. "Architectural Distortion-Based Digital Mammograms Classification Using Depth Wise Convolutional Neural Network" Biology 11, no. 1: 15. https://doi.org/10.3390/biology11010015
APA StyleRehman, K. u., Li, J., Pei, Y., Yasin, A., Ali, S., & Saeed, Y. (2022). Architectural Distortion-Based Digital Mammograms Classification Using Depth Wise Convolutional Neural Network. Biology, 11(1), 15. https://doi.org/10.3390/biology11010015







