The Mitogenome of Sedum plumbizincicola (Crassulaceae): Insights into RNA Editing, Lateral Gene Transfer, and Phylogenetic Implications
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling, DNA Extraction, and Sequencing
2.2. Mitogenome Assembly, and Gene Annotation
2.3. RNA Editing Sites Identification and Codon Usage Analysis
2.4. Identification of Gene Transfer
2.5. Structure Prediction of tRNAs and rRNAs
2.6. Mitophylogenetic Analysis
3. Results
3.1. Genome Features of Mitogenome
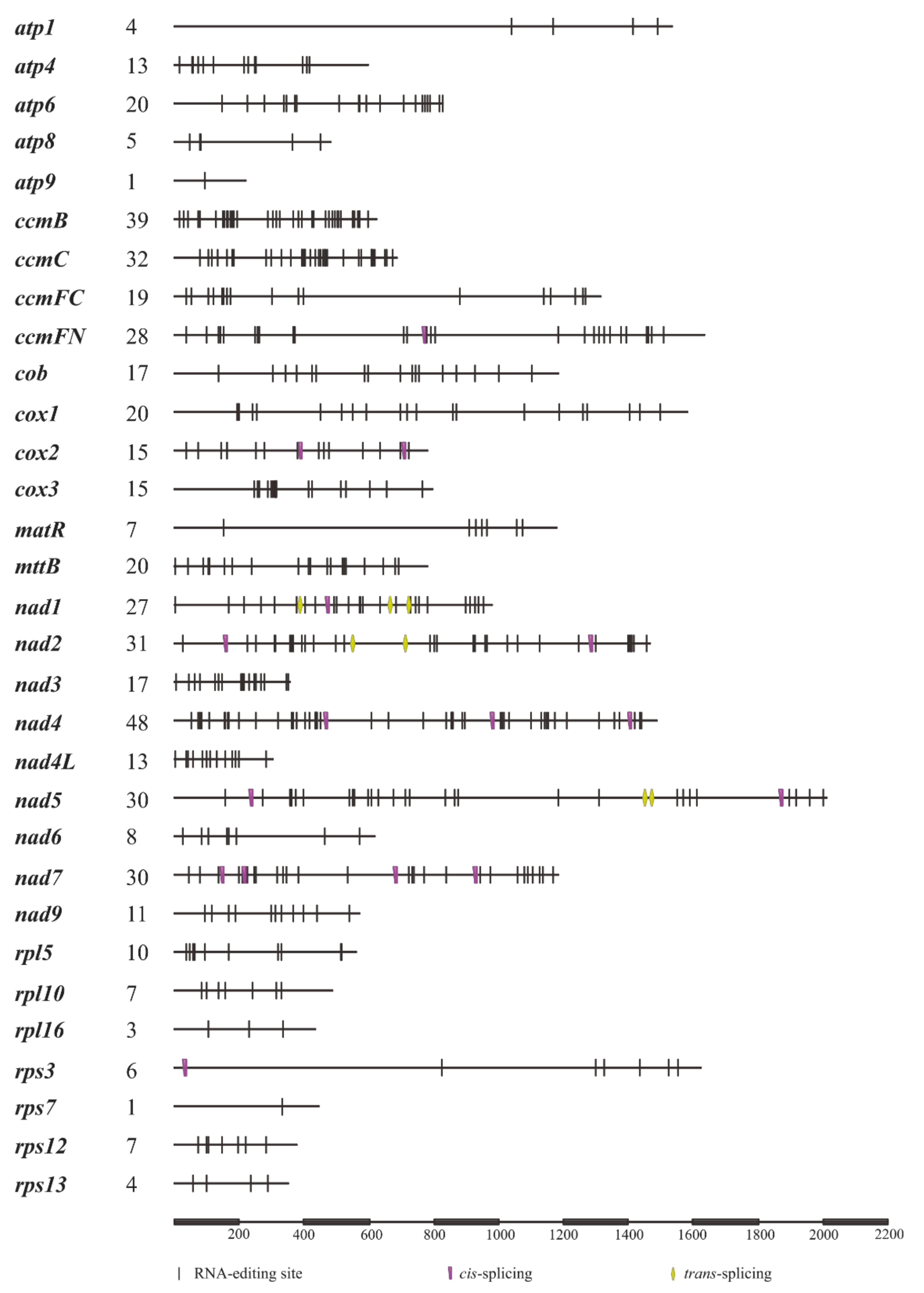
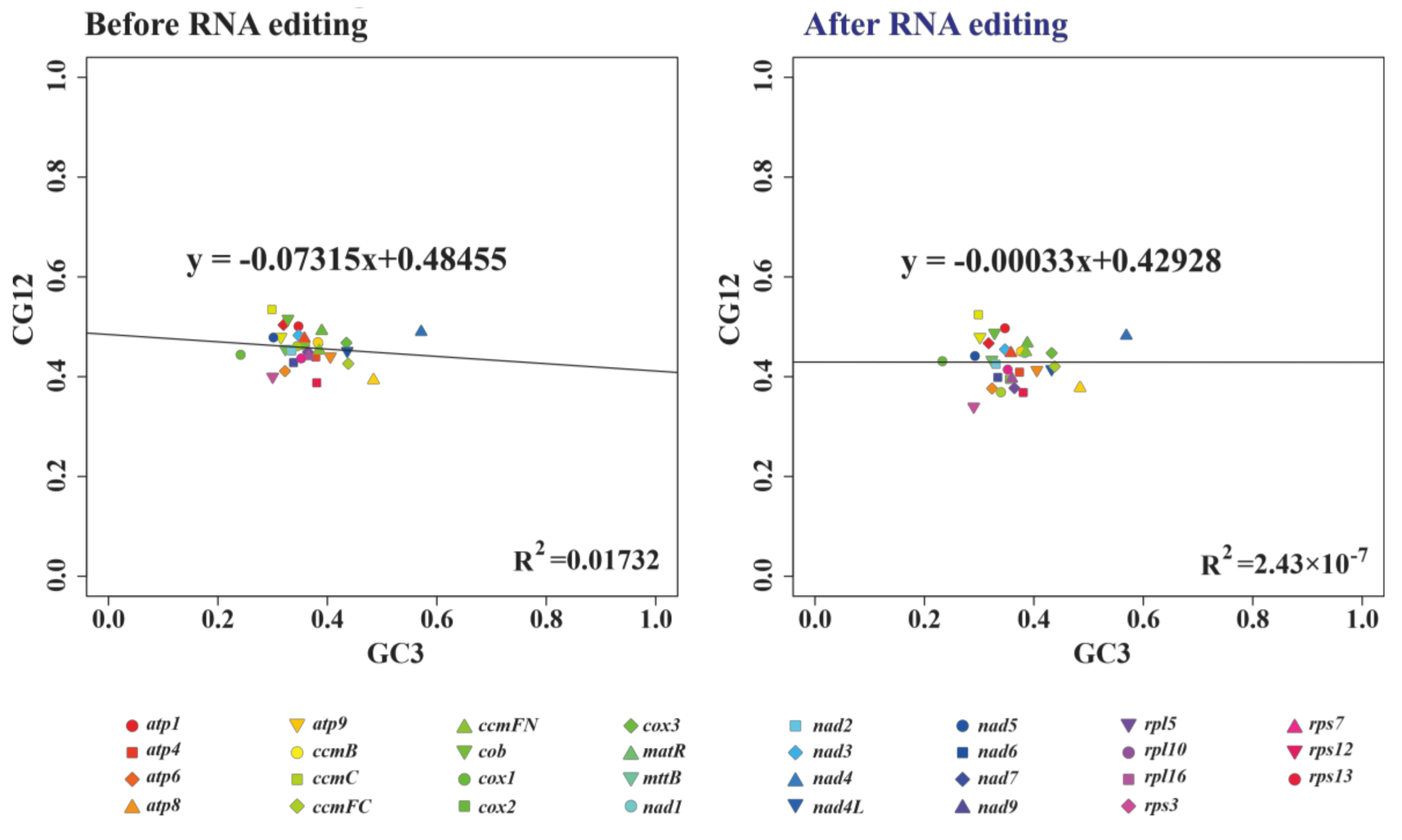
3.2. RNA Editing Sites and Codon Usage Pattern
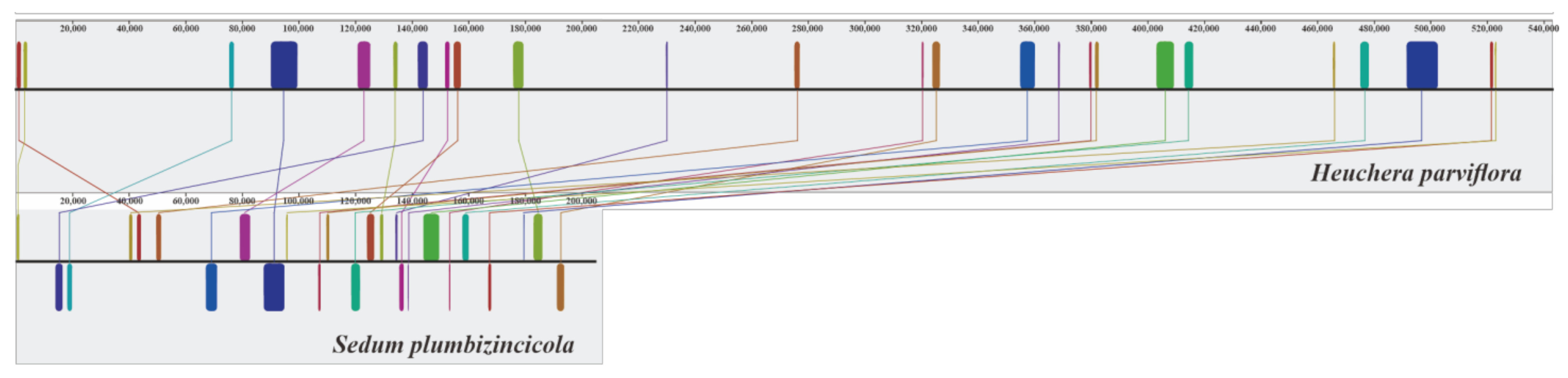
3.3. Identification of Gene Transfer
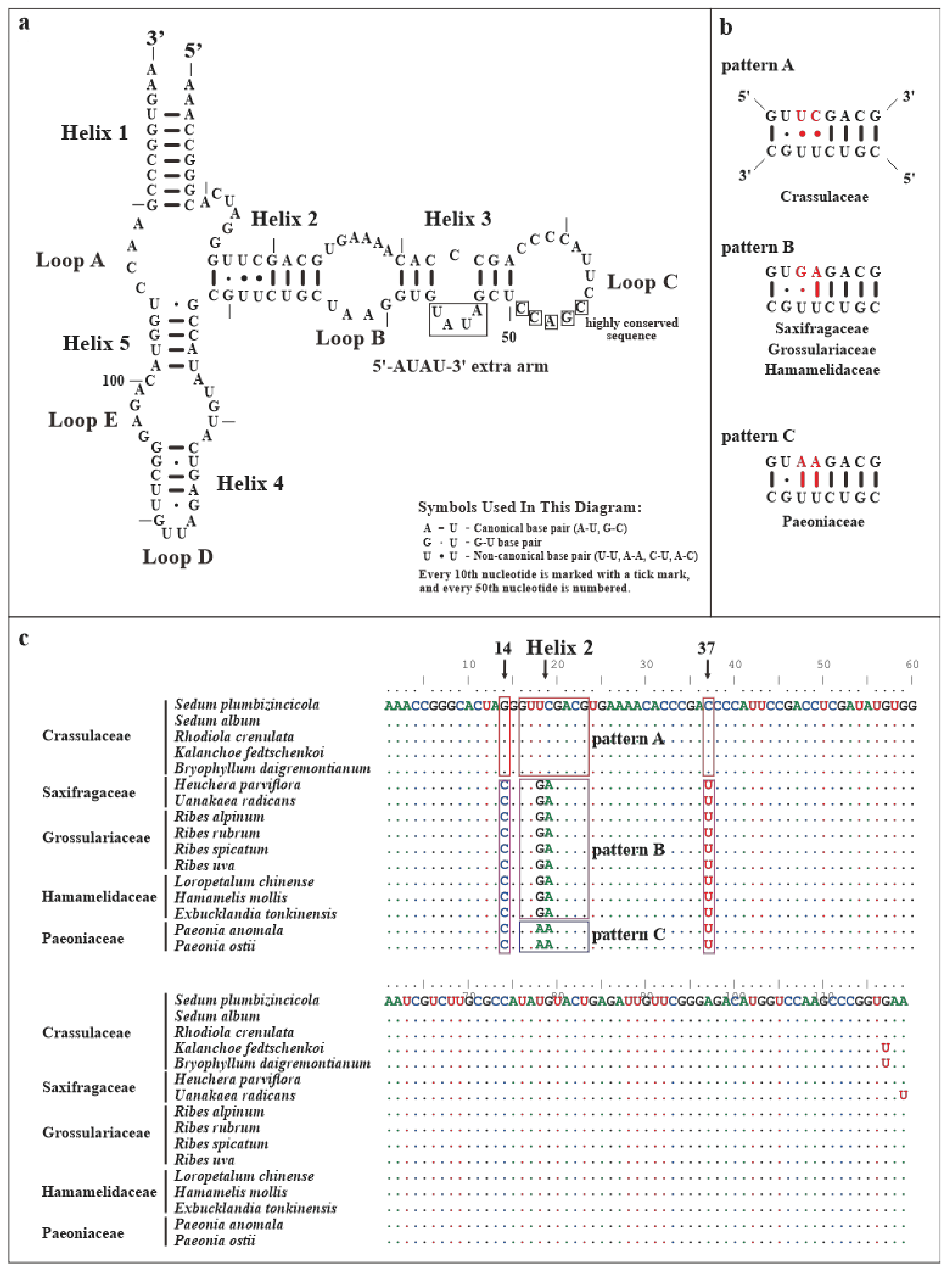
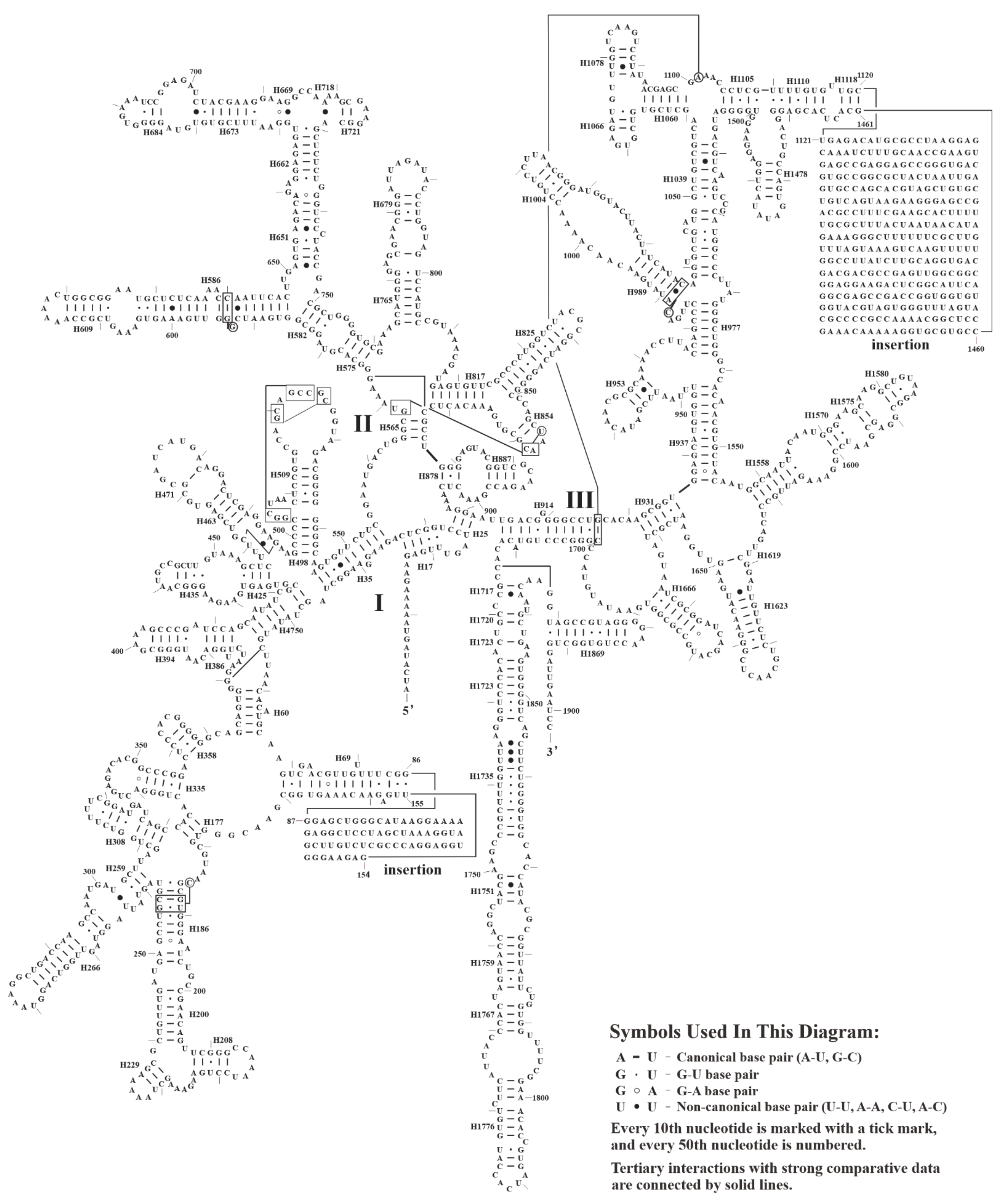
3.4. Secondary Structures of Mitochondrial RNAs
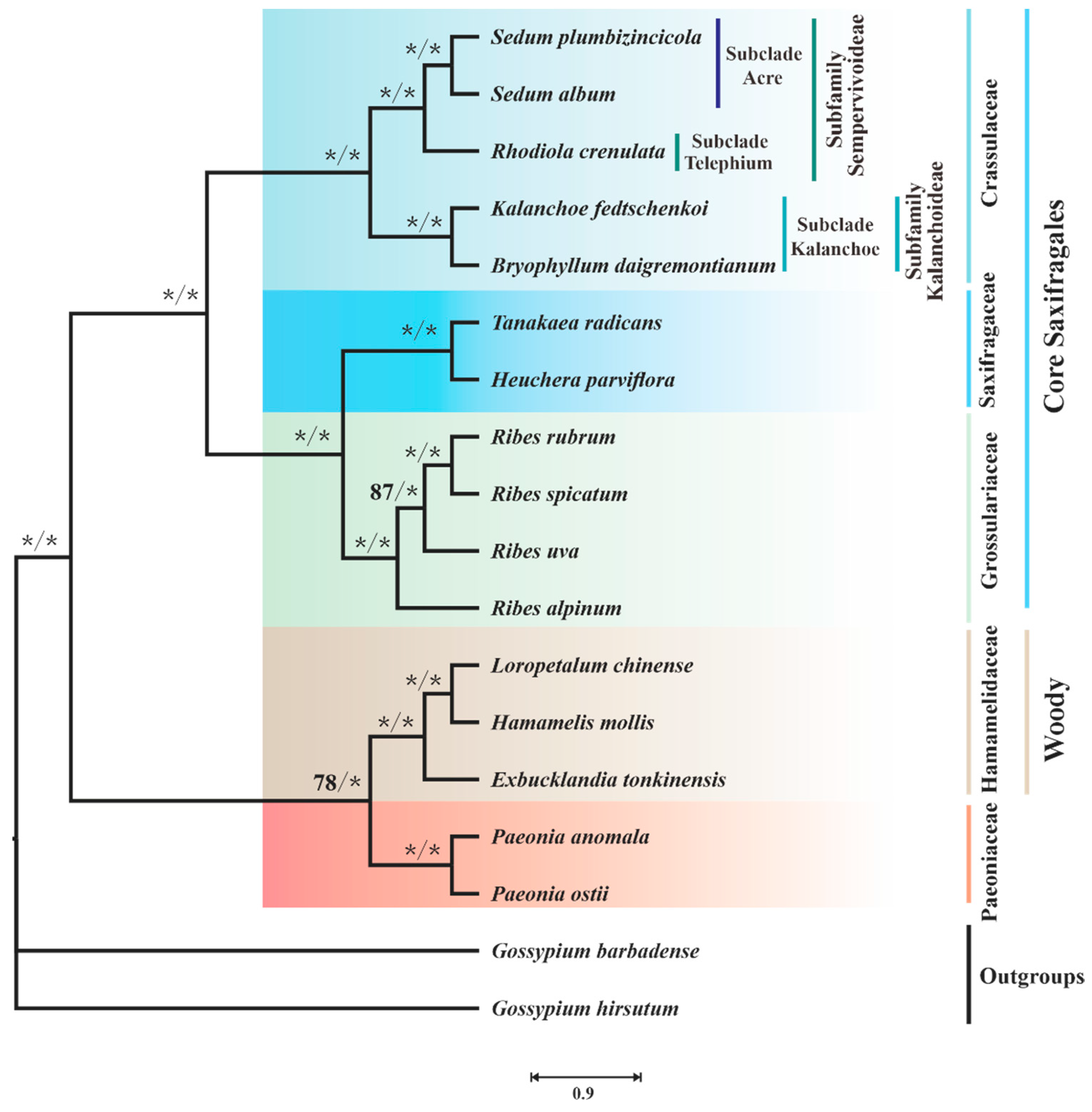
3.5. Mitophylogenetic Implications
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ren, Q.; Yuan, J.; Ren, L.; Zhang, L.; Zhang, L.; Jiang, L.; Chen, D.; Kan, X.; Zhang, B. The complete mitochondrial genome of the yellow-browed bunting, Emberiza chrysophrys (Passeriformes: Emberizidae), and phylogenetic relationships within the genus Emberiza. J. Genet. 2014, 93, 699–707. [Google Scholar] [CrossRef] [PubMed]
- Formaggioni, A.; Luchetti, A.; Plazzi, F. Mitochondrial genomic landscape: A portrait of the mitochondrial genome 40 years after the first complete sequence. Life 2021, 11, 663. [Google Scholar] [CrossRef] [PubMed]
- Bi, D.; Ding, H.; Wang, Q.; Jiang, L.; Lu, W.; Wu, X.; Zhu, R.; Zeng, J.; Zhou, S.; Yang, X. Two new mitogenomes of Picidae (Aves, Piciformes): Sequence, structure and phylogenetic analyses. Int. J. Biol. Macromol. 2019, 133, 683–692. [Google Scholar] [CrossRef] [PubMed]
- Mackenzie, S.; McIntosh, L. Higher plant mitochondria. Plant Cell 1999, 11, 571–585. [Google Scholar] [CrossRef]
- Klein, M.; Eckert-Ossenkopp, U.; Schmiedeberg, I.; Brandt, P.; Unseld, M.; Brennicke, A.; Schuster, W. Physical mapping of the mitochondrial genome of Arabidopsis thaliana by cosmid and YAC clones. Plant J. 1994, 6, 447–455. [Google Scholar] [CrossRef]
- Kan, S.-L.; Shen, T.-T.; Gong, P.; Ran, J.-H.; Wang, X.-Q. The complete mitochondrial genome of Taxus cuspidata (Taxaceae): Eight protein-coding genes have transferred to the nuclear genome. BMC Evol. Biol. 2020, 20, 10. [Google Scholar] [CrossRef]
- Skippington, E.; Barkman, T.J.; Rice, D.W.; Palmer, J.D. Miniaturized mitogenome of the parasitic plant Viscum scurruloideum is extremely divergent and dynamic and has lost all nad genes. Proc. Natl. Acad. Sci. USA 2015, 112, E3515–E3524. [Google Scholar] [CrossRef]
- Putintseva, Y.A.; Bondar, E.I.; Simonov, E.P.; Sharov, V.V.; Oreshkova, N.V.; Kuzmin, D.A.; Konstantinov, Y.M.; Shmakov, V.N.; Belkov, V.I.; Sadovsky, M.G. Siberian larch (Larix sibirica Ledeb.) mitochondrial genome assembled using both short and long nucleotide sequence reads is currently the largest known mitogenome. BMC Genomics 2020, 21, 654. [Google Scholar] [CrossRef]
- Kozik, A.; Rowan, B.A.; Lavelle, D.; Berke, L.; Schranz, M.E.; Michelmore, R.W.; Christensen, A.C. The alternative reality of plant mitochondrial DNA: One ring does not rule them all. PLoS Genet. 2019, 15, e1008373. [Google Scholar] [CrossRef]
- Unseld, M.; Marienfeld, J.R.; Brandt, P.; Brennicke, A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997, 15, 57–61. [Google Scholar] [CrossRef]
- Wu, Z.Q.; Liao, X.Z.; Zhang, X.N.; Tembrock, L.R.; Broz, A. Genomic architectural variation of plant mitochondria—A review of multichromosomal structuring. J. Syst. Evol. 2022, 60, 160–168. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Chuckalovcak, J.P.; Wu, M.; McCauley, D.E.; Palmer, J.D.; Taylor, D.R. Rapid evolution of enormous, multichromosomal genomes in flowering plant mitochondria with exceptionally high mutation rates. PLoS Biol. 2012, 10, e1001241. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Liu, Y. The mitochondrial genomes of bryophytes. Bry. Div. Evol. 2021, 43, 112–126. [Google Scholar] [CrossRef]
- Dong, S.; Zhao, C.; Zhang, S.; Zhang, L.; Wu, H.; Liu, H.; Zhu, R.; Jia, Y.; Goffinet, B.; Liu, Y. Mitochondrial genomes of the early land plant lineage liverworts (Marchantiophyta): Conserved genome structure, and ongoing low frequency recombination. BMC Genom. 2019, 20, 953. [Google Scholar] [CrossRef] [PubMed]
- Zardoya, R. Recent advances in understanding mitochondrial genome diversity. F1000Research 2020, 9. [Google Scholar] [CrossRef]
- Benne, R.; Van Den Burg, J.; Brakenhoff, J.P.; Sloof, P.; Van Boom, J.H.; Tromp, M.C. Major transcript of the frameshifted coxll gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell 1986, 46, 819–826. [Google Scholar] [CrossRef]
- Brenner, W.G.; Mader, M.; Müller, N.A.; Hoenicka, H.; Schroeder, H.; Zorn, I.; Fladung, M.; Kersten, B. High level of conservation of mitochondrial RNA editing sites among four populus species. G3 2019, 9, 709–717. [Google Scholar] [CrossRef]
- Ichinose, M.; Sugita, M. RNA editing and its molecular mechanism in plant organelles. Genes 2017, 8, 5. [Google Scholar] [CrossRef]
- Takenaka, M.; Zehrmann, A.; Verbitskiy, D.; Härtel, B.; Brennicke, A. RNA editing in plants and its evolution. Annu. Rev. Genet. 2013, 47, 335–352. [Google Scholar] [CrossRef]
- Sloan, D.B.; Taylor, D.R. Testing for selection on synonymous sites in plant mitochondrial DNA: The role of codon bias and RNA editing. J. Mol. Biol. 2010, 70, 479–491. [Google Scholar] [CrossRef]
- Ye, N.; Wang, X.; Li, J.; Bi, C.; Xu, Y.; Wu, D.; Ye, Q. Assembly and comparative analysis of complete mitochondrial genome sequence of an economic plant Salix suchowensis. PeerJ 2017, 5, e3148. [Google Scholar] [CrossRef] [PubMed]
- Michalovová, M.; Vyskot, B.; Kejnovsky, E. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: Size, relative age and chromosomal localization. Heredity 2013, 111, 314–320. [Google Scholar] [CrossRef] [PubMed]
- Gandini, C.L.; Sanchez-Puerta, M.V. Foreign plastid sequences in plant mitochondria are frequently acquired via mitochondrion-to-mitochondrion horizontal transfer. Sci. Rep. 2017, 7, 43402. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.-C.; Chen, H.; Yang, D.; Liu, C. Diversity of mitochondrial plastid DNAs (MTPTs) in seed plants. Mitochondrial DNA PartA 2018, 29, 635–642. [Google Scholar] [CrossRef]
- Leister, D. Origin, evolution and genetic effects of nuclear insertions of organelle DNA. Trends Genet. 2005, 21, 655–663. [Google Scholar] [CrossRef]
- Blanchard, J.L.; Schmidt, G.W. Pervasive migration of organellar DNA to the nucleus in plants. J. Mol. Biol. 1995, 41, 397–406. [Google Scholar] [CrossRef]
- Knoop, V.; Brennicke, A. Promiscuous mitochondrial group II intron sequences in plant nuclear genomes. J. Mol. Biol. 1994, 39, 144–150. [Google Scholar] [CrossRef]
- Kubo, N.; Takano, M.; Nishiguchi, M.; Kadowaki, K.-i. Mitochondrial sequence migrated downstream to a nuclear V-ATPase B gene is transcribed but non-functional. Gene 2001, 271, 193–201. [Google Scholar] [CrossRef]
- Lata, S.; Watts, A.; Bhat, S. Characterization of Arabidopsis thaliana lines with T-DNA insertions in the mitochondrial ribosomal protein genes Rps14 and Rps19. Indian J. Genet. Pl. Br. 2019, 79, 467–473. [Google Scholar]
- Choi, I.S.; Wojciechowski, M.F.; Steele, K.P.; Hunter, S.G.; Ruhlman, T.A.; Jansen, R.K. Born in the mitochondrion and raised in the nucleus: Evolution of a novel tandem repeat family in Medicago polymorpha (Fabaceae). Plant J. 2022, 110, 389–406. [Google Scholar] [CrossRef]
- Adams, K.L.; Daley, D.O.; Qiu, Y.-L.; Whelan, J.; Palmer, J.D. Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 2000, 408, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Thiede, J.; Eggli, U. Crassulaceae. In The Families and Genera of Vascular Plants; Kubitzki, K., Ed.; Springer: Hamburg, Germany, 2007; pp. 83–118. [Google Scholar]
- Messerschmid, T.F.; Klein, J.T.; Kadereit, G.; Kadereit, J.W. Linnaeus’s folly–phylogeny, evolution and classification of Sedum (Crassulaceae) and Crassulaceae subfamily Sempervivoideae. Taxon 2020, 69, 892–926. [Google Scholar] [CrossRef]
- Wu, L.; Liu, Y.; Zhou, S.; Guo, F.; Bi, D.; Guo, X.; Baker, A.; Smith, J.; Luo, Y. Sedum plumbizincicola XH Guo et SB Zhou ex LH Wu (Crassulaceae): A new species from Zhejiang Province, China. Plant Syst. Evol. 2013, 299, 487–498. [Google Scholar] [CrossRef]
- Ma, Y.; Oliveira, R.S.; Nai, F.; Rajkumar, M.; Luo, Y.; Rocha, I.; Freitas, H. The hyperaccumulator Sedum plumbizincicola harbors metal-resistant endophytic bacteria that improve its phytoextraction capacity in multi-metal contaminated soil. J. Environ. Manag. 2015, 156, 62–69. [Google Scholar] [CrossRef]
- Sun, L.; Cao, X.; Tan, C.; Deng, Y.; Cai, R.; Peng, X.; Bai, J. Analysis of the effect of cadmium stress on root exudates of Sedum plumbizincicola based on metabolomics. Ecotoxicol. Environ. Saf. 2020, 205, 111152. [Google Scholar] [CrossRef]
- Peng, J.-S.; Wang, Y.-J.; Ding, G.; Ma, H.-L.; Zhang, Y.-J.; Gong, J.-M. A pivotal role of cell wall in cadmium accumulation in the Crassulaceae hyperaccumulator Sedum plumbizincicola. Mol. Plant 2017, 10, 771–774. [Google Scholar] [CrossRef]
- Li, J.-t.; Gurajala, H.K.; Wu, L.-h.; van der Ent, A.; Qiu, R.-l.; Baker, A.J.; Tang, Y.-t.; Yang, X.-e.; Shu, W.-s. Hyperaccumulator plants from China: A synthesis of the current state of knowledge. Environ. Sci. Technol. 2018, 52, 11980–11994. [Google Scholar] [CrossRef]
- Duminil, J.; Besnard, G. Utility of the Mitochondrial Genome in Plant Taxonomic Studies. In Methods in Molecular Biology; Springer: New York, NY, USA, 2021; Volume 2222, pp. 107–118. [Google Scholar]
- Koren, S.; Walenz, B.P.; Berlin, K.; Miller, J.R.; Bergman, N.H.; Phillippy, A.M. Canu: Scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017, 27, 722–736. [Google Scholar] [CrossRef]
- Liu, H.; Wu, S.; Li, A.; Ruan, J. SMARTdenovo: A de novo assembler using long noisy reads. Gigabyte 2021, 2021, 1–9. [Google Scholar] [CrossRef]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Jin, J.-J.; Yu, W.-B.; Yang, J.-B.; Song, Y.; Depamphilis, C.W.; Yi, T.-S.; Li, D.-Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef] [PubMed]
- Tillich, M.; Lehwark, P.; Pellizzer, T.; Ulbricht-Jones, E.S.; Fischer, A.; Bock, R.; Greiner, S. GeSeq–versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 2017, 45, W6–W11. [Google Scholar] [CrossRef] [PubMed]
- Wynn, E.L.; Christensen, A.C. Repeats of unusual size in plant mitochondrial genomes: Identification, incidence and evolution. G3 2019, 9, 549–559. [Google Scholar] [CrossRef] [PubMed]
- Benson, G. Tandem repeats finder: A program to analyze DNA sequences. Nucleic Acids Res. 1999, 27, 573–580. [Google Scholar] [CrossRef]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef]
- Ding, H.; Zhu, R.; Dong, J.; Bi, D.; Jiang, L.; Zeng, J.; Huang, Q.; Liu, H.; Xu, W.; Wu, L. Next-Generation Genome Sequencing of Sedum plumbizincicola Sheds Light on the Structural Evolution of Plastid rRNA Operon and Phylogenetic Implications within Saxifragales. Plants 2019, 8, 386. [Google Scholar] [CrossRef]
- Yang, Z. PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 2007, 24, 1586–1591. [Google Scholar] [CrossRef]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sánchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sánchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, J.; Qiao, J.; Li, W.; Li, J.; Xu, R.; Wang, H.; Liu, Z.; Xing, B.; Wendel, J.F. Comparative analysis of codon usage between Gossypium hirsutum and G. barbadense mitochondrial genomes. Mitochondrial DNA PartB 2020, 5, 2500–2506. [Google Scholar] [CrossRef] [PubMed]
- Ismail, S.N.F.B.; Baharum, S.N.; Fazry, S.; Low, C.F. Comparative genome analysis reveals a distinct influence of nucleotide composition on virus–host species-specific interaction of prawn-infecting nodavirus. J. Fish Dis. 2019, 42, 1761–1772. [Google Scholar] [CrossRef]
- Sarkar, I.; Dey, P.; Sharma, S.K.; Ray, S.D.; Kochiganti, V.H.S.; Singh, R.; Pramod, P.; Singh, R.P. Turdoides affinis mitogenome reveals the translational efficiency and importance of NADH dehydrogenase complex-I in the Leiothrichidae family. Sci. Rep. 2020, 10, 16202. [Google Scholar] [CrossRef]
- Wright, F. The ‘effective number of codons’ used in a gene. Gene 1990, 87, 23–29. [Google Scholar] [CrossRef]
- Yuan, M.L.; Zhang, L.J.; Zhang, Q.L.; Zhang, L.; Li, M.; Wang, X.T.; Feng, R.Q.; Tang, P.A. Mitogenome evolution in ladybirds: Potential association with dietary adaptation. Ecol. Evol. 2020, 10, 1042–1053. [Google Scholar] [CrossRef]
- Sueoka, N. Directional mutation pressure and neutral molecular evolution. Proc. Natl. Acad. Sci. USA 1988, 85, 2653–2657. [Google Scholar] [CrossRef]
- Sueoka, N. Two aspects of DNA base composition: G+ C content and translation-coupled deviation from intra-strand rule of A= T and G= C. J. Mol. Biol. 1999, 49, 49–62. [Google Scholar] [CrossRef]
- Folk, R.A.; Mandel, J.R.; Freudenstein, J.V. A protocol for targeted enrichment of intron-containing sequence markers for recent radiations: A phylogenomic example from Heuchera (Saxifragaceae). Appl. Plant Sci. 2015, 3, 1500039. [Google Scholar] [CrossRef] [PubMed]
- Goremykin, V.V.; Salamini, F.; Velasco, R.; Viola, R. Mitochondrial DNA of Vitis vinifera and the issue of rampant horizontal gene transfer. Mol. Biol. Evol. 2009, 26, 99–110. [Google Scholar] [CrossRef] [PubMed]
- Rice, D.W.; Alverson, A.J.; Richardson, A.O.; Young, G.J.; Sanchez-Puerta, M.V.; Munzinger, J.; Barry, K.; Boore, J.L.; Zhang, Y.; DePamphilis, C.W. Horizontal transfer of entire genomes via mitochondrial fusion in the angiosperm Amborella. Science 2013, 342, 1468–1473. [Google Scholar] [CrossRef] [PubMed]
- Chan, P.P.; Lin, B.Y.; Mak, A.J.; Lowe, T.M. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucleic Acids Res. 2021, 49, 9077–9096. [Google Scholar] [CrossRef]
- Delihas, N.; Andersen, J. Generalized structures of the 5S ribosomal RNAs. Nucleic Acids Res. 1982, 10, 7323–7344. [Google Scholar] [CrossRef][Green Version]
- Cannone, J.J.; Subramanian, S.; Schnare, M.N.; Collett, J.R.; D’Souza, L.M.; Du, Y.; Feng, B.; Lin, N.; Madabusi, L.V.; Müller, K.M. The comparative RNA web (CRW) site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinform. 2002, 3, 2. [Google Scholar]
- Zeng, L.; Zhang, N.; Zhang, Q.; Endress, P.K.; Huang, J.; Ma, H. Resolution of deep eudicot phylogeny and their temporal diversification using nuclear genes from transcriptomic and genomic datasets. New Phytol. 2017, 214, 1338–1354. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312–1313. [Google Scholar] [CrossRef]
- Darriba, D.; Posada, D.; Kozlov, A.M.; Stamatakis, A.; Morel, B.; Flouri, T. ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 2020, 37, 291–294. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901–904. [Google Scholar] [CrossRef] [PubMed]
- Matsunaga, M.; Takahashi, Y.; Yui-Kurino, R.; Mikami, T.; Kubo, T. Evolutionary aspects of a unique internal mitochondrial targeting signal in nuclear-migrated rps19 of sugar beet (Beta vulgaris L.). Gene 2013, 517, 19–26. [Google Scholar] [CrossRef]
- Adams, K.L.; Palmer, J.D. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
- Adams, K.L.; Qiu, Y.-L.; Stoutemyer, M.; Palmer, J.D. Punctuated evolution of mitochondrial gene content: High and variable rates of mitochondrial gene loss and transfer to the nucleus during angiosperm evolution. Proc. Natl. Acad. Sci. USA 2002, 99, 9905–9912. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.-L.; Zhuang, Y.; Zhang, P.; Adams, K.L. Comparative analysis of structural diversity and sequence evolution in plant mitochondrial genes transferred to the nucleus. Mol. Biol. Evol. 2009, 26, 875–891. [Google Scholar] [CrossRef]
- Brennicke, A.; Möller, S.; Blanz, P.A. The 18S and 5S ribosomal RNA genes in Oenothera mitochondria: Sequence rearrangments in the 18S and 5S rRNA genes of higher plants. Mol. Gen. Genet. 1985, 198, 404–410. [Google Scholar] [CrossRef]
- Spencer, D.; Bonen, L.; Gray, M. Primary sequence of wheat mitochondrial 5S ribosomal ribonucleic acid: Functional and evolutionary implications. Biochemistry 1981, 20, 4022–4029. [Google Scholar] [CrossRef]
- Sloan, D.B.; Alverson, A.J.; Štorchová, H.; Palmer, J.D.; Taylor, D.R. Extensive loss of translational genes in the structurally dynamic mitochondrial genome of the angiosperm Silene latifolia. BMC Evol. Biol. 2010, 10, 274. [Google Scholar] [CrossRef]
- Chao, S.; Sederoff, R.; Levings, C.S. Nucleotide sequence and evolution of the 18S ribosomal RNA gene in maize mitochondria. Nucleic Acids Res. 1984, 12, 6629–6644. [Google Scholar] [CrossRef][Green Version]
- Sloan, D.B.; Müller, K.; McCauley, D.E.; Taylor, D.R.; Štorchová, H. Intraspecific variation in mitochondrial genome sequence, structure, and gene content in Silene vulgaris, an angiosperm with pervasive cytoplasmic male sterility. New Phytol. 2012, 196, 1228–1239. [Google Scholar] [CrossRef]
- Wu, Z.; Cuthbert, J.M.; Taylor, D.R.; Sloan, D.B. The massive mitochondrial genome of the angiosperm Silene noctiflora is evolving by gain or loss of entire chromosomes. Proc. Natl. Acad. Sci. USA 2015, 112, 10185–10191. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.; Zhang, M.; Chen, X.; Liu, Y.; Liu, B.; Li, J.; Wang, R.; Zhao, K.; Wu, J. Rearrangement and domestication as drivers of Rosaceae mitogenome plasticity. BMC Biol. 2022, 20, 181. [Google Scholar] [CrossRef] [PubMed]
- Wolfe, K.H.; Li, W.-H.; Sharp, P.M. Rates of nucleotide substitution vary greatly among plant mitochondrial, chloroplast, and nuclear DNAs. Proc. Natl. Acad. Sci. USA 1987, 84, 9054–9058. [Google Scholar] [CrossRef] [PubMed]
- Cho, Y.; Mower, J.P.; Qiu, Y.-L.; Palmer, J.D. Mitochondrial substitution rates are extraordinarily elevated and variable in a genus of flowering plants. Proc. Natl. Acad. Sci. USA 2004, 101, 17741–17746. [Google Scholar] [CrossRef] [PubMed]
- Drouin, G.; Daoud, H.; Xia, J. Relative rates of synonymous substitutions in the mitochondrial, chloroplast and nuclear genomes of seed plants. Mol. Phylogenet. Evol. 2008, 49, 827–831. [Google Scholar] [CrossRef]
- Kan, S.; Liao, X.; Wu, Z. The Roles of Mutation and Selection Acting on Mitochondrial Genomes Inferred from Intraspecific Variation in Seed Plants. Genes 2022, 13, 1036. [Google Scholar] [CrossRef]
- Christensen, A.C. Genes and junk in plant mitochondria—Repair mechanisms and selection. Genome Biol. Evol. 2014, 6, 1448–1453. [Google Scholar] [CrossRef]
- Christensen, A.C. Mitochondrial DNA repair and genome evolution. Annu. Rev. Plant Biol. 2017, 50, 11–31. [Google Scholar]
- Li, J.; Xu, Y.; Shan, Y.; Pei, X.; Yong, S.; Liu, C.; Yu, J. Assembly of the complete mitochondrial genome of an endemic plant, Scutellaria tsinyunensis, revealed the existence of two conformations generated by a repeat-mediated recombination. Planta 2021, 254, 36. [Google Scholar] [CrossRef]
- Backert, S.; Nielsen, B.L.; Börner, T. The mystery of the rings: Structure and replication of mitochondrial genomes from higher plants. Trends Genet. 1997, 2, 477–483. [Google Scholar] [CrossRef]
- Ogihara, Y.; Yamazaki, Y.; Murai, K.; Kanno, A.; Terachi, T.; Shiina, T.; Miyashita, N.; Nasuda, S.; Nakamura, C.; Mori, N. Structural dynamics of cereal mitochondrial genomes as revealed by complete nucleotide sequencing of the wheat mitochondrial genome. Nucleic Acids Res. 2005, 33, 6235–6250. [Google Scholar] [CrossRef] [PubMed]
- Gualberto, J.M.; Mileshina, D.; Wallet, C.; Niazi, A.K.; Weber-Lotfi, F.; Dietrich, A. The plant mitochondrial genome: Dynamics and maintenance. Biochimie 2014, 100, 107–120. [Google Scholar] [CrossRef] [PubMed]
- Fauron, C.; Casper, M.; Gao, Y.; Moore, B. The maize mitochondrial genome: Dynamic, yet functional. Trends Genet. 1995, 11, 228–235. [Google Scholar] [CrossRef]
- Kazama, T.; Toriyama, K. Whole mitochondrial genome sequencing and re-examination of a cytoplasmic male sterility-associated gene in Boro-taichung-type cytoplasmic male sterile rice. PLoS ONE 2016, 11, e0159379. [Google Scholar] [CrossRef]
- Best, C.; Mizrahi, R.; Ostersetzer-Biran, O. Why so complex? The intricacy of genome structure and gene expression, associated with angiosperm mitochondria, may relate to the regulation of embryo quiescence or dormancy—Intrinsic blocks to early plant life. Plants 2020, 9, 598. [Google Scholar] [CrossRef]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2020, 101, 1040–1056. [Google Scholar] [CrossRef]
- Yan, J.; Zhang, Q.; Yin, P. RNA editing machinery in plant organelles. Sci. China Life Sci. 2018, 61, 162–169. [Google Scholar] [CrossRef]
- Giegé, P.; Brennicke, A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl. Acad. Sci. USA 1999, 96, 15324–15329. [Google Scholar] [CrossRef]
- Notsu, Y.; Masood, S.; Nishikawa, T.; Kubo, N.; Akiduki, G.; Nakazono, M.; Hirai, A.; Kadowaki, K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: Frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genom. 2002, 268, 434–445. [Google Scholar] [CrossRef]
- Handa, H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): Comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003, 31, 5907–5916. [Google Scholar] [CrossRef]
- Mower, J.P.; Palmer, J.D. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol. Genet. Genom. 2006, 276, 285–293. [Google Scholar] [CrossRef] [PubMed]
- Picardi, E.; Horner, D.S.; Chiara, M.; Schiavon, R.; Valle, G.; Pesole, G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 2010, 38, 4755–4767. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wu, H.; Zhang, T.; Yang, M.; Yin, Y.; Pan, L.; Yu, X.; Zhang, X.; Hu, S.; Al-Mssallem, I.S. A complete sequence and transcriptomic analyses of date palm (Phoenix dactylifera L.) mitochondrial genome. PLoS ONE 2012, 7, e37164. [Google Scholar] [CrossRef] [PubMed]
- Bentolila, S.; Oh, J.; Hanson, M.R.; Bukowski, R. Comprehensive high-resolution analysis of the role of an Arabidopsis gene family in RNA editing. PLoS Genet. 2013, 9, e1003584. [Google Scholar] [CrossRef]
- Grimes, B.T.; Sisay, A.K.; Carroll, H.D.; Cahoon, A.B. Deep sequencing of the tobacco mitochondrial transcriptome reveals expressed ORFs and numerous editing sites outside coding regions. BMC Genomics 2014, 15, 31. [Google Scholar] [CrossRef]
- Edera, A.A.; Gandini, C.L.; Sanchez-Puerta, M. Towards a comprehensive picture of C-to-U RNA editing sites in angiosperm mitochondria. Plant Mol. Biol. 2018, 97, 215–231. [Google Scholar] [CrossRef]
- Grewe, F.; Herres, S.; Viehöver, P.; Polsakiewicz, M.; Weisshaar, B.; Knoop, V. A unique transcriptome: 1782 positions of RNA editing alter 1406 codon identities in mitochondrial mRNAs of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2011, 39, 2890–2902. [Google Scholar] [CrossRef]
- Guo, W.; Zhu, A.; Fan, W.; Mower, J.P. Complete mitochondrial genomes from the ferns Ophioglossum californicum and Psilotum nudum are highly repetitive with the largest organellar introns. New Phytol. 2017, 213, 391–403. [Google Scholar] [CrossRef]
- Gualberto, J.M.; Lamattina, L.; Bonnard, G.; Weil, J.-H.; Grienenberger, J.-M. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature 1989, 341, 660–662. [Google Scholar] [CrossRef]
- Chateigner-Boutin, A.-L.; Small, I. Plant RNA editing. RNA Biol. 2010, 7, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Li, X. Analysis of synonymous codon usage patterns in different plant mitochondrial genomes. Mol. Biol. Rep. 2009, 36, 2039–2046. [Google Scholar] [CrossRef] [PubMed]
- Chang, S.; Wang, Y.; Lu, J.; Gai, J.; Li, J.; Chu, P.; Guan, R.; Zhao, T. The mitochondrial genome of soybean reveals complex genome structures and gene evolution at intercellular and phylogenetic levels. PLoS ONE 2013, 8, e56502. [Google Scholar]
- Yang, H.; Li, W.; Yu, X.; Zhang, X.; Zhang, Z.; Liu, Y.; Wang, W.; Tian, X. Insights into molecular structure, genome evolution and phylogenetic implication through mitochondrial genome sequence of Gleditsia sinensis. Sci. Rep. 2021, 11, 14850. [Google Scholar] [CrossRef] [PubMed]
- Liao, X.; Zhao, Y.; Kong, X.; Khan, A.; Zhou, B.; Liu, D.; Kashif, M.H.; Chen, P.; Wang, H.; Zhou, R. Complete sequence of kenaf (Hibiscus cannabinus) mitochondrial genome and comparative analysis with the mitochondrial genomes of other plants. Sci. Rep. 2018, 8, 12714. [Google Scholar] [CrossRef]
- Group, A.P.; Chase, M.W.; Christenhusz, M.J.; Fay, M.F.; Byng, J.; Judd, W.; Soltis, D.; Mabberley, D.; Sennikov, A.; Soltis, P. An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc. 2016, 181, 1–20. [Google Scholar]
- Folk, R.A.; Stubbs, R.L.; Mort, M.E.; Cellinese, N.; Allen, J.M.; Soltis, P.S.; Soltis, D.E.; Guralnick, R.P. Rates of niche and phenotype evolution lag behind diversification in a temperate radiation. Proc. Natl. Acad. Sci. USA 2019, 116, 10874–10882. [Google Scholar] [CrossRef]
- Han, S.; Bi, D.; Yi, R.; Ding, H.; Wu, L.; Kan, X. Plastome evolution of Aeonium and Monanthes (Crassulaceae): Insights into the variation of plastomic tRNAs, and the patterns of codon usage and aversion. Planta 2022, 256, 35. [Google Scholar] [CrossRef]



| Species | Sedum plumbizincicola | Heuchera parviflora |
|---|---|---|
| Accession | OP588116 | KR559021 |
| Size (bp) | 212,159 | 542,954 |
| GC content | 44.51% | 45.75% |
| Functional genes | 48 | 77 |
| tRNAs | 14 | 28 |
| rRNAs | 3 | 7 |
| PCGs | 31 | 42 |
| ORFs | 2 | 0 |
| Pseudogenes | 11 | 7 |
| Coding region (bp) | 33,814 (15.93%) | 45,189 (8.32%) |
| Noncoding region (bp) | 178,345 (84.07%) | 497,765 (91.68%) |
| Dispersed repeats (bp) | 12,884 (6.07%) | 119,727 (22.05%) |
| Tandem repeats (bp) | 129 (0.06%) | 1459 (0.27%) |
| Plastid-derived sequences (bp) | 10,394 (4.90%) | 25,562 (4.71%) |
| Gene | ω | dN | dS |
|---|---|---|---|
| Concatenated mitochondrial genes | 0.4188 | 0.0292 | 0.0697 |
| Concatenated plastid genes | 0.1763 | 0.0562 | 0.3190 |
| AA Conversion | Count (Ratio) | Conversion Type |
|---|---|---|
| Ser → Leu | 114 (22.98%) | Nonsynonymous |
| Pro → Leu | 112 (22.58%) | Nonsynonymous |
| Ser → Phe | 73 (14.72%) | Nonsynonymous |
| Pro → Ser | 42 (8.47%) | Nonsynonymous |
| Arg → Cys | 37 (7.46%) | Nonsynonymous |
| Arg → Trp | 32 (6.45%) | Nonsynonymous |
| His → Tyr | 21 (4.23%) | Nonsynonymous |
| Leu → Phe | 15 (3.02%) | Nonsynonymous |
| Thr → Ile | 9 (1.81%) | Nonsynonymous |
| Ala → Val | 6 (1.21%) | Nonsynonymous |
| Thr → Met | 6 (1.21%) | Nonsynonymous |
| Pro → Phe | 6 (1.21%) | Nonsynonymous |
| Gln → Termination | 1 (0.20%) | Nonsynonymous |
| Leu → Leu | 6 (1.21%) | Synonymous |
| Phe → Phe | 5 (1.01%) | Synonymous |
| Ile → Ile | 3 (0.60%) | Synonymous |
| Tyr → Tyr | 3 (0.60%) | Synonymous |
| Pro → Pro | 2 (0.40%) | Synonymous |
| Val → Val | 2 (0.40%) | Synonymous |
| Ser → Ser | 1 (0.20%) | Synonymous |
| Amino Acid | Codon | Genomic DNA | Change after Editing | ||
|---|---|---|---|---|---|
| Count | Ratio | Count | Ratio | ||
| Ala | GCA | 151 | 1.65% | −1 | −0.01% |
| GCC | 126 | 1.38% | −1 | −0.01% | |
| GCG | 74 | 0.81% | −3 | −0.03% | |
| GCU | 248 | 2.71% | −1 | −0.01% | |
| Arg | AGA | 115 | 1.26% | 0 | 0 |
| AGG | 66 | 0.72% | 0 | 0 | |
| CGU | 128 | 1.40% | −28 | −0.31% | |
| CGC | 58 | 0.63% | −9 | −0.10% | |
| CGA | 119 | 1.30% | 0 | 0 | |
| CGG | 85 | 0.93% | −32 | −0.35% | |
| Asn | AAC | 81 | 0.88% | 0 | 0 |
| AAU | 203 | 2.22% | 0 | 0 | |
| Asp | GAC | 90 | 0.98% | 0 | 0 |
| GAU | 202 | 2.20% | 0 | 0 | |
| Cys | UGC | 56 | 0.61% | +9 | +0.10% |
| UGU | 79 | 0.86% | +28 | +0.31% | |
| Gln | CAA | 206 | 2.25% | −1 | −0.01% |
| CAG | 57 | 0.62% | 0 | 0 | |
| Glu | GAA | 253 | 2.76% | 0 | 0 |
| GAG | 111 | 1.21% | 0 | 0 | |
| Gly | GGA | 240 | 2.62% | 0 | 0 |
| GGC | 89 | 0.97% | 0 | 0 | |
| GGG | 123 | 1.34% | 0 | 0 | |
| GGU | 212 | 2.31% | 0 | 0 | |
| His | CAC | 42 | 0.46% | −6 | −0.07% |
| CAU | 175 | 1.91% | −15 | −0.16% | |
| Ile | AUA | 202 | 2.20% | +4 | +0.04% |
| AUC | 194 | 2.12% | -2 | -0.02% | |
| AUU | 333 | 3.63% | +7 | +0.08% | |
| Leu | CUA | 131 | 1.43% | +34 | +0.37% |
| CUC | 98 | 1.07% | +4 | +0.04% | |
| CUG | 88 | 0.96% | +32 | +0.35% | |
| CUU | 205 | 2.24% | +20 | +0.22% | |
| UUA | 250 | 2.73% | +75 | +0.82% | |
| UUG | 183 | 2.00% | +46 | +0.50% | |
| Lys | AAA | 233 | 2.54% | 0 | 0 |
| AAG | 125 | 1.36% | 0 | 0 | |
| Met | AUG | 244 | 2.66% | +6 | 0.07% |
| Phe | UUC | 247 | 2.70% | +25 | 0.27% |
| UUU | 348 | 3.80% | +69 | 0.75% | |
| Pro | CCA | 144 | 1.57% | −45 | −0.49% |
| CCC | 118 | 1.29% | −26 | −0.28% | |
| CCG | 87 | 0.95% | −41 | −0.45% | |
| CCU | 184 | 2.01% | −48 | −0.52% | |
| Ser | AGC | 89 | 0.97% | 0 | 0 |
| AGU | 149 | 1.63% | 0 | 0 | |
| UCA | 175 | 1.91% | −64 | −0.70% | |
| UCC | 133 | 1.45% | −15 | −0.16% | |
| UCG | 110 | 1.20% | −38 | −0.41% | |
| UCU | 189 | 2.06% | −28 | −0.31% | |
| Thr | ACA | 117 | 1.28% | −4 | −0.04% |
| ACC | 123 | 1.34% | −1 | −0.01% | |
| ACG | 72 | 0.79% | −6 | −0.07% | |
| ACU | 157 | 1.71% | −4 | −0.04% | |
| Trp | UGG | 141 | 1.54% | +32 | 0.35% |
| Tyr | UAC | 69 | 0.75% | +3 | 0.03% |
| UAU | 226 | 2.47% | +18 | 0.20% | |
| Val | GUA | 172 | 1.88% | +1 | 0.01% |
| GUC | 103 | 1.12% | −1 | −0.01% | |
| GUG | 127 | 1.39% | +3 | 0.03% | |
| GUU | 178 | 1.94% | +3 | 0.03% | |
| Termination | UAA | 19 | 0.21% | +1 | 0.01% |
| UAG | 6 | 0.07% | 0 | 0 | |
| UGA | 5 | 0.05% | 0 | 0 | |
| MTPT Regions | Mitogenome Coordinates | MTPT Size (bp) | MTPT GC Content (%) | Plastome Coordinates | Plastomic Sequence Size (bp) | Plastomic Sequence GC Content (%) | Identity (%) | MTPT Annotations |
|---|---|---|---|---|---|---|---|---|
| MTPT1 | 43,609–44,639 | 1031 | 36.86 | 88,171–89,250 (−) | 1080 | 37.31 | 86.38 | ycf2-partial |
| MTPT2 | 83,645–84,202 | 558 | 38.71 | 66,564–67,129 (−) | 566 | 38.52 | 89.02 | rps12-partial |
| MTPT3 | 96,864–105,274 | 8411 | 37.00 | 45,747–54,551 (+) | 8805 | 36.72 | 93.72 | trnF-GAA, ndhJ, ndhK, ndhC, trnV-UAC, trnM-CAU, atpE, atpB, rbcL |
| MTPT4 | 186,562–186,955 | 394 | 44.42 | 31,555–31,958 (+) | 404 | 42.08 | 90.59 | psbD-partial |
| Domain | Identity (%) | ||
|---|---|---|---|
| 5S mtrRNA | 18S mtrRNA | 26S mtrRNA | |
| overall | 96.64 | 97.16 | 88.16 |
| domain I (insertion) | 98.94 (97.09) | 79.91 (40.56) | |
| domain II | 99.14 | 98.70 | |
| domain III (insertion) | 95.50 (88.24) | 76.13 (65.66) | |
| domain IV | 98.64 | ||
| domain V | 98.96 | ||
| domain VI | 99.59 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, H.; Bi, D.; Zhang, S.; Han, S.; Ye, Y.; Yi, R.; Yang, J.; Liu, B.; Wu, L.; Zhuo, R.; et al. The Mitogenome of Sedum plumbizincicola (Crassulaceae): Insights into RNA Editing, Lateral Gene Transfer, and Phylogenetic Implications. Biology 2022, 11, 1661. https://doi.org/10.3390/biology11111661
Ding H, Bi D, Zhang S, Han S, Ye Y, Yi R, Yang J, Liu B, Wu L, Zhuo R, et al. The Mitogenome of Sedum plumbizincicola (Crassulaceae): Insights into RNA Editing, Lateral Gene Transfer, and Phylogenetic Implications. Biology. 2022; 11(11):1661. https://doi.org/10.3390/biology11111661
Chicago/Turabian StyleDing, Hengwu, De Bi, Sijia Zhang, Shiyun Han, Yuanxin Ye, Ran Yi, Jianke Yang, Birong Liu, Longhua Wu, Renying Zhuo, and et al. 2022. "The Mitogenome of Sedum plumbizincicola (Crassulaceae): Insights into RNA Editing, Lateral Gene Transfer, and Phylogenetic Implications" Biology 11, no. 11: 1661. https://doi.org/10.3390/biology11111661
APA StyleDing, H., Bi, D., Zhang, S., Han, S., Ye, Y., Yi, R., Yang, J., Liu, B., Wu, L., Zhuo, R., & Kan, X. (2022). The Mitogenome of Sedum plumbizincicola (Crassulaceae): Insights into RNA Editing, Lateral Gene Transfer, and Phylogenetic Implications. Biology, 11(11), 1661. https://doi.org/10.3390/biology11111661





