DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders
Abstract
:Simple Summary
Abstract
1. Introduction
2. History and Development of DNA Methylation
3. Methylation Detection Method
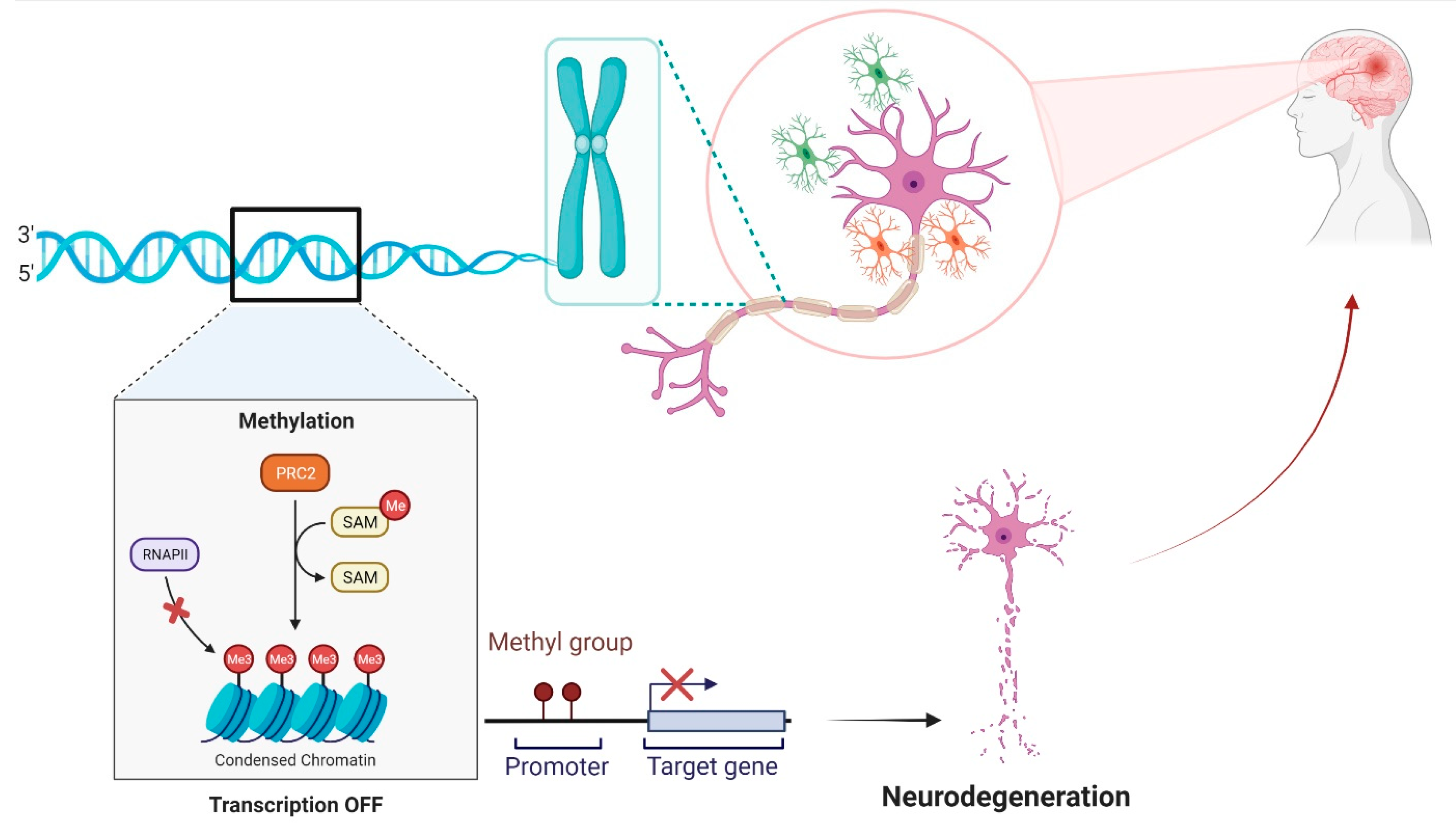
4. DNA Methylation in Premature and Mature Brain
5. Role of DNA Methylation in Neurological Disorders
5.1. Alzheimer’s Disease
5.2. Parkinson Disease
5.3. Huntington Disease
5.4. Amyotrophic Lateral Sclerosis
6. Targeting DNA Methylation in Management of AD and Other Neurodegenerative Diseases
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Jin, B.; Li, Y.; Robertson, K.D. DNA Methylation: Superior or Subordinate in the Epigenetic Hierarchy? Genes Cancer 2011, 2, 607–617. [Google Scholar] [CrossRef] [Green Version]
- Robertson, K.D. DNA methylation and human disease. Nat. Rev. Genet. 2005, 6, 597–610. [Google Scholar] [CrossRef]
- Lister, R.; Pelizzola, M.; Dowen, R.H.; Hawkins, R.D.; Hon, G.; Tonti-Filippini, J.; Nery, J.R.; Lee, L.; Ye, Z.; Ngo, Q.-M.; et al. Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 2009, 462, 315–322. [Google Scholar] [CrossRef] [Green Version]
- Zhu, J.-K. Active DNA Demethylation Mediated by DNA Glycosylases. Annu. Rev. Genet. 2009, 43, 143–166. [Google Scholar] [CrossRef] [Green Version]
- Holliday, R.; Pugh, J.E. DNA Modification Mechanisms and Gene Activity during Development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef] [PubMed]
- Compere, S.J.; Palmiter, R.D. DNA methylation controls the inducibility of the mouse metallothionein-I gene in lymphoid cells. Cell 1981, 25, 233–240. [Google Scholar] [CrossRef]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, J.; Szyf, M. Downregulation of DNA (cytosine-5-) methyltransferase is a late event in NGF-induced PC12 cell differen-tiation. Brain Res. Mol. Brain Res. 1999, 71, 23–31. [Google Scholar] [CrossRef]
- Goto, K.; Numata, M.; Komura, J.-I.; Ono, T.; Bestor, T.H.; Kondo, H. Expression of DNA methyltransferase gene in mature and immature neurons as well as proliferating cells in mice. Differentiation 1994, 56, 39–44. [Google Scholar] [CrossRef]
- Suzuki, M.M.; Kerr, A.R.; De Sousa, D.; Bird, A. CpG methylation is targeted to transcription units in an invertebrate genome. Genome Res. 2007, 17, 625–631. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schübeler, D. Function and information content of DNA methylation. Nature 2015, 517, 321–326. [Google Scholar] [CrossRef] [PubMed]
- Cedar, H.; Bergman, Y. Linking DNA methylation and histone modification: Patterns and paradigms. Nat. Rev. Genet. 2009, 10, 295–304. [Google Scholar] [CrossRef] [PubMed]
- Lehnertz, B.; Ueda, Y.; Derijck, A.A.; Braunschweig, U.; Perez-Burgos, L.; Kubicek, S.; Chen, T.; Li, E.; Jenuwein, T.; Peters, A.H. Suv39h-Mediated Histone H3 Lysine 9 Methylation Directs DNA Methylation to Major Satellite Repeats at Pericentric Heterochromatin. Curr. Biol. 2003, 13, 1192–1200. [Google Scholar] [CrossRef] [Green Version]
- Vire, E.; Brenner, C.; Deplus, R.; Blanchon, L.; Fraga, M.; Didelot, C.M.; Morey, L.; Van Eynde, A.; Bernard, D.; Vanderwinden, J.-M.; et al. The Polycomb group protein EZH2 directly controls DNA methylation. Nature 2005, 439, 871–874. [Google Scholar] [CrossRef] [PubMed]
- Saxonov, S.; Berg, P.; Brutlag, D.L. A genome-wide analysis of CpG dinucleotides in the human genome distinguishes two distinct classes of promoters. Proc. Natl. Acad. Sci. USA 2006, 103, 1412–1417. [Google Scholar] [CrossRef] [Green Version]
- Tazi, J.; Bird, A. Alternative chromatin structure at CpG islands. Cell 1990, 60, 909–920. [Google Scholar] [CrossRef]
- Ramirez-Carrozzi, V.R.; Braas, D.; Bhatt, D.M.; Cheng, C.S.; Hong, C.; Doty, K.R.; Black, J.; Hoffmann, A.; Carey, M.; Smale, S.T. A Unifying Model for the Selective Regulation of Inducible Transcription by CpG Islands and Nucleosome Remodeling. Cell 2009, 138, 114–128. [Google Scholar] [CrossRef] [Green Version]
- Carninci, P.; Sandelin, A.; Lenhard, B.; Katayama, S.; Shimokawa, K.; Ponjavic, J.; Semple, C.; Taylor, M.; Engström, P.; Frith, M.; et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006, 38, 626–635. [Google Scholar] [CrossRef]
- Iyer, L.M.; Abhiman, S.; Aravind, L. Natural History of Eukaryotic DNA Methylation Systems. Prog. Mol. Biol. Transl. Sci. 2011, 101, 25–104. [Google Scholar] [CrossRef]
- Avery, O.T.; MacLeod, C.M.; Mccarty, M. Studies on the chemical nature of the substance inducing transformation of pneumococcal types: Induction of transformation by a desoxyribonucleic acid fraction isolated from pneumococcus type III. J. Exp. Med. 1944, 79, 137–158. [Google Scholar] [CrossRef]
- Gerlach, E.; Dreisbach, R.; Deuticke, B. Paper chromatographic separation of nucleotides, nucleosides, purines, and pyrimidines. J. Chromatogr. A 1965, 18, 81–85. [Google Scholar] [CrossRef]
- MacDonald, J.L.; Roskams, A.J. Epigenetic regulation of nervous system development by DNA methylation and histone deacetylation. Prog. Neurobiol. 2009, 88, 170–183. [Google Scholar] [CrossRef]
- Iyer, L.M.; Abhiman, S.; De Souza, R.F.; Aravind, L. Origin and evolution of peptide-modifying dioxygenases and identification of the wybutosine hydroxylase/hydroperoxidase. Nucleic Acids Res. 2010, 38, 5261–5279. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Laird, P.W. Principles and challenges of genome-wide DNA methylation analysis. Nat. Rev. Genet. 2010, 11, 191–203. [Google Scholar] [CrossRef] [PubMed]
- Kurdyukov, S.; Bullock, M. DNA Methylation Analysis: Choosing the Right Method. Biology 2016, 5, 3. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; James, S.R.; Kazim, L.; Karpf, A.R. Specific Method for the Determination of Genomic DNA Methylation by Liquid Chromatography-Electrospray Ionization Tandem Mass Spectrometry. Anal. Chem. 2004, 77, 504–510. [Google Scholar] [CrossRef]
- Hur, K.; Cejas, P.; Feliu, J.; Moreno-Rubio, J.; Burgos, E.; Boland, C.R.; Goel, A. Hypomethylation of long interspersed nuclear element-1 (LINE-1) leads to activation of proto-oncogenes in human colorectal cancer metastasis. Gut 2013, 63, 635–646. [Google Scholar] [CrossRef] [Green Version]
- Jaligot, E.; Beulé, T.; Rival, A. Methylation-sensitive RFLPs: Characterisation of two oil palm markers showing somaclonal variation-associated polymorphism. Theor. Appl. Genet. 2002, 104, 1263–1269. [Google Scholar] [CrossRef]
- Karimi, M.; Johansson, S.; Stach, D.; Corcoran, M.; Grandér, D.; Schalling, M.; Bakalkin, G.; Lyko, F.; Larsson, C.; Ekström, T.J. LUMA (LUminometric Methylation Assay)—A high throughput method to the analysis of genomic DNA methylation. Exp. Cell Res. 2006, 312, 1989–1995. [Google Scholar] [CrossRef]
- Miura, F.; Fumihito, M. Highly sensitive targeted methylome sequencing by post-bisulfite adaptor tagging. DNA Res. 2014, 22, 13–18. [Google Scholar] [CrossRef] [Green Version]
- Bibikova, M.; Fan, J.-B. GoldenGate® Assay for DNA Methylation Profiling. Methods Mol. Biol. 2009, 507, 149–163. [Google Scholar] [CrossRef]
- Herman, J.G.; Graff, J.R.; Myohanen, S.; Nelkin, B.D.; Baylin, S.B. Methylation-specific PCR: A novel PCR assay for methylation status of CpG islands. Proc. Natl. Acad. Sci. USA 1996, 93, 9821–9826. [Google Scholar] [CrossRef] [Green Version]
- Mahapatra, S.; Klee, E.W.; Young, C.Y.; Sun, Z.; Jimenez, R.E.; Klee, G.G.; Tindall, D.J.; Donkena, K.V. Global Methylation Profiling for Risk Prediction of Prostate Cancer. Clin. Cancer Res. 2012, 18, 2882–2895. [Google Scholar] [CrossRef] [Green Version]
- Qian, X.; Shen, Q.; Goderie, S.K.; He, W.; Capela, A.; Davis, A.A.; Temple, S. Timing of CNS Cell Generation. Neuron 2000, 28, 69–80. [Google Scholar] [CrossRef] [Green Version]
- Teter, B.; Osterburg, H.H.; Anderson, C.P.; Finch, C.E. Methylation of the rat glial fibrillary acidic protein gene shows tissue-specific domains. J. Neurosci. Res. 1994, 39, 680–693. [Google Scholar] [CrossRef] [PubMed]
- Yokoyama, A.S.; Rutledge, J.C.; Medici, V. DNA methylation alterations in Alzheimer’s disease. Environ. Epigenet. 2017, 3, dvx008. [Google Scholar] [CrossRef]
- Feng, J.; Zhou, Y.; Campbell, S.L.; Le, T.; Li, E.; Sweatt, J.D.; Silva, A.; Fan, G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Lubin, F.D.; Roth, T.L.; Sweatt, J.D. Epigenetic Regulation of bdnf Gene Transcription in the Consolidation of Fear Memory. J. Neurosci. 2008, 28, 10576–10586. [Google Scholar] [CrossRef] [PubMed]
- Martinowich, K.; Hattori, D.; Wu, H.; Fouse, S.; He, F.; Hu, Y.; Fan, G.; Sun, Y.E. DNA Methylation-Related Chromatin Remodeling in Activity-Dependent Bdnf Gene Regulation. Science 2003, 302, 890–893. [Google Scholar] [CrossRef] [Green Version]
- Guo, J.U.; Ma, D.K.; Mo, H.; Ball, M.; Jang, M.-H.; Bonaguidi, M.A.; Balazer, J.A.; Eaves, H.L.; Xie, B.; Ford, E.; et al. Neuronal activity modifies the DNA methylation landscape in the adult brain. Nat. Neurosci. 2011, 14, 1345–1351. [Google Scholar] [CrossRef]
- Nguyen, S.; Meletis, K.; Fu, D.; Jhaveri, S.; Jaenisch, R. Ablation of de novo DNA methyltransferase Dnmt3a in the nervous system leads to neuromuscular defects and shortened lifespan. Dev. Dyn. 2007, 236, 1663–1676. [Google Scholar] [CrossRef] [PubMed]
- LaSalle, J.M.; Goldstine, J.; Balmer, D.; Greco, C.M. Quantitative localization of heterogeneous methyl-CpG-binding protein 2 (MeCP2) expression phenotypes in normal and Rett syndrome brain by laser scanning cytometry. Hum. Mol. Genet. 2001, 10, 1729–1740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nan, X.; Ng, H.-H.; Johnson, C.A.; Laherty, C.D.; Turner, B.M.; Eisenman, R.N.; Bird, A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 1998, 393, 386–389. [Google Scholar] [CrossRef] [PubMed]
- Sarraf, S.A.; Stancheva, I. RETRACTED: Methyl-CpG Binding Protein MBD1 Couples Histone H3 Methylation at Lysine 9 by SETDB1 to DNA Replication and Chromatin Assembly. Mol. Cell 2004, 15, 595–605. [Google Scholar] [CrossRef]
- Chen, R.Z.; Akbarian, S.; Tudor, M.; Jaenisch, R. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat. Genet. 2001, 27, 327–331. [Google Scholar] [CrossRef]
- Moretti, P.; Levenson, J.M.; Battaglia, F.; Atkinson, R.; Teague, R.; Antalffy, B.; Armstrong, D.; Arancio, O.; Sweatt, J.D.; Zoghbi, H. Learning and Memory and Synaptic Plasticity Are Impaired in a Mouse Model of Rett Syndrome. J. Neurosci. 2006, 26, 319–327. [Google Scholar] [CrossRef] [Green Version]
- Nelson, E.D.; Kavalali, E.T.; Monteggia, L.M. MeCP2-Dependent Transcriptional Repression Regulates Excitatory Neurotransmission. Curr. Biol. 2006, 16, 710–716. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ng, H.-H.; Zhang, Y.; Hendrich, B.; Johnson, C.A.; Turner, B.M.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D.; Bird, A. MBD2 is a transcriptional repressor belonging to the MeCP1 histone deacetylase complex. Nat. Genet. 1999, 23, 58–61. [Google Scholar] [CrossRef]
- Cohen, S.; Gabel, H.W.; Hemberg, M.; Hutchinson, A.N.; Sadacca, L.A.; Ebert, D.H.; Harmin, D.A.; Greenberg, R.S.; Verdine, V.K.; Zhou, Z.; et al. Genome-Wide Activity-Dependent MeCP2 Phosphorylation Regulates Nervous System Development and Function. Neuron 2011, 72, 72–85. [Google Scholar] [CrossRef] [Green Version]
- Li, B.-Z.; Huang, Z.; Cui, Q.; Song, X.-H.; Du, L.; Jeltsch, A.; Chen, P.; Li, G.; Li, E.; Xu, G.-L. Histone tails regulate DNA methylation by allosterically activating de novo methyltransferase. Cell Res. 2011, 21, 1172–1181. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Zhong, X.; Chau, K.F.; Williams, E.C.; Chang, Q. Loss of activity-induced phosphorylation of MeCP2 enhances synaptogenesis, LTP and spatial memory. Nat. Neurosci. 2011, 14, 1001–1008. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Emperle, M.; Guo, Y.; Grimm, S.A.; Ren, W.; Adam, S.; Uryu, H.; Zhang, Z.M.; Chen, D.; Yin, J.; et al. Comprehensive struc-ture-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms. Nat. Commun. 2020, 11, 3355. [Google Scholar] [CrossRef]
- Vignini, A.; Morganti, S.; Salvolini, E.; Sartini, D.; Luzzi, S.; Fiorini, R.; Provinciali, L.; Di Primio, R.; Mazzanti, L.; Emanuelli, M. Amyloid precursor protein expression is enhanced in human platelets from subjects with Alzheimer’s disease and frontotemporal lobar degeneration: A real-time PCR study. Exp. Gerontol. 2013, 48, 1505–1508. [Google Scholar] [CrossRef] [PubMed]
- Traynor, B.J.; Renton, A.E. Exploring the epigenetics of Alzheimer disease. JAMA Neurol. 2015, 72, 8–9. [Google Scholar] [CrossRef] [PubMed]
- Altuna, M.; Urdánoz-Casado, A.; Sánchez-Ruiz de Gordoa, J.; Zelaya, M.V.; Labarga, A.; Lepesant, J.; Roldán, M.; Blanco-Luquin, I.; Per-dones, Á.; Larumbe, R.; et al. DNA methylation signature of human hippocampus in Alzheimer’s disease is linked to neurogenesis. Clin. Epigenet. 2019, 11, 91. [Google Scholar] [CrossRef] [Green Version]
- Huo, Z.; Zhu, Y.; Yu, L.; Yang, J.; De Jager, P.; Bennett, D.A.; Zhao, J. DNA methylation variability in Alzheimer’s disease. Neurobiol. Aging 2019, 76, 35–44. [Google Scholar] [CrossRef]
- Tan, Y.J.; Ng, A.; Vipin, A.; Lim, J.; Chander, R.J.; Ji, F.; Qiu, Y.; Ting, S.; Hameed, S.; Lee, T.S.; et al. Higher Pe-ripheral TREM2 mRNA Levels Relate to Cognitive Deficits and Hippocampal Atrophy in Alzheimer’s Disease and Amnestic Mild Cognitive Impairment. J. Alzheimers Dis. 2017, 58, 413–423. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Ji, H.; Liu, J.; Xu, C.; Chang, L.; Cui, W.; Ye, C.; Hu, H.; Chen, Y.; Zhou, X.; et al. Association of OPRK1 and OPRM1 methylation with mild cognitive impairment in Xinjiang Han and Uygur populations. Neurosci. Lett. 2017, 636, 170–176. [Google Scholar] [CrossRef]
- Mitsumori, R.; Sakaguchi, K.; Shigemizu, D.; Mori, T.; Akiyama, S.; Ozaki, K.; Niida, S.; Shimoda, N. Lower DNA methylation levels in CpG island shores of CR1, CLU, and PICALM in the blood of Japanese Alzheimer’s disease patients. PLoS ONE 2020, 15, e0239196. [Google Scholar] [CrossRef]
- Smith, A.R.; Smith, R.G.; Macdonald, R.; Marzi, S.J.; Burrage, J.; Troakes, C.; Al-Sarraj, S.; Mill, J.; Lunnon, K. The histone modification H3K4me3 is al-tered at the ANK1 locus in Alzheimer’s disease brain. Future Sci. OA 2021, 7, FSO665. [Google Scholar] [CrossRef]
- Wang, J.; Yu, J.T.; Tan, L. PLD3 in Alzheimer’s disease. Mol. Neurobiol. 2015, 51, 480–486. [Google Scholar] [CrossRef] [PubMed]
- Masliah, E.; Dumaop, W.; Galasko, U.; Desplats, P. Distinctive patterns of DNA methylation associated with Parkinson disease: Identification of concordant epigenetic changes in brain and peripheral blood leukocytes. Epigenetics 2013, 8, 1030–1038. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Barrera, J.; Yun, Y.; Murphy, S.K.; Beach, T.G.; Woltjer, R.L.; Serrano, G.E.; Kantor, B.; Chiba-Falek, O. Cell-Type Specific Changes in DNA Methylation of SNCA Intron 1 in Synucleinopathy Brains. Front. Neurosci. 2021, 15, 652226. [Google Scholar] [CrossRef] [PubMed]
- Nasamran, C.A.; Sachan, A.N.S.; Mott, J.; Kuras, Y.I.; Scherzer, C.R.; Harvard Biomarkers Study; Ricciardelli, E.; Jepsen, K.; Edland, S.D.; Fisch, K.M.; et al. Differential blood DNA methylation across Lewy body dementias. Alzheimer’s Dement. Diagn. Assess. Dis. Monit. 2021, 13, e12156. [Google Scholar] [CrossRef] [PubMed]
- Chuang, Y.-H.; Paul, K.C.; Bronstein, J.M.; Bordelon, Y.; Horvath, S.; Ritz, B. Parkinson’s disease is associated with DNA methylation levels in human blood and saliva. Genome Med. 2017, 9, 76. [Google Scholar] [CrossRef] [PubMed]
- Young, J.I.; Sivasankaran, S.K.; Wang, L.; Ali, A.; Mehta, A.; Davis, D.A.; Dykxhoorn, D.M.; Petito, C.K.; Beecham, G.W.; Martin, E.R.; et al. Genome-wide brain DNA methylation analysis suggests epigenetic reprogramming in Parkinson disease. Neurol. Genet. 2019, 5, e342. [Google Scholar] [CrossRef] [Green Version]
- Mao, W.; Zhao, C.; Ding, H.; Liang, K.; Xue, J.; Chan, P.; Cai, Y. Pyrosequencing analysis of methylation levels of clock genes in leukocytes from Parkinson’s disease patients. Neurosci. Lett. 2018, 668, 115–119. [Google Scholar] [CrossRef]
- Mudò, G.; Mäkelä, J.; Di Liberto, V.; Tselykh, T.V.; Olivieri, M.; Piepponen, P.; Eriksson, O.; Mälkiä, A.; Bonomo, A.; Kairisalo, M.; et al. Transgenic expression and activation of PGC-1α protect dopaminergic neurons in the MPTP mouse model of Parkinson’s disease. Cell. Mol. Life Sci. 2012, 69, 1153–1165. [Google Scholar] [CrossRef]
- Kaut, O.; Schmitt, I.; Wüllner, U. Genome-scale methylation analysis of Parkinson’s disease patients’ brains reveals DNA hypomethylation and increased mRNA expression of cytochrome P450 2E1. Neurogenetics 2012, 13, 87–91. [Google Scholar] [CrossRef]
- Racette, B.A.; Searles Nielsen, S.; Criswell, S.R.; Sheppard, L.; Seixas, N.; Warden, M.N.; Checkoway, H. Dose-dependent progression of parkinsonism in manganese-exposed welders. Neurology 2017, 88, 344–351. [Google Scholar] [CrossRef] [Green Version]
- Coupland, K.G.; Kim, W.S.; Halliday, G.M.; Hallupp, M.; Dobson-Stone, C.; Kwok, J.B. Role of the Long Non-Coding RNA MAPT-AS1 in Regulation of Microtubule Associated Protein Tau (MAPT) Expression in Parkinson’s Disease. PLoS ONE 2016, 11, e0157924. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Go, R.; Corley, M.J.; Ross, G.W.; Petrovitch, H.; Masaki, K.H.; Maunakea, A.K.; He, Q.; Tiirikainen, M.I. Ge-nome-wide epigenetic analyses in Japanese immigrant plantation workers with Parkinson’s disease and exposure to orga-nochlorines reveal possible involvement of glial genes and pathways involved in neurotoxicity. BMC Neurosci. 2020, 21, 31. [Google Scholar] [CrossRef] [PubMed]
- Wood, H. Neurodegenerative disease: Altered DNA methylation and RNA splicing could be key mechanisms in Huntington dis-ease. Nature reviews. Neurology 2013, 9, 119. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Tsuji, J.; Labadorf, A.; Roussos, P.; Chen, J.F.; Myers, R.H.; Akbarian, S.; Weng, Z. The Role of H3K4me3 in Transcrip-tional Regulation Is Altered in Huntington’s Disease. PLoS ONE 2015, 10, e0144398. [Google Scholar] [CrossRef]
- Villar-Menéndez, I.; Nuñez, F.; Díaz-Sánchez, S.; Albasanz, J.L.; Taura, J.; Fernández-Dueñas, V.; Ferrer, I.; Martín, M.; Ciruela, F.; Barrachina, M. Striatal adenosine A2A receptor expression is controlled by S-adenosyl-L-methionine-mediated methylation. Purinergic Signal. 2014, 10, 523–528. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zsindely, N.; Siági, F.; Bodai, L. DNA Methylation in Huntington’s Disease. Int. J. Mol. Sci. 2021, 22, 12736. [Google Scholar] [CrossRef]
- Ng, C.W.; Yildirim, F.; Yap, Y.S.; Dalin, S.; Matthews, B.J.; Velez, P.J.; Labadorf, A.; Housman, D.E.; Fraenkel, E. Extensive changes in DNA methylation are associated with expression of mutant huntingtin. Proc. Natl. Acad. Sci. USA 2013, 110, 2354–2359. [Google Scholar] [CrossRef] [Green Version]
- Lederer, C.W.; Torrisi, A.; Pantelidou, M.; Santama, N.; Cavallaro, S. Pathways and genes differentially expressed in the motor cortex of patients with sporadic amyotrophic lateral sclerosis. BMC Genom. 2007, 8, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hurd, M.D.; Martorell, P.; Delavande, A.; Mullen, K.J.; Langa, K. Monetary Costs of Dementia in the United States. N. Engl. J. Med. 2013, 368, 1326–1334. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Yu, J.T.; Tan, M.S.; Jiang, T.; Tan, L. Epigenetic mechanisms in Alzheimer’s disease: Implications for pathogenesis and therapy. Ageing Res. Rev. 2013, 12, 1024–1041. [Google Scholar] [CrossRef] [PubMed]
- Lybartseva, G.; Smith, J.L.; Markesbery, W.R.; Lovell, M.A. Alterations of zinc transporter proteins ZnT-1, ZnT-4 and ZnT-6 in preclinical Alzheimer’s disease brain. Brain Pathol. 2010, 20, 343–350. [Google Scholar] [CrossRef] [Green Version]
- Lashley, T.; Gami, P.; Valizadeh, N.; Li, A.; Revesz, T.; Balazs, R. Alterations in global DNA methylation and hydroxymethylation are not detected in Alzheimer’s disease. Neuropathol. Appl. Neurobiol. 2015, 41, 497–506. [Google Scholar] [CrossRef]
- Wang, S.-C.; Oelze, B.; Schumacher, A. Age-Specific Epigenetic Drift in Late-Onset Alzheimer’s Disease. PLoS ONE 2008, 3, e2698. [Google Scholar] [CrossRef] [Green Version]
- Hou, Y.; Chen, H.; He, Q.; Jiang, W.; Luo, T.; Duan, J.; Mu, N.; He, Y.; Wang, H. Changes in methylation patterns of multiple genes from peripheral blood leucocytes of Alzheimer’s disease patients. Acta Neuropsychiatr. 2013, 25, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Iglesias, O.; Carrera, I.; Carril, J.C.; Fernández-Novoa, L.; Cacabelos, N.; Cacabelos, R. DNA Methylation in Neurodegenerative and Cerebrovascular Disorders. Int. J. Mol. Sci. 2020, 21, 2220. [Google Scholar] [CrossRef] [Green Version]
- LaFerla, F.M.; Green, K.N.; Oddo, S. Intracellular amyloid-β in Alzheimer’s disease. Nat. Rev. Neurosci. 2007, 8, 499–509. [Google Scholar] [CrossRef] [PubMed]
- Rogaeva, E.; Meng, Y.; Lee, J.H.; Gu, Y.; Kawarai, T.; Zou, F.; Katayama, T.; Baldwin, C.T.; Cheng, R.; Hasegawa, H.; et al. The neuronal sortilin-related receptor SORL1 is genetically associated with Alzheimer disease. Nat. Genet. 2007, 39, 168–177. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Karl, T.; Garner, B. Understanding the function of ABCA7 in Alzheimer’s disease. Biochem. Soc. Trans. 2015, 43, 920–923. [Google Scholar] [CrossRef]
- Tan, M.-S.; Yu, J.-T.; Tan, L. Bridging integrator 1 (BIN1): Form, function, and Alzheimer’s disease. Trends Mol. Med. 2013, 19, 594–603. [Google Scholar] [CrossRef]
- West, R.L.; Lee, J.M.; Maroun, L.E. Hypomethylation of the amyloid precursor protein gene in the brain of an alzheimer’s disease patient. J. Mol. Neurosci. 1995, 6, 141–146. [Google Scholar] [CrossRef] [PubMed]
- Monti, N.; Cavallaro, R.A.; Stoccoro, A.; Nicolia, V.; Scarpa, S.; Kovacs, G.G.; Fiorenza, M.T.; Lucarelli, M.; Aronica, E.; Ferrer, I.; et al. CpG and non-CpG Presenilin1 methylation pattern in course of neurodevelopment and neurodegeneration is associated with gene expression in human and murine brain. Epigenetics 2020, 15, 781–799. [Google Scholar] [CrossRef] [Green Version]
- Globisch, D.; Münzel, M.; Müller, M.; Michalakis, S.; Wagner, M.; Koch, S.; Brückl, T.; Biel, M.; Carell, T. Tissue Distribution of 5-Hydroxymethylcytosine and Search for Active Demethylation Intermediates. PLoS ONE 2010, 5, e15367. [Google Scholar] [CrossRef] [Green Version]
- Lonze, B.; Ginty, D.D. Function and Regulation of CREB Family Transcription Factors in the Nervous System. Neuron 2002, 35, 605–623. [Google Scholar] [CrossRef] [Green Version]
- Mendioroz, M.; Celarain, N.; Altuna, M.; De Gordoa, J.S.-R.; Zelaya, M.V.; Roldán, M.; Rubio, I.; Larumbe, R.; Erro, M.E.; Méndez, I.; et al. CRTC1 gene is differentially methylated in the human hippocampus in Alzheimer’s disease. Alzheimer’s Res. Ther. 2016, 8, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tekpli, X.; Landvik, N.E.; Anmarkud, K.H.; Skaug, V.; Haugen, A.; Zienolddiny, S. DNA methylation at promoter regions of interleukin 1B, interleukin 6, and interleukin 8 in non-small cell lung cancer. Cancer Immunol. Immunother. 2012, 62, 337–345. [Google Scholar] [CrossRef] [PubMed]
- Connor, B.; Young, D.; Yan, Q.; Faull, R.; Synek, B.; Dragunow, M. Brain-derived neurotrophic factor is reduced in Alzheimer’s disease. Mol. Brain Res. 1997, 49, 71–81. [Google Scholar] [CrossRef]
- Tanila, H. The role of BDNF in Alzheimer’s disease. Neurobiol. Dis. 2017, 97, 114–118. [Google Scholar] [CrossRef] [Green Version]
- Chang, L.; Wang, Y.; Danjie, J.; Dai, D.; Xu, X.; Jiang, D.; Zhongming, C.; Ye, H.; Zhang, X.; Zhou, X.; et al. Elevation of Peripheral BDNF Promoter Methylation Links to the Risk of Alzheimer’s Disease. PLoS ONE 2014, 9, e110773. [Google Scholar] [CrossRef]
- Nagata, T.; Kobayashi, N.; Ishii, J.; Shinagawa, S.; Nakayama, R.; Shibata, N.; Kuerban, B.; Ohnuma, T.; Kondo, K.; Arai, H.; et al. Association between DNA Methylation of the BDNF Promoter Region and Clinical Presentation in Alzheimer’s Disease. Dement. Geriatr. Cogn. Disord. Extra 2015, 5, 64–73. [Google Scholar] [CrossRef]
- Berson, A.; Nativio, R.; Berger, S.L.; Bonini, N.M. Epigenetic Regulation in Neurodegenerative Diseases. Trends Neurosci. 2018, 41, 587–598. [Google Scholar] [CrossRef]
- Xiao, X.; Liu, X.; Jiao, B. Epigenetics: Recent Advances and Its Role in the Treatment of Alzheimer’s Disease. Front. Neurol. 2020, 11, 538301. [Google Scholar] [CrossRef] [PubMed]
- An, S.; Khanna, K.K.; Wu, J.M. mRNA levels and methylation patterns of the 2-5A synthetase gene in control and Alzheimer’s disease (AD) fibroblasts. Biochem. Mol. Biol. Int. 1994, 33, 835–840. [Google Scholar]
- JingYun, Y.; Chibnik, L.B.; Srivastava, G.P.; Pochet, N.; Yang, J.; Xu, J.; Kozubek, J.; Obholzer, N.; Leurgans, S.E.; Schneider, J.A.; et al. Association of Brain DNA Methylation in SORL1, ABCA7, HLA-DRB5, SLC24A4, and BIN1 With Pathological Diagnosis of Alzheimer Disease. JAMA Neurol. 2015, 72, 15–24. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.L.; Tang, N.L.S.; Lam, L.C.W. Association of gene expression and methylation of UQCRC1 to the predisposition of Alzheimer’s disease in a Chinese population. J. Psychiatr. Res. 2016, 76, 143–147. [Google Scholar] [CrossRef]
- Celarain, N.; De Gordoa, J.S.-R.; Zelaya, M.V.; Roldán, M.; Larumbe, R.; Pulido, L.; Echavarri, C.; Mendioroz, M. TREM2 upregulation correlates with 5-hydroxymethycytosine enrichment in Alzheimer’s disease hippocampus. Clin. Epigenet. 2016, 8, 37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, C.; Liu, G.; Ji, H.; Chen, W.; Dai, D.; Chen, Z.; Zhou, D.; Xu, L.; Hu, H.; Cui, W.; et al. Elevated methylation of OPRM1 and OPRL1 genes in Alzheimer’s disease. Mol. Med. Rep. 2018, 18, 4297–4302. [Google Scholar] [CrossRef] [Green Version]
- Mercorio, R.; Pergoli, L.; Galimberti, D.; Favero, C.; Carugno, M.; Valle, E.D.; Barretta, F.; Cortini, F.; Scarpini, E.; Valentina, V.B.; et al. PICALM Gene Methylation in Blood of Alzheimer’s Disease Patients Is Associated with Cognitive Decline. J. Alzheimer’s Dis. 2018, 65, 283–292. [Google Scholar] [CrossRef]
- Smith, A.R.; Smith, R.G.; Burrage, J.; Troakes, C.; Al-Sarraj, S.; Kalaria, R.N.; Sloan, C.; Robinson, A.; Mill, J.; Lunnon, K. A cross-brain regions study of ANK1 DNA methylation in different neurodegenerative diseases. Neurobiol. Aging 2018, 74, 70–76. [Google Scholar] [CrossRef] [Green Version]
- Blanco-Luquin, I.; Altuna, M.; De Gordoa, J.S.-R.; Casado, A.U.; Roldán, M.; Cámara, M.; Zelaya, V.; Erro, M.E.; Echavarri, C.; Mendioroz, M. PLD3 epigenetic changes in the hippocampus of Alzheimer’s disease. Clin. Epigenet. 2018, 10, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, R.G.; Hannon, E.; De Jager, P.L.; Chibnik, L.; Lott, S.J.; Condliffe, D.; Smith, A.R.; Haroutunian, V.; Troakes, C.; Al-Sarraj, S.; et al. Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimer’s Dement. J. Alzheimer’s Assoc. 2018, 14, 1580–1588. [Google Scholar] [CrossRef]
- Bernstein, A.; Lin, Y.; Street, R.C.; Lin, L.; Dai, Q.; Yu, L.; Bao, H.; Gearing, M.; Lah, J.J.; Nelson, P.T.; et al. 5-Hydroxymethylation-associated epigenetic modifiers of Alzheimer’s disease modulate Tau-induced neurotoxicity. Hum. Mol. Genet. 2016, 25, 2437–2450. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Zhu, Y.; Yang, J.; Li, L.; Wu, H.; De Jager, P.L.; Jin, P.; Bennett, D.A. A genome-wide profiling of brain DNA hydroxymethylation in Alzheimer’s disease. Alzheimer’s Dement. 2017, 13, 674–688. [Google Scholar] [CrossRef]
- Blanch, M.; Mosquera, J.L.; Ansoleaga, B.; Ferrer, I.; Barrachina, M. Altered Mitochondrial DNA Methylation Pattern in Alzheimer Disease–Related Pathology and in Parkinson Disease. Am. J. Pathol. 2016, 186, 385–397. [Google Scholar] [CrossRef] [Green Version]
- Miranda-Morales, E.; Meier, K.; Carrillo, A.S.; Pacheco, J.S.; Vazquez, P.; Arias-Carrión, O. Implications of DNA Methylation in Parkinson’s Disease. Front. Mol. Neurosci. 2017, 10, 225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mishra, A.; Mishra, P.S.; Bandopadhyay, R.; Khurana, N.; Angelopoulou, E.; Paudel, Y.N.; Piperi, C. Neuroprotective Potential of Chrysin: Mechanistic Insights and Therapeutic Potential for Neurological Disorders. Molecules 2021, 26, 6456. [Google Scholar] [CrossRef] [PubMed]
- Angelopoulou, E.; Paudel, Y.N.; Piperi, C.; Mishra, A. Neuroprotective potential of cinnamon and its metabolites in Parkinson’s disease: Mechanistic insights, limitations, and novel therapeutic opportunities. J. Biochem. Mol. Toxicol. 2021, 35, e22711. [Google Scholar] [CrossRef]
- Singleton, A.B.; Farrer, M.; Johnson, J.; Singleton, A.; Hague, S.; Kachergus, J.; Hulihan, M.; Peuralinna, T.; Dutra, A.; Nussbaum, R.; et al. alpha-Synuclein Locus Triplication Causes Parkinson’s Disease. Science 2003, 302, 841. [Google Scholar] [CrossRef] [Green Version]
- Chartier-Harlin, M.-C.; Kachergus, J.; Roumier, C.; Mouroux, V.; Douay, X.; Lincoln, S.; Levecque, C.; Larvor, L.; Andrieux, J.; Hulihan, M.; et al. α-synuclein locus duplication as a cause of familial Parkinson’s disease. Lancet 2004, 364, 1167–1169. [Google Scholar] [CrossRef]
- Matsumoto, L.; Takuma, H.; Tamaoka, A.; Kurisaki, H.; Date, H.; Tsuji, S.; Iwata, A. CpG Demethylation Enhances Alpha-Synuclein Expression and Affects the Pathogenesis of Parkinson’s Disease. PLoS ONE 2010, 5, e15522. [Google Scholar] [CrossRef] [Green Version]
- Guhathakurta, S.; Evangelista, B.A.; Ghosh, S.; Basu, S.; Kim, Y.S. Hypomethylation of intron1 of α-synuclein gene does not correlate with Parkinson’s disease. Mol. Brain 2017, 10, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Boni, L.; Tierling, S.; Roeber, S.; Walter, J.; Giese, A.; Kretzschmar, H.A. Next-Generation Sequencing Reveals Regional Differences of the α-Synuclein Methylation State Independent of Lewy Body Disease. Neuromol. Med. 2011, 13, 310–320. [Google Scholar] [CrossRef]
- Funahashi, Y.; Yoshino, Y.; Yamazaki, K.; Mori, Y.; Mori, T.; Ozaki, Y.; Sao, T.; Ochi, S.; Iga, J.-I.; Ueno, S.-I. DNA methylation changes atSNCAintron 1 in patients with dementia with Lewy bodies. Psychiatry Clin. Neurosci. 2016, 71, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshino, Y.; Mori, T.; Yoshida, T.; Yamazaki, K.; Ozaki, Y.; Sao, T.; Funahashi, Y.; Iga, J.-I.; Ueno, S.-I. Elevated mRNA Expression and Low Methylation of SNCA in Japanese Alzheimer’s Disease Subjects. J. Alzheimer’s Dis. 2016, 54, 1349–1357. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-Y.; Lansbury, J.P.T. β-Synuclein Inhibits Formation of α-Synuclein Protofibrils: A Possible Therapeutic Strategy against Parkinson’s Disease. Biochemistry 2003, 42, 3696–3700. [Google Scholar] [CrossRef]
- Beyer, K.; Domingo-Sàbat, M.; Santos, C.; Tolosa, E.; Ferrer, I.; Ariza, A. The decrease of β-synuclein in cortical brain areas defines a molecular subgroup of dementia with Lewy bodies. Brain 2010, 133, 3724–3733. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jang, B.; Ishigami, A.; Maruyama, N.; Carp, R.I.; Kim, Y.-S.; Choi, E.-K. Peptidylarginine deiminase and protein citrullination in prion diseases. Prion 2013, 7, 42–46. [Google Scholar] [CrossRef] [Green Version]
- Barrachina, M.; Ferrer, I. DNA Methylation of Alzheimer Disease and Tauopathy-Related Genes in Postmortem Brain. J. Neuropathol. Exp. Neurol. 2009, 68, 880–891. [Google Scholar] [CrossRef]
- Behrens, M.I.; Brüggemann, N.; Chana, P.; Venegas, P.; Kägi, M.; Parrao, T.; Orellana, P.; Garrido, C.; Rojas, C.V.; Hauke, J.; et al. Clinical spectrum of Kufor-Rakeb syndrome in the Chilean kindred with ATP13A2 mutations. Mov. Disord. 2010, 25, 1929–1937. [Google Scholar] [CrossRef]
- Cai, M.; Tian, J.; Zhao, G.-H.; Luo, W.; Zhang, B.-R. Study of Methylation Levels of Parkin Gene Promoter in Parkinson’s Disease Patients. Int. J. Neurosci. 2011, 121, 497–502. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Liu, S.; Sothern, R.B.; Xu, S.; Chan, P. Expression of clock genesPer1andBmal1in total leukocytes in health and Parkinson’s disease. Eur. J. Neurol. 2009, 17, 550–554. [Google Scholar] [CrossRef]
- Lin, Q.; Ding, H.; Zheng, Z.; Gu, Z.; Ma, J.; Chen, L.; Chan, P.; Cai, Y. Promoter methylation analysis of seven clock genes in Parkinson’s disease. Neurosci. Lett. 2012, 507, 147–150. [Google Scholar] [CrossRef]
- Simón-Sánchez, J.; Schulte, C.; Bras, J.M.; Sharma, M.; Gibbs, J.R.; Berg, D.; Paisan-Ruiz, C.; Lichtner, P.; Scholz, S.W.; Hernandez, D.G.; et al. Genome-wide association study reveals genetic risk underlying Parkinson’s disease. Nat. Genet. 2009, 41, 1308–1312. [Google Scholar] [CrossRef]
- Bsc, K.G.C.; Mellick, G.; Silburn, P.A.; Mather, K.; Armstrong, N.J.; Sachdev, P.; Brodaty, H.; Huang, Y.; Halliday, G.; Hallupp, M.; et al. DNA methylation of theMAPTgene in Parkinson’s disease cohorts and modulation by vitamin EIn Vitro. Mov. Disord. 2013, 29, 1606–1614. [Google Scholar] [CrossRef] [Green Version]
- Kwok, J.B.J.; Teber, E.T.; Loy, C.; Hallupp, M.; Nicholson, G.; Mellick, G.D.; Buchanan, D.D.; Silburn, P.A.; Schofield, P.R. Tau haplotypes regulate transcription and are associated with Parkinson’s disease. Ann. Neurol. 2004, 55, 329–334. [Google Scholar] [CrossRef]
- Tsunemi, T.; La Spada, A.R. PGC-1α at the intersection of bioenergetics regulation and neuron function: From Huntington’s disease to Parkinson’s disease and beyond. Prog. Neurobiol. 2012, 97, 142–151. [Google Scholar] [CrossRef] [Green Version]
- Tan, Y.; Wu, L.; Li, D.; Liu, X.; Ding, J.; Chen, S. Methylation status of DJ-1 in leukocyte DNA of Parkinson’s disease patients. Transl. Neurodegener. 2016, 5, 5. [Google Scholar] [CrossRef] [Green Version]
- Moore, K.; McKnight, A.J.; Craig, D.; O’Neill, F.A. Epigenome-Wide Association Study for Parkinson’s Disease. Neuromol. Med. 2014, 16, 845–855. [Google Scholar] [CrossRef] [PubMed]
- Wüllner, U.; Kaut, O.; Deboni, L.; Piston, D.; Schmitt, I. DNA methylation in Parkinson’s disease. J. Neurochem. 2016, 139, 108–120. [Google Scholar] [CrossRef] [Green Version]
- Madrid, A.; Hogan, K.J.; Papale, L.A.; Clark, L.R.; Asthana, S.; Johnson, S.C.; Alisch, R.S. DNA Hypomethylation in Blood Links B3GALT4 and ZADH2 to Alzheimer’s Disease. J. Alzheimer’s Dis. 2018, 66, 927–934. [Google Scholar] [CrossRef] [PubMed]
- Hernández, H.G.; Sandoval-Hernández, A.G.; Garrido-Gil, P.; Labandeira-Garcia, J.L.; Zelaya, M.V.; Bayon, G.F.; Fernández, A.F.; Fraga, M.F.; Arboleda, G.; Arboleda, H. Alzheimer’s disease DNA methylome of pyramidal layers in frontal cortex: Laser-assisted microdissection study. Epigenomics 2018, 10, 1365–1382. [Google Scholar] [CrossRef]
- Andrés-Benito, P.; Delgado-Morales, R.; Ferrer, I. Altered Regulation of KIAA0566, and Katanin Signaling Expression in the Locus Coeruleus With Neurofibrillary Tangle Pathology. Front. Cell. Neurosci. 2018, 12, 131. [Google Scholar] [CrossRef]
- Miao, L.; Yin, R.-X.; Zhang, Q.-H.; Hu, X.-J.; Huang, F.; Chen, W.-X.; Cao, X.-L.; Wu, J.-Z. Integrated DNA methylation and gene expression analysis in the pathogenesis of coronary artery disease. Aging 2019, 11, 1486–1500. [Google Scholar] [CrossRef]
- Smith, A.; Smith, R.G.; Pishva, E.; Hannon, E.; Roubroeks, J.A.Y.; Burrage, J.; Troakes, C.; Al-Sarraj, S.; Sloan, C.; Mill, J.; et al. Parallel profiling of DNA methylation and hydroxymethylation highlights neuropathology-associated epigenetic variation in Alzheimer’s disease. Clin. Epigenet. 2019, 11, 52. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Marshall, L.; Oh, G.; Jakubowski, J.L.; Groot, D.; He, Y.; Wang, T.; Petronis, A.; Labrie, V. Epigenetic dysregulation of enhancers in neurons is associated with Alzheimer’s disease pathology and cognitive symptoms. Nat. Commun. 2019, 10, 2246. [Google Scholar] [CrossRef] [Green Version]
- Chouliaras, L.; Mastroeni, D.; Delvaux, E.; Grover, A.; Kenis, G.; Hof, P.R.; Steinbusch, H.W.; Coleman, P.D.; Rutten, B.; Hove, D.L.V.D. Consistent decrease in global DNA methylation and hydroxymethylation in the hippocampus of Alzheimer’s disease patients. Neurobiol. Aging 2013, 34, 2091–2099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradley-Whitman, M.; Lovell, M. Epigenetic changes in the progression of Alzheimer’s disease. Mech. Ageing Dev. 2013, 134, 486–495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coppieters, N.; Dieriks, B.; Lill, C.; Faull, R.; Curtis, M.; Dragunow, M. Global changes in DNA methylation and hydroxymethylation in Alzheimer’s disease human brain. Neurobiol. Aging 2014, 35, 1334–1344. [Google Scholar] [CrossRef] [PubMed]
- Pishva, E.; Creese, B.; Smith, A.R.; Viechtbauer, W.; Proitsi, P.; Hove, D.L.V.D.; Ballard, C.; Mill, J.; Lunnon, K. Psychosis-associated DNA methylomic variation in Alzheimer’s disease cortex. Neurobiol. Aging 2020, 89, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Phipps, A.J.; Vickers, J.C.; Taberlay, P.C.; Woodhouse, A. Neurofilament-labeled pyramidal neurons and astrocytes are deficient in DNA methylation marks in Alzheimer’s disease. Neurobiol. Aging 2016, 45, 30–42. [Google Scholar] [CrossRef] [PubMed]
- Henry, C.J.; Huang, Y.; Wynne, A.M.; Godbout, J.P. Peripheral lipopolysaccharide (LPS) challenge promotes microglial hyperactivity in aged mice that is associated with exaggerated induction of both pro-inflammatory IL-1β and anti-inflammatory IL-10 cytokines. Brain Behav. Immun. 2009, 23, 309–317. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jowaed, A.; Schmitt, I.; Kaut, O.; Wüllner, U. Methylation Regulates Alpha-Synuclein Expression and Is Decreased in Parkinson’s Disease Patients’ Brains. J. Neurosci. 2010, 30, 6355–6359. [Google Scholar] [CrossRef]
- Su, X.; Chu, Y.; Kordower, J.H.; Li, B.; Cao, H.; Huang, L.; Nishida, M.; Song, L.; Wang, D.; Federoff, H.J. PGC−1α Promoter Methylation in Parkinson’s Disease. PLoS ONE 2015, 10, e0134087. [Google Scholar] [CrossRef] [PubMed]
- Pieper, H.C.; Evert, B.O.; Kaut, O.; Riederer, P.F.; Waha, A.; Wüllner, U. Different methylation of the TNF-alpha promoter in cortex and substantia nigra: Implications for selective neuronal vulnerability. Neurobiol. Dis. 2008, 32, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, S.S.; Checkoway, H.; Criswell, S.R.; Farin, F.M.; Stapleton, P.L.; Sheppard, L.; Racette, B.A. Inducible nitric oxide synthase gene methylation and parkinsonism in manganese-exposed welders. Park. Relat. Disord. 2015, 21, 355–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.; Hwang, Y.J.; Kim, K.Y.; Kowall, N.W.; Ryu, H. Epigenetic Mechanisms of Neurodegeneration in Huntington’s Disease. Neurotherapeutics 2013, 10, 664–676. [Google Scholar] [CrossRef] [PubMed]
- Thomas, E.A. DNA methylation in Huntington’s disease: Implications for transgenerational effects. Neurosci. Lett. 2015, 625, 34–39. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sugars, K.L.; Rubinsztein, D.C. Transcriptional abnormalities in Huntington disease. Trends Genet. 2003, 19, 233–238. [Google Scholar] [CrossRef]
- Stack, E.C.; Del Signore, S.J.; Luthi-Carter, R.; Soh, B.Y.; Goldstein, D.R.; Matson, S.; Goodrich, S.; Markey, A.L.; Cormier, K.; Hagerty, S.W.; et al. Modulation of nucleosome dynamics in Huntington’s disease. Hum. Mol. Genet. 2007, 16, 1164–1175. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Yang, Y.; Lin, X.; Wang, J.-Q.; Wu, Y.-S.; Xie, W.; Wang, D.; Zhu, S.; Liao, Y.-Q.; Sun, Q.; et al. Genome-wide loss of 5-hmC is a novel epigenetic feature of Huntington’s disease. Hum. Mol. Genet. 2013, 22, 3641–3653. [Google Scholar] [CrossRef]
- Bai, G.; Cheung, I.; Shulha, H.P.; Coelho, J.; Li, P.; Dong, X.; Jakovcevski, M.; Wang, Y.; Grigorenko, A.; Jiang, Y.; et al. Epigenetic dysregulation of hairy and enhancer of split 4 (HES4) is associated with striatal degeneration in postmortem Huntington brains. Hum. Mol. Genet. 2014, 24, 1441–1456. [Google Scholar] [CrossRef] [Green Version]
- Villar-Menéndez, I.; Blanch, M.; Tyebji, S.; Pereira-Veiga, T.; Albasanz, J.L.; Martín, M.; Ferrer, I.; Perez-Navarro, E.; Barrachina, M. Increased 5-Methylcytosine and Decreased 5-Hydroxymethylcytosine Levels are Associated with Reduced Striatal A2AR Levels in Huntington’s Disease. Neuromol. Med. 2013, 15, 295–309. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Seredenina, T.; Coppola, G.; Kuhn, A.; Geschwind, D.H.; Luthi-Carter, R.; Thomas, E.A. Gene expression profiling of R6/2 transgenic mice with different CAG repeat lengths reveals genes associated with disease onset and progression in Huntington’s disease. Neurobiol. Dis. 2011, 42, 459–467. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zadel, M.; Maver, A.; Kovanda, A.; Peterlin, B. DNA Methylation Profiles in Whole Blood of Huntington’s Disease Patients. Front. Neurol. 2018, 9, 655. [Google Scholar] [CrossRef]
- Figueroa-Romero, C.; Hur, J.; Bender, D.E.; Delaney, C.E.; Cataldo, M.D.; Smith, A.L.; Yung, R.; Ruden, D.M.; Callaghan, B.C.; Feldman, E.L. Identification of Epigenetically Altered Genes in Sporadic Amyotrophic Lateral Sclerosis. PLoS ONE 2012, 7, e52672. [Google Scholar] [CrossRef]
- Chestnut, B.A.; Chang, Q.; Price, A.; Lesuisse, C.; Wong, M.; Martin, L.J. Epigenetic Regulation of Motor Neuron Cell Death through DNA Methylation. J. Neurosci. 2011, 31, 16619–16636. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, H.; Dzitoyeva, S.; Manev, H. Effect of aging on 5-hydroxymethylcytosine in the mouse hippocampus. Restor. Neurol. Neurosci. 2012, 30, 237–245. [Google Scholar] [CrossRef] [Green Version]
- Eklow, C.; Makrygiannakis, D.; Backdahl, L.; Padyukov, L.; Ulfgren, A.-K.; Lorentzen, J.C.; Malmstrom, V. Cellular distribution of the C-type II lectin dendritic cell immunoreceptor (DCIR) and its expression in the rheumatic joint: Identification of a subpopulation of DCIR+ T cells. Ann. Rheum. Dis. 2008, 67, 1742–1749. [Google Scholar] [CrossRef] [PubMed]
- Piccio, L.; Buonsanti, C.; Mariani, M.; Cella, M.; Gilfillan, S.; Cross, A.H.; Colonna, M.; Panina-Bordignon, P. Blockade of TREM-2 exacerbates experimental autoimmune encephalomyelitis. Eur. J. Immunol. 2007, 37, 1290–1301. [Google Scholar] [CrossRef]
- Ramji, D.; Vitelli, A.; Tronche, F.; Cortese, R.; Ciliberto, G. The two C/EBP isoforms, IL6DBP/NFIL6 and CEBP6δ/NFIL63, are induced by IL6β to promote acute phase gene transcription via different mechanisms. Nucleic Acids Res. 1993, 21, 289–294. [Google Scholar] [CrossRef] [Green Version]
- Bacon, C.; Endris, V.; Rappold, G. Dynamic expression of the Slit-Robo GTPase activating protein genes during development of the murine nervous system. J. Comp. Neurol. 2009, 513, 224–236. [Google Scholar] [CrossRef]
- Pellikka, M.; Tanentzapf, G.; Pinto, M.; Smith, C.T.; McGlade, C.J.; Ready, D.F.; Tepass, U. Crumbs, the Drosophila homologue of human CRB1/RP12, is essential for photoreceptor morphogenesis. Nature 2002, 416, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Inoue, K.-I.; Shiga, T.; Ito, Y. Runx transcription factors in neuronal development. Neural Dev. 2008, 3, 20. [Google Scholar] [CrossRef] [Green Version]
- Stoccoro, A.; Mosca, L.; Carnicelli, V.; Cavallari, U.; Lunetta, C.; Marocchi, A.; Migliore, L.; Coppedè, F. Mitochondrial DNA copy number and D-loop region methylation in carriers of amyotrophic lateral sclerosis gene mutations. Epigenomics 2018, 10, 1431–1443. [Google Scholar] [CrossRef] [PubMed]
- Teijido, O.; Cacabelos, R. Pharmacoepigenomic Interventions as Novel Potential Treatments for Alzheimer’s and Parkinson’s Diseases. Int. J. Mol. Sci. 2018, 19, 3199. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, X.; Li, R.; Yang, Z.-F.; Wang, Y.-Z.; Gong, X.-L.; Wang, X.-M. A DNA Methyltransferase Inhibitor, 5-Aza-2′-Deoxycytidine, Exacerbates Neurotoxicity and Upregulates Parkinson’s Disease-Related Genes in Dopaminergic Neurons. CNS Neurosci. Ther. 2013, 19, 183–190. [Google Scholar] [CrossRef]
- Pan, Y.; Daito, T.; Sasaki, Y.; Chung, Y.H.; Xing, X.; Pondugula, S.; Swamidass, S.J.; Wang, T.; Kim, A.H.; Yano, H. Inhibition of DNA Methyltransferases Blocks Mutant Huntingtin-Induced Neurotoxicity. Sci. Rep. 2016, 6, 31022. [Google Scholar] [CrossRef] [PubMed]
- Martin, L.J.; Wong, M. Aberrant Regulation of DNA Methylation in Amyotrophic Lateral Sclerosis: A New Target of Disease Mechanisms. Neurotherapeutics 2013, 10, 722–733. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Santiago, R.; Ezquerra, M. Epigenetic Research of Neurodegenerative Disorders Using Patient iPSC-Based Models. Stem Cells Int. 2015, 2016, 9464591. [Google Scholar] [CrossRef]
- Salameh, Y.; Bejaoui, Y.; El Hajj, N. DNA Methylation Biomarkers in Aging and Age-Related Diseases. Front. Genet. 2020, 11, 171. [Google Scholar] [CrossRef]
- Delgado-Morales, R.; Esteller, M. Opening up the DNA methylome of dementia. Mol. Psychiatry 2017, 22, 485–496. [Google Scholar] [CrossRef]
- Armstrong, M.J.; Jin, Y.; Allen, E.G.; Jin, P. Diverse and dynamic DNA modifications in brain and diseases. Hum. Mol. Genet. 2019, 28, R241–R253. [Google Scholar] [PubMed]

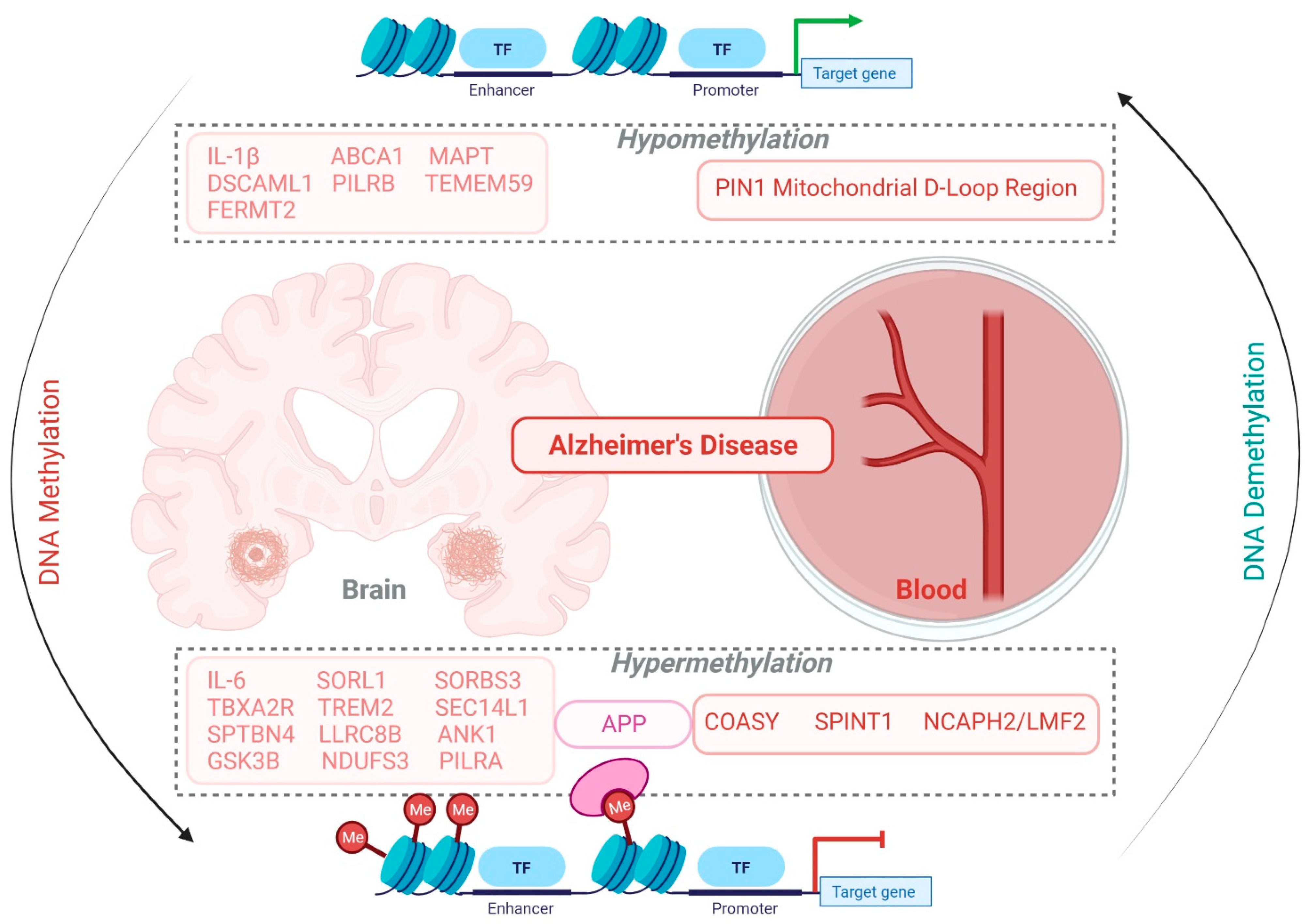
| Disease | Sample | Methylation (Hyper/Hypo) | Experimental Method | Gene | Ref. |
|---|---|---|---|---|---|
| AD | Blood | Hypermethylation | Bisulphite sequencing PCR and methylation-specific PCR are used | SIRT1 | [53] |
| AD | Dorsolateral prefrontal cortex tissue | Differently methylated | CpG sites generated using a bead assay | SORL1, ABCA7, HLA-DRB5, SLC24A4, BIN1. | [54] |
| AD | Hippocampus | Hypomethylation | Bisulfite cloning sequencing of CpG sites in two promoter regions Prom1 and Prom2 | CREB-regulated transcription factor 1 | [55] |
| AD | Blood | Hypermethylation | Bisulfite treated DNA was analyzed by melting curve analysis-methylation assay | UQCRC1 | [56] |
| AD | Hippocampus | Hypermethylation | Bisulfite cloning sequencing and further measured by 5-hydroxymethycytosine (5hmC) | TREM2 | [57] |
| AD | Blood | Hypermethylation | Dual-luciferase assays | OPRM1, OPRL1 | [58] |
| AD | Blood | Hypomethylation | Quantitative bisulfite-PCR pyrosequencing | PICALM | [59] |
| AD | Brain | Hypermethylation | Bisulfite pro-sequencing | ANK1 gene | [60] |
| AD | Hippocampus | Hypermethylation | RT-qPCR | PLD3 gene | [61] |
| PD | Postmortem human brain samples (frontal cortex) | Hypermethylation | Illumina Infinium array | MRI1, TMEM9 | [62] |
| PD | Postmortem human brain samples (frontal cortex) | Hypomethylation | Illumina Infinium array | GSST1, TUBA3E, KCNH1 | [62] |
| PD | Brain tissue | Hypomethylation | Fluorescence-activated nuclei sorting and bisulfite pro-sequencing | CpGs located in SNCA intron 1 | [63] |
| PD | Blood | Differently methylated | Cross-sectional analysis of blood methylation | SRSF7, ADNP, GDNF, SYN3, CPLX1, SNCA, TREM2. | [64] |
| PD | Blood and saliva | Altered methylation | Illumina Infinium array | ABCB9, C1orf200, AZU1, LARS2, PARK2, LRRK2, APC, AXIN1 | [65] |
| PD | Brain | Differently methylated | Genome wide screening and RNA sequencing | ARFGAP1, DUSP22 promoter, SNCA | [66] |
| PD | Leukocytes | Hypomethylation | Methylation-specific PCR | NPAS2 | [67] |
| PD | Brain | Hypermethylation | Bisulfite sequencing and micro array gene expression analysis | PGC1-α | [68] |
| PD | Brain | Hypomethylation | Genome wide methylation | CYP2E1 | [69] |
| PD | Blood | Hypomethylation | - | NOS2 | [70] |
| PD | Leukocytes, Brain | Hypermethylation | Bisulfite pyrosequencing and MAPT promoter methylation assay | MAPT | [71] |
| PD | Brain | Hypermethylation | Illumina Infinium array | FANCC/TNKS2 | [72] |
| HD | Striatal cells carrying polyglutamine-expanded HTT (STHdhQ111/Q111) and wild-type cells (STHdhQ7/Q7) | Altered DNA methylation | mRNA-Seq, ChIP-Seq assay and Motif Scanning | Htt | [73] |
| HD | Prefrontal cortex | Differently methylated | Fluorescence-based nuclei sorting (FACS)-ChIP-seq | HES4 | [74] |
| HD | Putamen of HD patients and striatum of mice | Differently methylated | Bisulfite sequencing and TaqMan PCR | ADORA2A | [75] |
| HD | Blood | Differently methylated | Microarray methylation | CLDN16, NXT2, DDC. | [76] |
| HD | Blood | Differently methylated | mRNA-Seq, ChIP-Seq assay and motif scanning | FBXL5, S100P, PRDX1, COPS7B, SP1, SEC24C, PDIA6, USP5, GRAP, POP5, WRB, PCSK7. | [77] |
| ALS | Postmortem spinal cord tissue | Hypomethylation | Bisulfite pyrosequencing, genome-wide expression profiling, and RT-PCR | MLC1, CRB1, CTNND2, FURIN, SLC31A1, CMTM3, STAT5A, SRGAP1, LPXN, PLD4, OBFC2A, TXNIP, PSAP, SLC35E1, RBM38, CLEC4A, HMHA1, PLSCR1, AXL, PHYHD1. | [78] |
| ALS | Postmortem spinal cord tissue | Hypermethylation | Bisulfite pyrosequencing, genome wide expression profiling, and RT-PCR | LUM, SLC13A4, GJB2, TYRP1, CLDN19, LINGO2, PLEKHA4, NNAT, TSPAN18, PLCB4, TMEM139, PNMAL1, DMBT1, TNFSF10, NNAT, PCP4, MAB21L2, PEG10, TMEM139, KCNJ12, FGF18. | [78] |
| Methylation of DNA | Gene/Target/Pathway Involved | Effect | Model | Experimental Method | Outcomes | Ref. |
|---|---|---|---|---|---|---|
| 5-mC | B3GALT4, ZADH2 | Decrease | AD and healthy patients | Rey Auditory Verbal Learning Test (RAVLT), Trail Making Test Part B (TMT-B), INNOTEST assays, and Triplex assay | Hypomethylation of B3GALT4, ZADH2 associated with the level of AB and tau in CSF | [139] |
| 5-mC | HOXA3, GSTP1, CXXC1-3, BIN1 | Increase | AD and healthy patients | Laser-assisted microdissection and Infinium DNA Methylation 450K analysis | 504 DMCs and 237 DMRs were identified and increased in the 5mC pyramidal layer, which is associated with oxidative stress | [140] |
| 5-mC | KIAA056 | Decrease | NFT pathology stages I-IV | Bisulfite sequencing and Infinium Human Methylation 450 BeadChip | Downregulation of 5mC in KIAA056 and in NFT pathology cases | [141] |
| 5-mC | ANKRD30B, ANK1, Cell adhesion | Increase | AD and neurotypical patients | Genome-wide DNA methylation, mRNA expression profiling, functional enrichment analysis, and differential methylation of genes | 856 DMCs were identified along with a correlation between 5-mC and gene expression | [142] |
| 5-mC | WNT5B, ANK1, ARD5B | Increase and decrease | AD patients | Illumina Infinium Human Methylation 450K microarray | Increased 5-mC level in WNT5B, ANK1, and decreased in ARD5Bz | [143] |
| 5-mC | Amyloid neuropathy and neurogenesis | Decrease | AD and healthy patients | RNA sequencing, aging analysis, gene annotation, and enrichment analysis | Identification of 1224 DMRs, enhancement in the DCSAML1 gene which targets BACE1 | [144] |
| 5-mC | - | Decrease | AD and healthy patients | Immunohistochemistry | Downregulation of 5-mC and negative correlation between 5mC and amyloid plaque level | [145] |
| 5-mC | - | Increase | AD patients and preclinical samples | Immunohistochemistry | Upregulation of 5-mC and hippocampus gyrus in both clinical and preclinical cases | [146] |
| 5-mC | - | Increase | Early and late-onset AD patients | Immunohistochemistry | Upregulation of 5mC in middle frontal gyrus and middle temporal gyrus in AD patients and shows a positive correlation with AD biomarkers | [147] |
| 5-mC | AS3MT, WTI, TBX15 | Decrease | AD with psychosis and without psychosis patients | Immunohistochemistry | Decrease level of AS3MT, WTI, TBX15 gene associated with AD patients | [148] |
| 5-mC | - | Decrease | Early and late AD patients | Immunohistochemistry | Genetic dysregulation may be occurring in astrocytes and NF-positive pyramidal neurons in AD | [149] |
| IL-1β Promoters | IL-1β | Decrease | BALB/c mice (3–4- and 18–20-month-old) | LPS-induced neuroinflammation and Quantitative PCR (qPCR) | Microglial transferred to M1 phenotype which causes neuroinflammation and neuronal cell damages | [150] |
| SNCA Promoters | SNCA | Decrease | Healthy and PD patients | qPCR | Aggregation of a-syn, neuronal damage of DA, and neuroinflammation is triggered by activating glial cells | [151] |
| PGC-1α Promoters | PGC-1α | Increase | Human brain of PD and healthy patients | Bisulfite sequencing, Microarray gene expression analysis, ELISA analysis | Up-regulation of neuroinflammation, ER stress, epigenetic modification, and ROS production | [152] |
| TNF-α Promoters | TNF-alpha | Decrease | PD and healthy patients | Bisulfite PCR and sequencing | SNpc cells could underlie the increased susceptibility of dopaminergic neurons to TNF-alpha-mediated inflammatory reactions. | [153] |
| NOS2 Promoters | NOS2 | Decrease | PD and healthy patients | Qiagen’s Assay | Down-regulation of NO production to deactivate the microglial | [154] |
| Neurodegenerative Disease | Drug | Class of Drug | Inference | Reference |
|---|---|---|---|---|
| AD | Epigallocatechin gallate, epigallocatechin 3-gallate, tea catechin, tea vigo, catechin deriv., | DNMT inhibitors | Improve memory, prevent cell death in Aβ-treated neurons, Aβ aggregation. | [174] |
| AD | Vitamin B6, folate, Folacin; Pteroylglutamic acid | SAMe methyl donors | Attenuate homocystine level | [174] |
| PD | 5-Aza-2′-Deoxycytidine | DNMTs inhibitor | Upregulate tyrosine hydroxylase, dopamine production, and alpha-synuclein expression | [175] |
| HD | decitabine and FdCyd | DNMTs inhibitors | Restore expression of Bndf | [176] |
| ALS | RG108 | DNMTs inhibitors | Block DNA methylation accumulation in motor neurons | [177] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaur, G.; Rathod, S.S.S.; Ghoneim, M.M.; Alshehri, S.; Ahmad, J.; Mishra, A.; Alhakamy, N.A. DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders. Biology 2022, 11, 90. https://doi.org/10.3390/biology11010090
Kaur G, Rathod SSS, Ghoneim MM, Alshehri S, Ahmad J, Mishra A, Alhakamy NA. DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders. Biology. 2022; 11(1):90. https://doi.org/10.3390/biology11010090
Chicago/Turabian StyleKaur, Gagandeep, Suraj Singh S. Rathod, Mohammed M. Ghoneim, Sultan Alshehri, Javed Ahmad, Awanish Mishra, and Nabil A. Alhakamy. 2022. "DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders" Biology 11, no. 1: 90. https://doi.org/10.3390/biology11010090
APA StyleKaur, G., Rathod, S. S. S., Ghoneim, M. M., Alshehri, S., Ahmad, J., Mishra, A., & Alhakamy, N. A. (2022). DNA Methylation: A Promising Approach in Management of Alzheimer’s Disease and Other Neurodegenerative Disorders. Biology, 11(1), 90. https://doi.org/10.3390/biology11010090








