Comprehensive Sequence Analysis of Parvalbumins in Fish and Their Comparison with Parvalbumins in Tetrapod Species
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Retrieving of Genes
2.2. Estimation of Transcription Levels
2.3. Sequence Alignment
2.4. Phylogenetic Tree Analysis
2.5. Analysis and Depiction of Molecular Structures
3. Results and Discussion
3.1. The Organization of Our Study to Compare Parvalbumin Sequences with a Focus on Teleost Fish
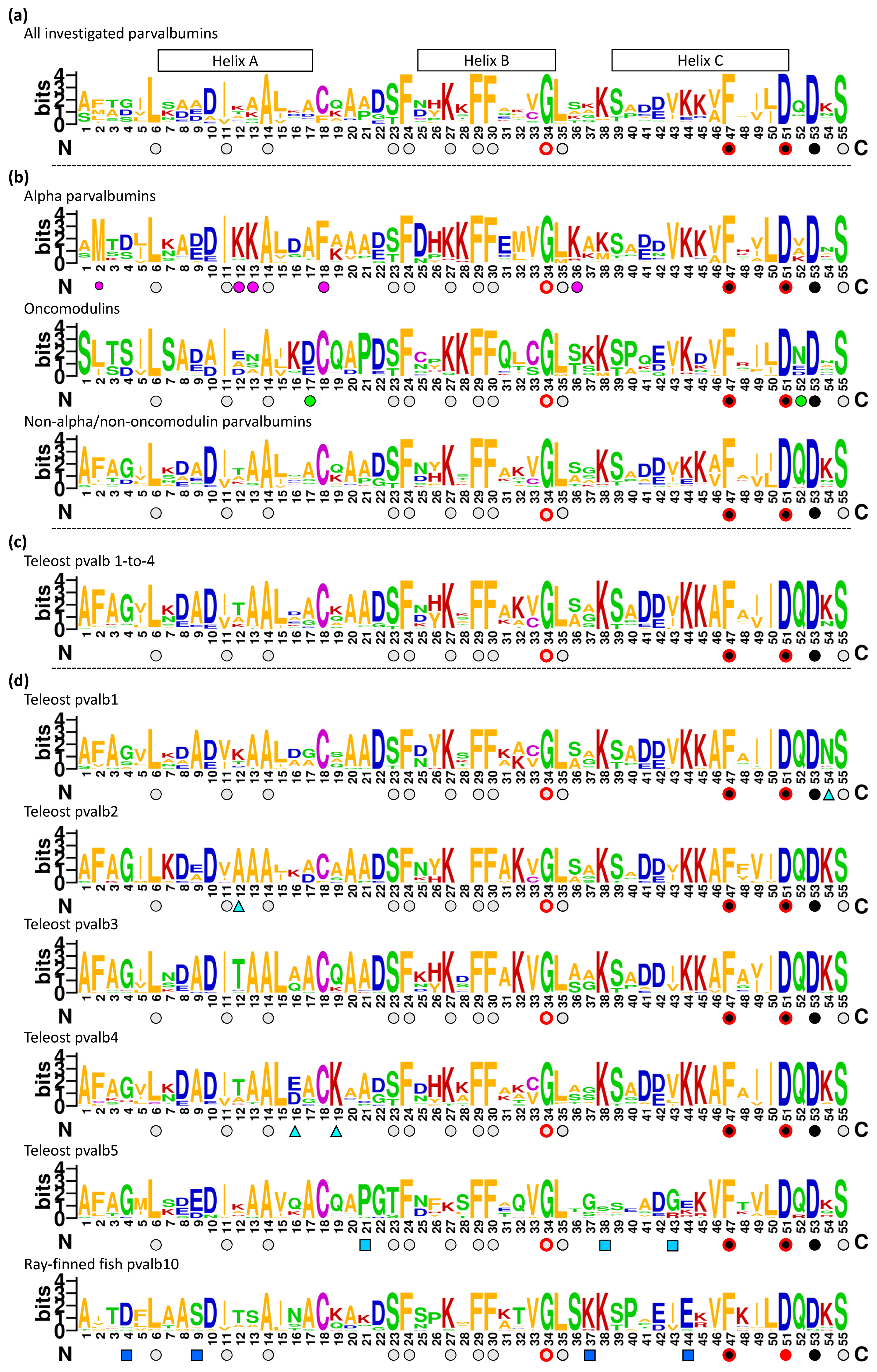
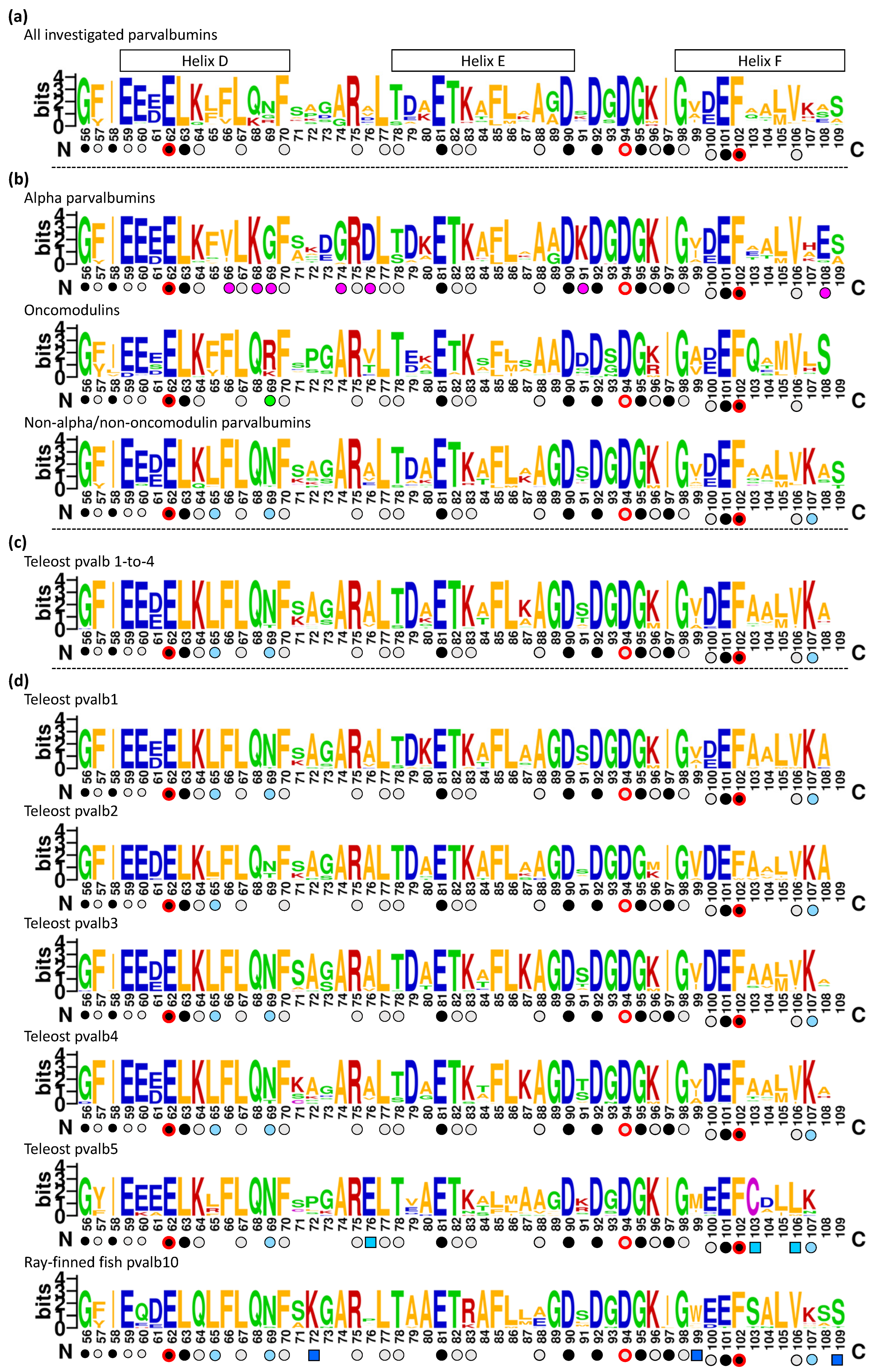
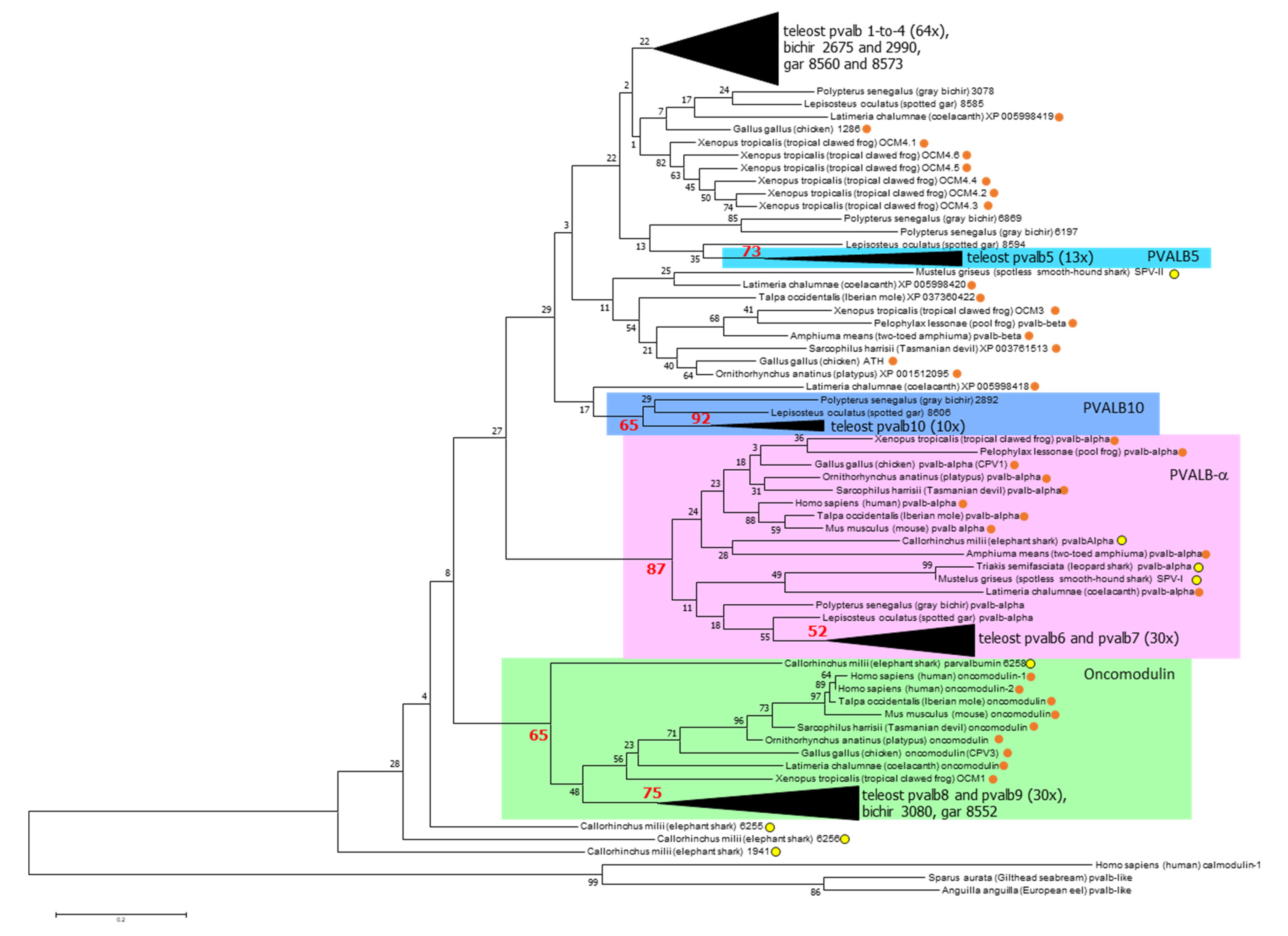
3.2. The Overall Phylogeny of Parvalbumin Sequences in Jawed Vertebrate Species; There Are Ancient Alpha and Oncomodulin Lineages, but Their Relationships to the Other Parvalbumins Cannot Be Assessed with Certainty
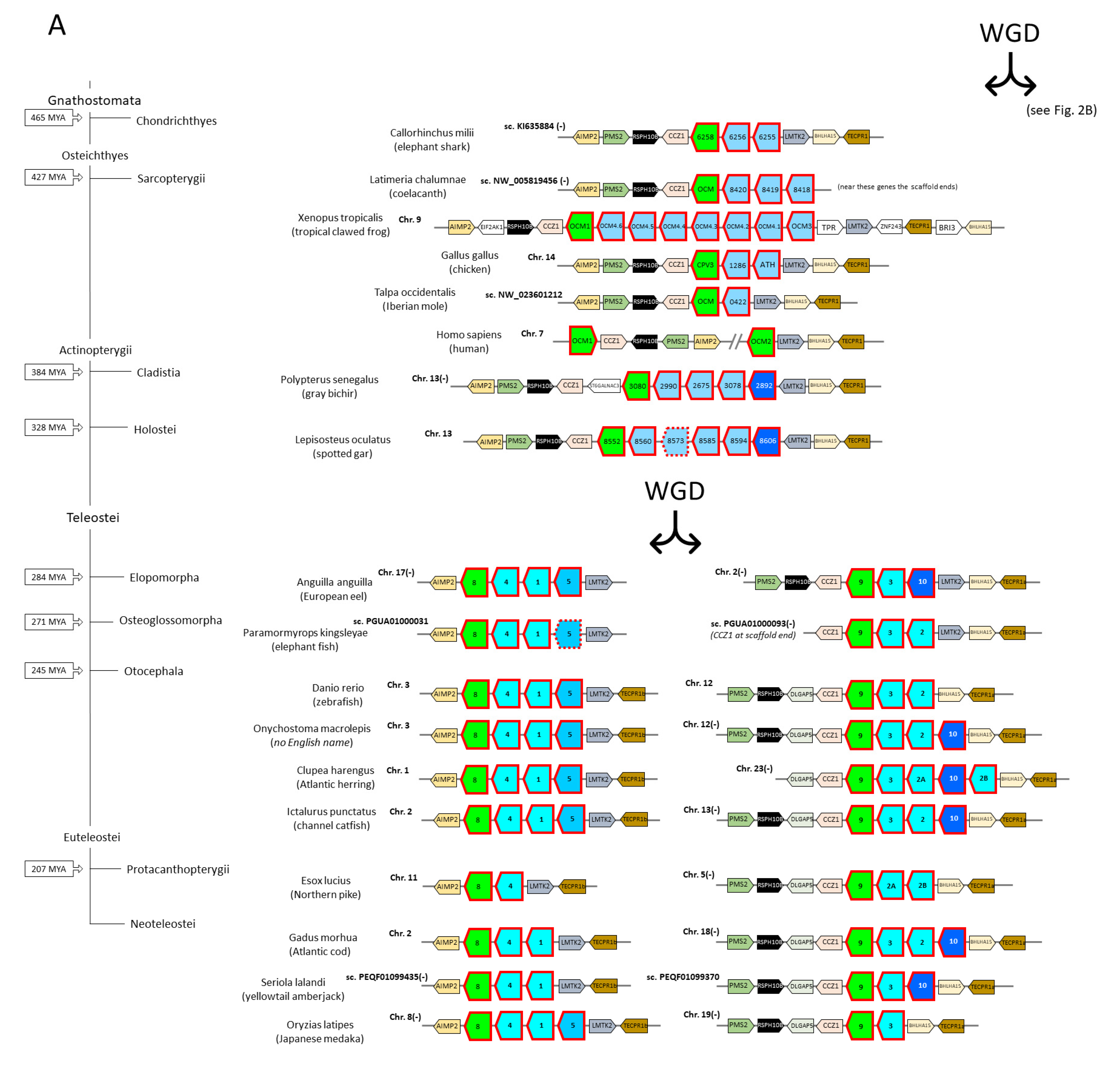
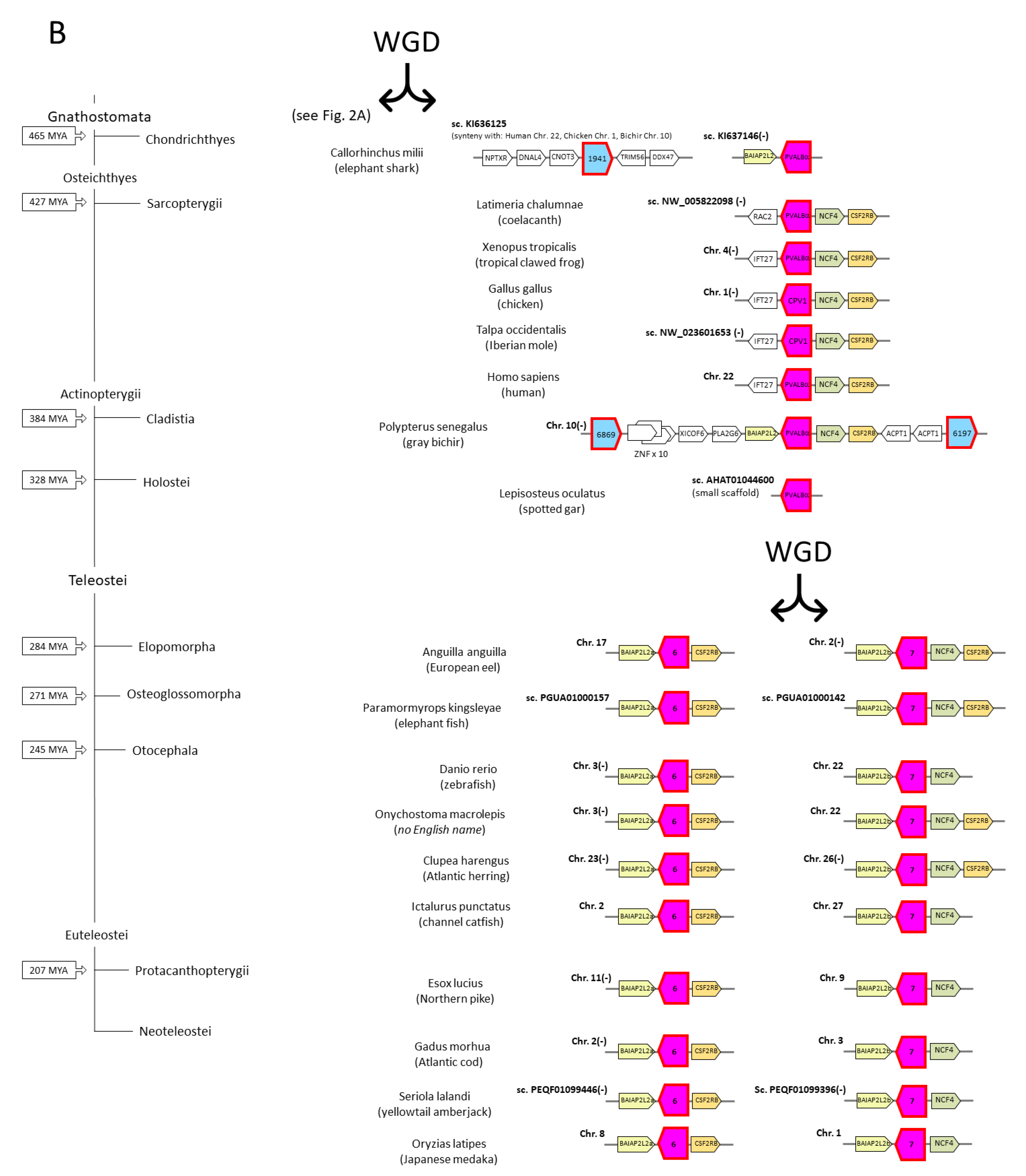
3.3. The Parvalbumin Genes Map to Paralogous Regions That Derived from Whole-Genome Duplication Events
3.4. Parvalbumin Nomenclature and Lineages in Teleost Fish; Delving beyond the Concept of “Microheterogeneity”
3.5. The Consensus Sequence of Parvalbumins Mapped to the Molecular Structures
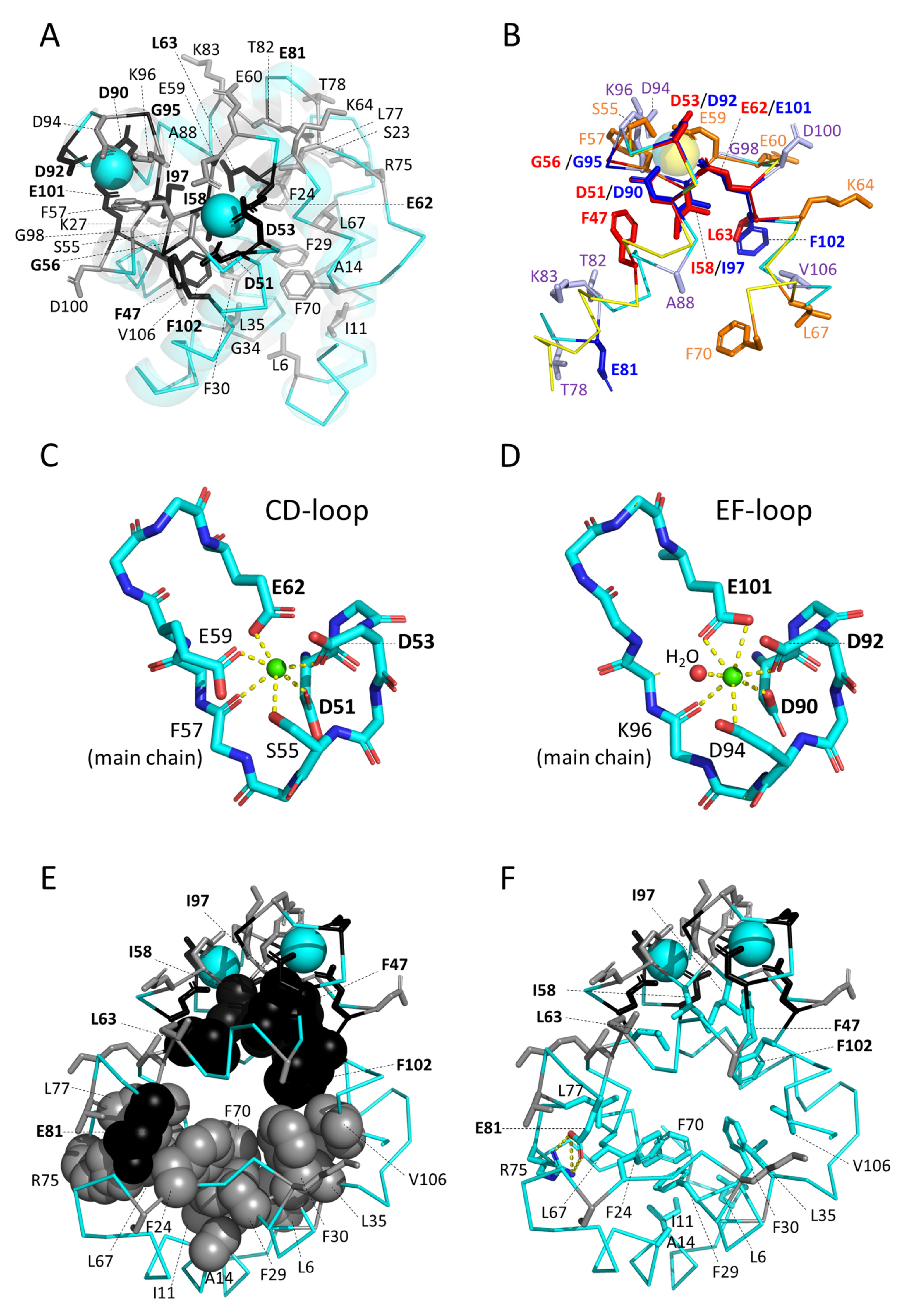
3.5.1. Similarities between and within Parvalbumins
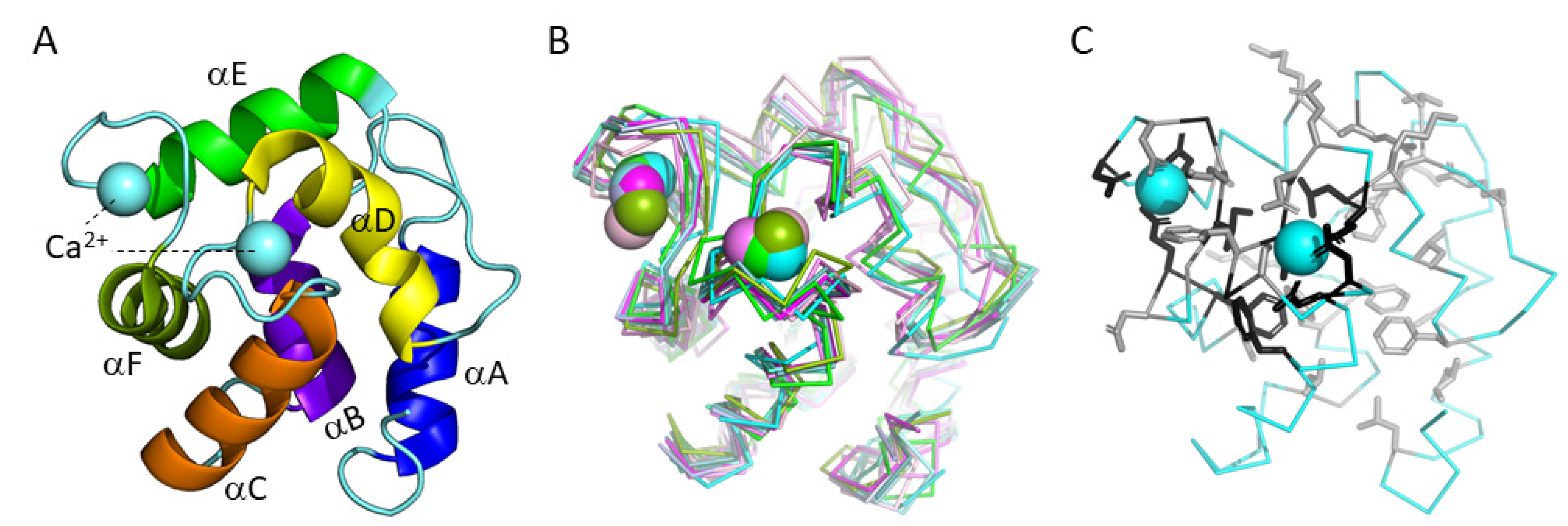
3.5.2. The Binding of Ca2+
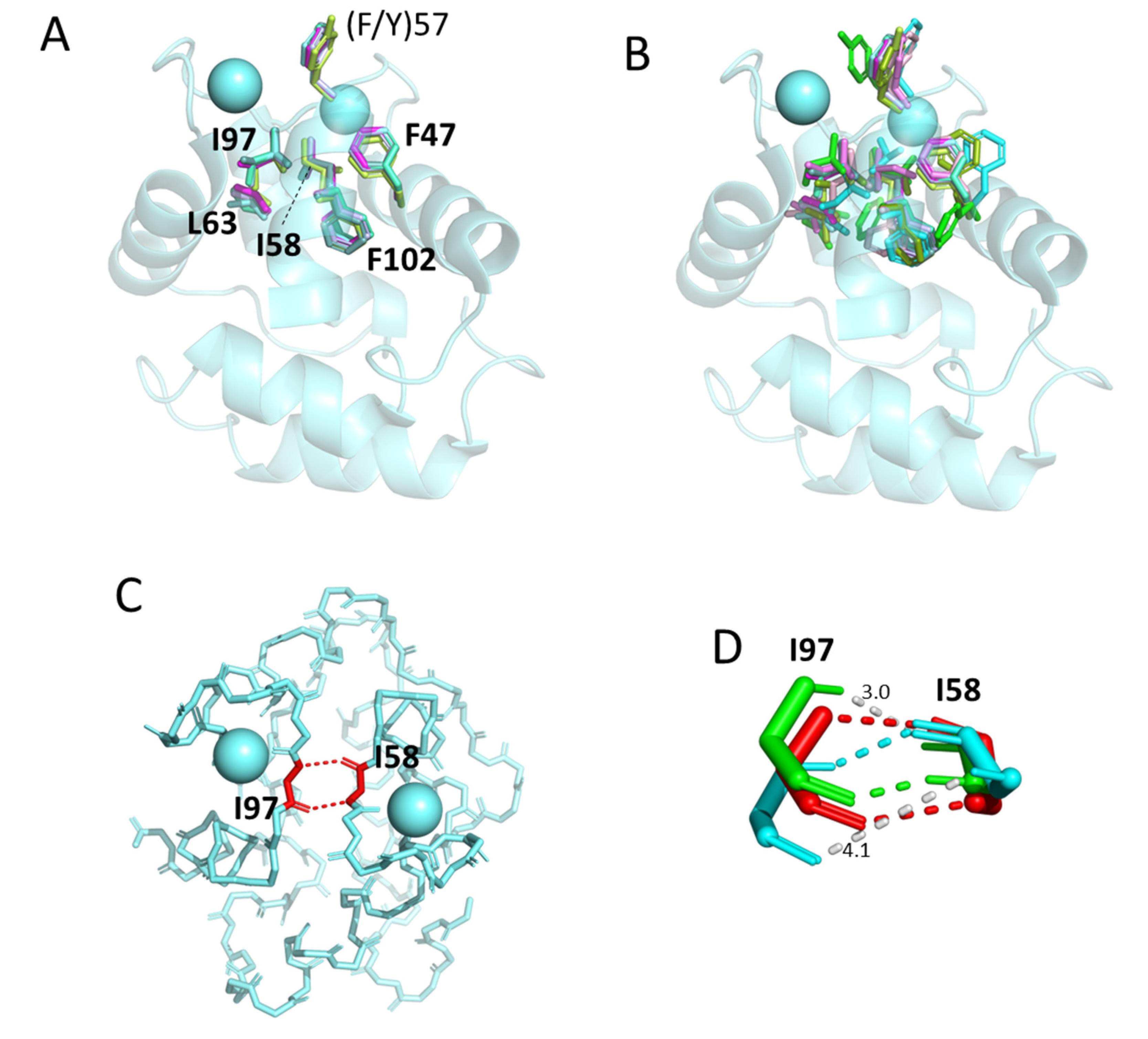
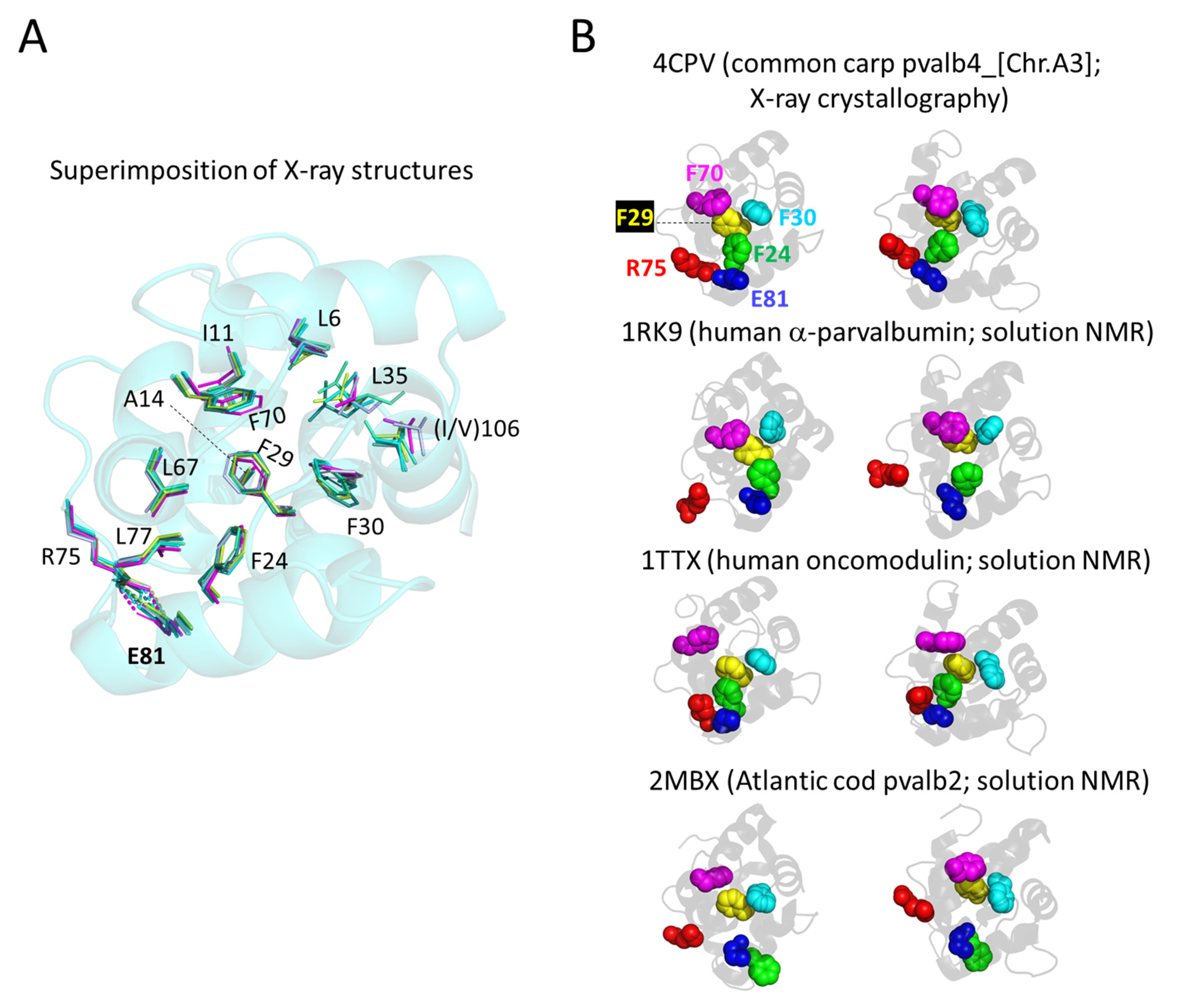
3.5.3. The Conserved Inner Core
3.5.4. The Conserved Core Residues Have Similar Positions in Those Structures of α-Parvalbumins, Oncomodulins, and Non-α/Non-Oncomodulin Parvalbumins That Are Determined by X-ray Crystallography
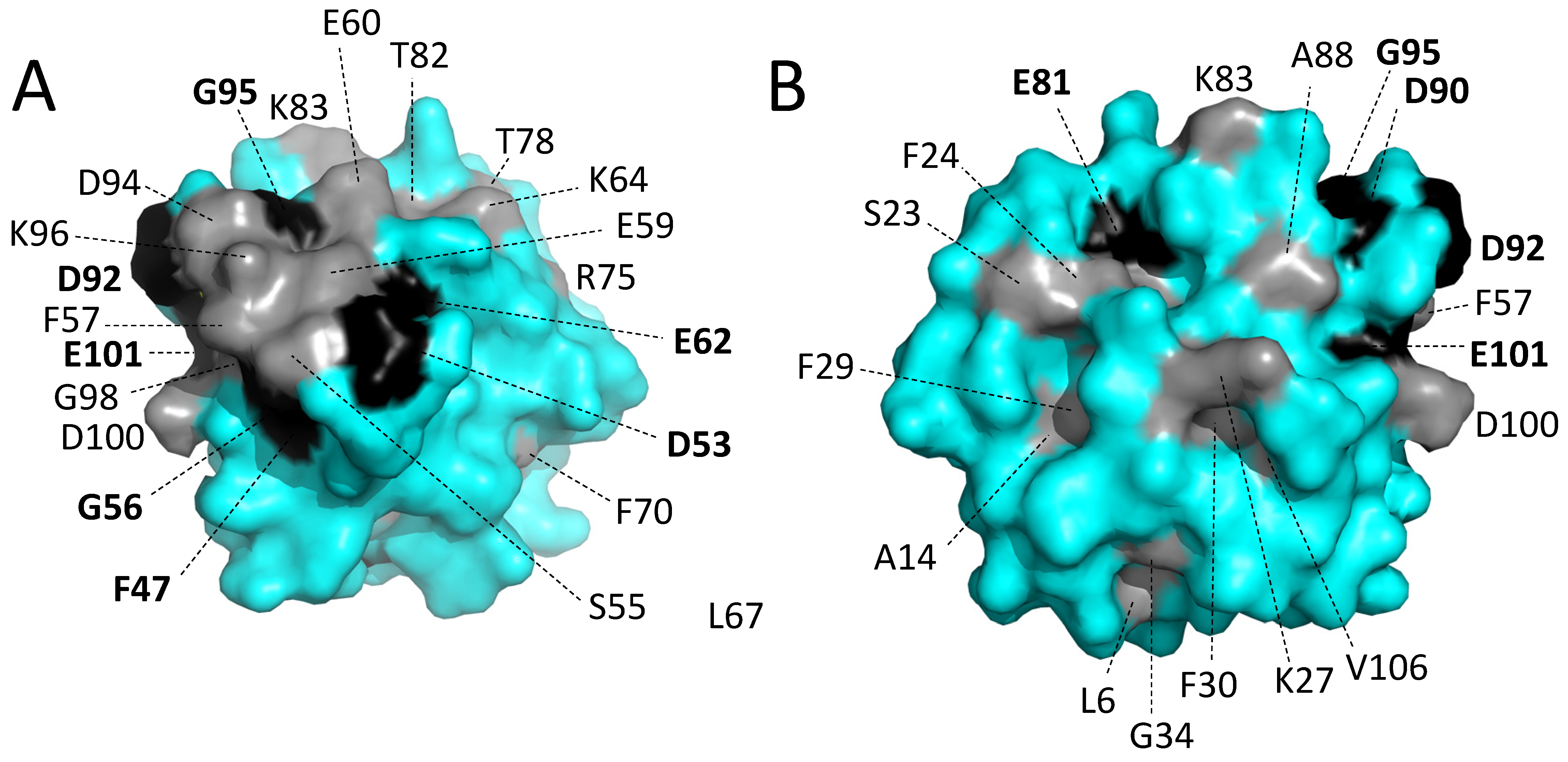
3.5.5. The Conserved Residues at the Molecular Surface
3.6. Comparing the α-Parvalbumins, Oncomodulins, and Other Parvalbumins
3.6.1. Characteristic Features of α-Parvalbumins
3.6.2. Characteristic Features of Oncomodulins
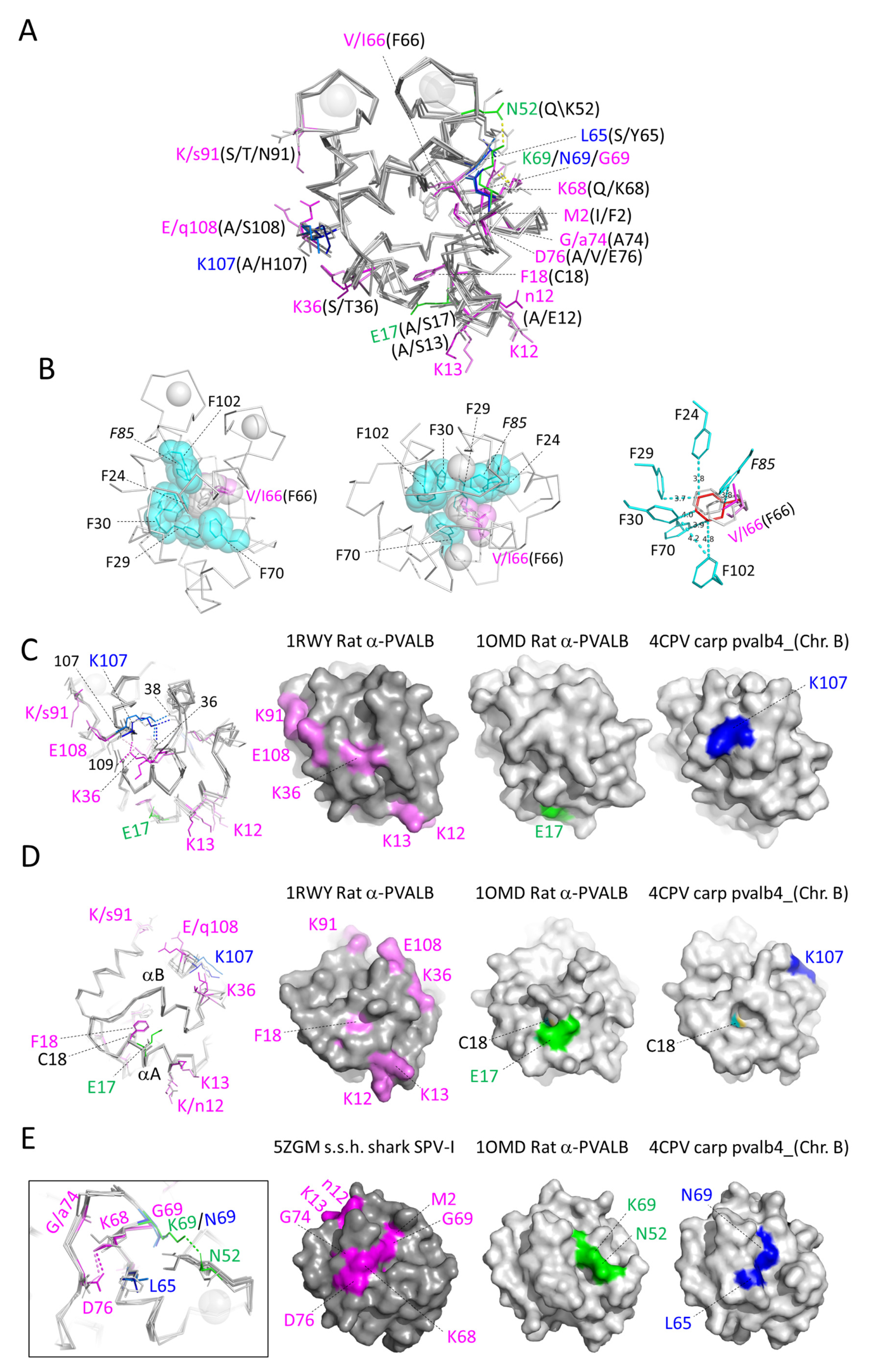
3.6.3. Characteristic Features of Non-α/Non-Oncomodulin Parvalbumins
3.7. Characteristic Features of the Individual Non-α/Non-Oncomodulin Parvalbumins in Teleost Fish
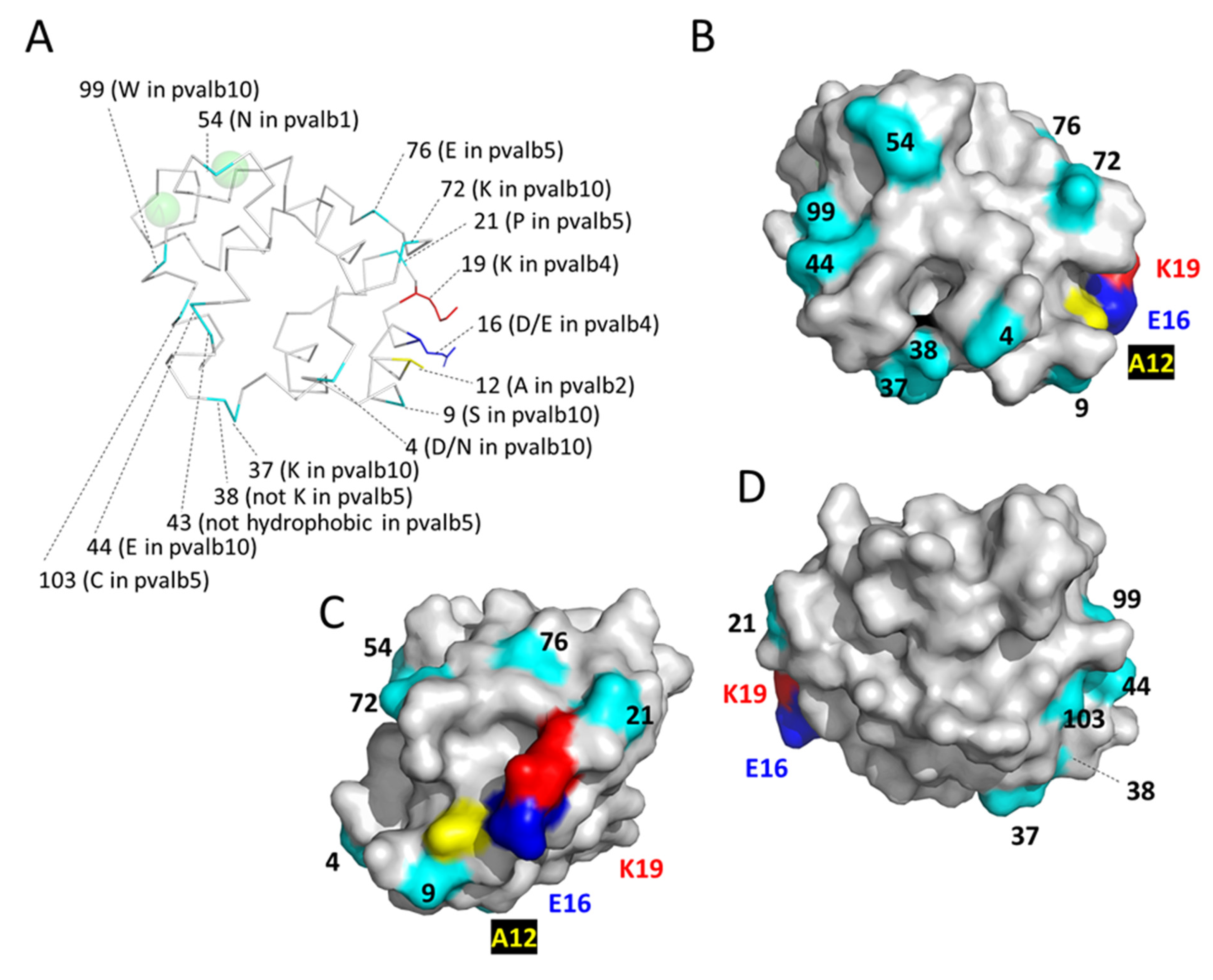
3.7.1. Characteristic Features of Teleost Fish Parvalbumins pvalb1-to-4
3.7.2. Characteristic Features of Teleost Fish pvalb5
3.7.3. Characteristic Features of Teleost Fish pvalb10
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Kuehn, A.; Swoboda, I.; Arumugam, K.; Hilger, C.; Hentges, F. Fish allergens at a glance: Variable allergenicity of parvalbumins, the major fish allergens. Front. Immunol. 2014, 5, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verkhratsky, A.; Parpura, V. Calcium signalling and calcium channels: Evolution and general principles. Eur. J. Pharmacol. 2014, 739, 1–3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaller, B. Cytosolic Ca2+ buffers. Cold Spring Harb. Perspect. Biol. 2010, 2, a004051. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baba, M.L.; Goodman, M.; Berger-Cohn, J.; Demaille, J.G.; Matsuda, G. The early adaptive evolution of calmodulin. Mol. Biol. Evol. 1984, 1, 442–455. [Google Scholar]
- Pauls, T.L.; Cox, J.A.; Berchtold, M.W. The Ca2+(-)binding proteins parvalbumin and oncomodulin and their genes: New structural and functional findings. Biochim. Biophys. Acta 1996, 1306, 39–54. [Google Scholar] [CrossRef]
- Girard, F.; Venail, J.; Schwaller, B.; Celio, M.R. The EF-hand Ca(2+)-binding protein super-family: A genome-wide analysis of gene expression patterns in the adult mouse brain. Neuroscience 2015, 294, 116–155. [Google Scholar] [CrossRef]
- Haiech, J.; Moreau, M.; Leclerc, C.; Kilhoffer, M.C. Facts and conjectures on calmodulin and its cousin proteins, parvalbumin and troponin C. Biochim. Biophys. Acta Mol. Cell Res. 2019, 1866, 1046–1053. [Google Scholar] [CrossRef]
- Raymackers, J.M.; Gailly, P.; Schoor, M.C.; Pette, D.; Schwaller, B.; Hunziker, W.; Celio, M.R.; Gillis, J.M. Tetanus relaxation of fast skeletal muscles of the mouse made parvalbumin deficient by gene inactivation. J. Physiol. 2000, 527, 355–364. [Google Scholar] [CrossRef]
- Schwaller, B. Cytosolic Ca2+ Buffers Are Inherently Ca2+ Signal Modulators. Cold Spring Harb. Perspect. Biol. 2020, 12, a035543. [Google Scholar] [CrossRef]
- Kumar, V.D.; Lee, L.; Edwards, B.F. Refined crystal structure of calcium-liganded carp parvalbumin 4.25 at 1.5-A resolution. Biochemistry 1990, 29, 1404–1412. [Google Scholar] [CrossRef]
- Kretsinger, R.H.; Nockolds, C.E. Carp muscle calcium-binding protein. II: Structure determination and general description. J. Biol. Chem. 1973, 248, 3313–3326. [Google Scholar] [CrossRef] [PubMed]
- Deuticke, H.J. Über die Sedimentationskonstante von Muskelproteinen. Phys. Chem. 1934, 224, 216–228. [Google Scholar] [CrossRef]
- Henrotte, J.G. A crystalline constituent from myogen of carp muscles. Nature 1952, 169, 968–969. [Google Scholar] [CrossRef] [PubMed]
- Pechère, J.F. Muscular parvalbumins as homologous proteins. Comp. Biochem. Physiol. 1968, 24, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Nockolds, C.E.; Kretsinger, R.H.; Coffee, C.J.; Bradshaw, R.A. Structure of a calcium-binding carp myogen. Proc. Natl. Acad. Sci. USA 1972, 69, 581–584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coffee, C.J.; Bradshaw, R.A. Carp muscle calcium-binding protein. I. Characterization of the tryptic peptides and the complete amino acid sequence of component B. J. Biol. Chem. 1973, 248, 3305–3312. [Google Scholar] [CrossRef] [PubMed]
- Capony, J.P.; Rydèn, L.; Demaille, J.; Pechère, J.F. The primary structure of the major parvalbumin from hake muscle. Overlapping peptides obtained with chemical and enzymatic methods. The complete amino-acid sequence. Eur. J. Biochem. 1973, 32, 97–108. [Google Scholar] [CrossRef]
- Rao, K.S.; Gerday, C. Low molecular weight proteins of pike (esox lucius) white muscles. II. Chemical and physical properties. Comp. Biochem. Physiol. B 1973, 44, 1113–1125. [Google Scholar]
- Frankenne, F.; Joassin, L.; Gerday, C. The amino acid sequence of the pike (Esox lucius) parvalbumin 3. FEBS Lett. 1973, 35, 145–147. [Google Scholar] [CrossRef] [Green Version]
- Lehky, P.; Blum, H.E.; Stein, E.A.; Fischer, E.H. Isolation and characterization of parvalbumins from the skeletal muscle of higher vertebrates. J. Biol. Chem. 1974, 249, 4332–4334. [Google Scholar] [CrossRef]
- Enfield, D.L.; Ericsson, L.H.; Blum, H.E.; Fischer, E.H.; Neurath, H. Amino-acid sequence of parvalbumin from rabbit skeletal muscle. Proc. Natl. Acad. Sci. USA 1975, 72, 1309–1313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pechère, J.P.; Capony, J. Demaille. Evolutionary Aspects of the Structure of Muscular Parvalbumins. Syst. Biol. 1973, 22, 533–548. [Google Scholar]
- Goodman, M.; Pechère, J.F. The evolution of muscular parvalbumins investigated by the maximum parsimony method. J. Mol. Evol. 1977, 9, 131–158. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, M.W. Structure and expression of genes encoding the three-domain Ca2+-binding proteins parvalbumin and oncomodulin. Biochim. Biophys. Acta 1989, 1009, 201–215. [Google Scholar] [CrossRef]
- Modrell, M.S.; Lyne, M.; Carr, A.R.; Zakon, H.H.; Buckley, D.; Campbell, A.S.; Davis, M.C.; Micklem, G.; Baker, C.V. Insights into electrosensory organ development, physiology and evolution from a lateral line-enriched transcriptome. eLife 2017, 6, e24197. [Google Scholar] [CrossRef]
- Mukherjee, S.; Bartoš, O.; Zdeňková, K.; Hanák, P.; Horká, P.; Musilova, Z. Evolution of the Parvalbumin Genes in Teleost Fishes after the Whole-Genome Duplication. Fishes 2021, 6, 70. [Google Scholar] [CrossRef]
- Celio, M.R.; Heizmann, C.W. Calcium-binding protein parvalbumin as a neuronal marker. Nature 1981, 293, 300–302. [Google Scholar] [CrossRef]
- Kawaguchi, Y.; Kubota, Y. Neurochemical features and synaptic connections of large physiologically-identified GABAergic cells in the rat frontal cortex. Neuroscience 1998, 85, 677–701. [Google Scholar] [CrossRef] [PubMed]
- Sohal, V.S.; Zhang, F.; Yizhar, O.; Deisseroth, K. Parvalbumin neurons and gamma rhythms enhance cortical circuit performance. Nature 2009, 459, 698–702. [Google Scholar] [CrossRef] [Green Version]
- Caillard, O.; Moreno, H.; Schwaller, B.; Llano, I.; Celio, M.R.; Marty, A. Role of the calcium-binding protein parvalbumin in short-term synaptic plasticity. Proc. Natl. Acad. Sci. USA 2000, 97, 13372–13377. [Google Scholar] [CrossRef] [Green Version]
- Eggermann, E.; Jonas, P. How the ‘slow’ Ca(2+) buffer parvalbumin affects transmitter release in nanodomain-coupling regimes. Nat. Neurosci. 2011, 15, 20–22. [Google Scholar] [CrossRef] [PubMed]
- Celio, M.R.; Heizmann, C.W. Calcium-binding protein parvalbumin is associated with fast contracting muscle fibres. Nature 1982, 297, 504–506. [Google Scholar] [CrossRef]
- Pauls, T.L.; Portis, F.; Macrì, E.; Belser, B.; Heitz, P.; Doglioni, C.; Celio, M.R. Parvalbumin is expressed in normal and pathological human parathyroid glands. J. Histochem. Cytochem. 2000, 48, 105–111. [Google Scholar] [CrossRef]
- Olinger, E.; Schwaller, B.; Loffing, J.; Gailly, P.; Devuyst, O. Parvalbumin: Calcium and magnesium buffering in the distal nephron. Nephrol. Dial. Transplant. 2012, 27, 3988–3994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Henzi, T.; Schwaller, B. Antagonistic Regulation of Parvalbumin Expression and Mitochondrial Calcium Handling Capacity in Renal Epithelial Cells. PLoS ONE 2015, 10, e0142005. [Google Scholar] [CrossRef] [Green Version]
- Heizmann, C.W.; Berchtold, M.W.; Rowlerson, A.M. Correlation of parvalbumin concentration with relaxation speed in mammalian muscles. Proc. Natl. Acad. Sci. USA 1982, 79, 7243–7247. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Müntener, M.; Käser, L.; Weber, J.; Berchtold, M.W. Increase of skeletal muscle relaxation speed by direct injection of parvalbumin cDNA. Proc. Natl. Acad. Sci. USA 1995, 92, 6504–6508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwaller, B.; Dick, J.; Dhoot, G.; Carroll, S.; Vrbova, G.; Nicotera, P.; Pette, D.; Wyss, A.; Bluethmann, H.; Hunziker, W.; et al. Prolonged contraction-relaxation cycle of fast-twitch muscles in parvalbumin knockout mice. Am. J. Physiol. 1999, 276, C395–C403. [Google Scholar] [CrossRef]
- Stavrou, D.; Haglid, K.G.; Weidenbach, W. The brain specific proteins S 100 and 14.3.2 in experimental brain tumors of the rat. Z. Gesamte Exp. Med. 1971, 156, 237–242. [Google Scholar] [CrossRef] [PubMed]
- MacManus, J.P. Occurrence of a low-molecular-weight calcium-binding protein in neoplastic liver. Cancer Res. 1979, 39, 3000–3005. [Google Scholar]
- Brewer, L.M.; MacManus, J.P. Localization and synthesis of the tumor protein oncomodulin in extraembryonic tissues of the fetal rat. Dev. Biol. 1985, 112, 49–58. [Google Scholar] [CrossRef] [PubMed]
- Sakaguchi, N.; Henzl, M.T.; Thalmann, I.; Thalmann, R.; Schulte, B.A. Oncomodulin is expressed exclusively by outer hair cells in the organ of Corti. J. Histochem. Cytochem. 1998, 46, 29–40. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yin, Y.; Henzl, M.T.; Lorber, B.; Nakazawa, T.; Thomas, T.T.; Jiang, F.; Langer, R.; Benowitz, L.I. Oncomodulin is a macrophage-derived signal for axon regeneration in retinal ganglion cells. Nat. Neurosci. 2006, 9, 843–852. [Google Scholar] [CrossRef] [PubMed]
- Tong, B.; Hornak, A.J.; Maison, S.F.; Ohlemiller, K.K.; Liberman, M.C.; Simmons, D.D. Oncomodulin, an EF-Hand Ca2+ Buffer, Is Critical for Maintaining Cochlear Function in Mice. J. Neurosci. 2016, 36, 1631–1635. [Google Scholar] [CrossRef]
- Niemi, J.P.; DeFrancesco-Oranburg, T.; Cox, A.; Lindborg, J.A.; Echevarria, F.D.; McCluskey, J.; Simmons, D.D.; Zigmond, R.E. The Conditioning Lesion Response in Dorsal Root Ganglion Neurons Is Inhibited in Oncomodulin Knock-Out Mice. eNeuro 2022, 9, ENEURO-0477. [Google Scholar] [CrossRef]
- Climer, L.K.; Cox, A.M.; Reynolds, T.J.; Simmons, D.D. Oncomodulin: The Enigmatic Parvalbumin Protein. Front. Mol. Neurosci. 2019, 12, 235. [Google Scholar] [CrossRef] [Green Version]
- Brewer, J.M.; Wunderlich, J.K.; Kim, D.H.; Carr, M.Y.; Beach, G.G.; Ragland, W.L. Avian thymic hormone (ATH) is a parvalbumin. Biochem. Biophys. Res. Commun. 1989, 160, 1155–1161. [Google Scholar] [CrossRef]
- Kuster, T.; Staudenmann, W.; Hughes, G.J.; Heizmann, C.W. Parvalbumin isoforms in chicken muscle and thymus. Amino acid sequence analysis of muscle parvalbumin by tandem mass spectrometry. Biochemistry 1991, 30, 8812–8816. [Google Scholar] [CrossRef]
- Henzl, M.T.; Serda, R.E.; Boschi, J.M. Identification of a novel parvalbumin in avian thymic tissue. Biochem. Biophys. Res. Commun. 1991, 177, 881–887. [Google Scholar] [CrossRef]
- Hapak, R.C.; Zhao, H.; Boschi, J.M.; Henzl, M.T. Novel avian thymic parvalbumin displays high degree of sequence homology to oncomodulin. J. Biol. Chem. 1994, 269, 5288–5296. [Google Scholar] [CrossRef]
- Vásquez, G.M.; Ragland, W.L. Avian thymic hormone treatment of peripheral blood mononuclear cells from young chicks stimulates acute graft-versus-host reaction in chicken embryos. Dev. Comp. Immunol. 2005, 29, 663–668. [Google Scholar] [CrossRef] [PubMed]
- Novak Kujundžić, R.; Steffens, W.L.; Brewer, J.M.; Henzl, M.T.; Ragland, W.L. Characterization of avian thymic hormone and chicken parvalbumin 3 target cells. Int. Immunopharmacol. 2013, 15, 282–288. [Google Scholar] [CrossRef] [PubMed]
- Heller, S.; Bell, A.M.; Denis, C.S.; Choe, Y.; Hudspeth, A.J. Parvalbumin 3 is an abundant Ca2+ buffer in hair cells. J. Assoc. Res. Otolaryngol. 2002, 3, 488–498. [Google Scholar] [CrossRef] [PubMed]
- Brewer, J.M.; Arnold, J.; Beach, G.G.; Ragland, W.L.; Wunderlich, J.K. Comparison of the amino acid sequences of tissue-specific parvalbumins from chicken muscle and thymus and possible evolutionary significance. Biochem. Biophys. Res. Commun. 1991, 181, 226–231. [Google Scholar] [CrossRef]
- Rada, J.A.; Wiechmann, A.F. Ocular expression of avian thymic hormone: Changes during the recovery from induced myopia. Mol. Vis. 2009, 15, 778–792. [Google Scholar]
- Roquet, F.; Declercq, J.P.; Tinant, B.; Rambaud, J.; Parello, J. Crystal structure of the unique parvalbumin component from muscle of the leopard shark (Triakis semifasciata). The first X-ray study of an alpha-parvalbumin. J. Mol. Biol. 1992, 223, 705–720. [Google Scholar] [CrossRef]
- Stephen, J.N.; Sharp, M.F.; Ruethers, T.; Taki, A.; Campbell, D.E.; Lopata, A.L. Allergenicity of bony and cartilaginous fish—Molecular and immunological properties. Clin. Exp. Allergy 2017, 47, 300–312. [Google Scholar] [CrossRef]
- Capony, J.P.; Demaille, J.; Pina, C.; Pechère, J.F. The amino-acid sequence of the most acidic major parvalbumin from frog muscle. Eur. J. Biochem. 1975, 56, 215–227. [Google Scholar] [CrossRef]
- Jauregui-Adell, J.; Pechere, J.F.; Briand, G.; Richet, C.; Demaille, J.G. Amino-acid sequence of an alpha-parvalbumin, pI = 4.88, from frog skeletal muscle. Eur. J. Biochem. 1982, 123, 337–345. [Google Scholar] [CrossRef]
- Sasaki, T.; Tanokura, M.; Asaoka, K. The complete amino acid sequence of bullfrog (Rana catesbeiana) parvalbumin pI4.97. FEBS Lett. 1990, 268, 249–251. [Google Scholar] [CrossRef] [Green Version]
- Taka, H.; Kaga, N.; Fujimura, T.; Mineki, R.; Imaizumi, M.; Suzuki, Y.; Suzuki, R.; Tanokura, M.; Shindo, N.; Murayama, K. Rapid determination of parvalbumin amino acid sequence from Rana catesbeiana (pI 4.78) by combination of ESI mass spectrometry, protein sequencing, and amino acid analysis. J. Biochem. 2000, 127, 723–729. [Google Scholar] [CrossRef] [PubMed]
- Hamada, Y.; Nagashima, Y.; Shiomi, K. Reactivity of serum immunoglobulin E to bullfrog Rana catesbeiana parvalbumins in fish-allergic patients. Fish. Sci. 2004, 70, 1137–1143. [Google Scholar] [CrossRef]
- Gillis, J.M.; Piront, A.; Gosselin-Rey, C. Parvalbumins. Distribution and physical state inside the muscle cell. Biochim. Biophys. Acta 1979, 585, 444–450. [Google Scholar] [CrossRef] [PubMed]
- Friedberg, F. Parvalbumin isoforms in zebrafish. Mol. Biol. Rep. 2005, 32, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Swoboda, I.; Bugajska-Schretter, A.; Verdino, P.; Keller, W.; Sperr, W.R.; Valent, P.; Valenta, R.; Spitzauer, S. Recombinant carp parvalbumin; the major cross-reactive fish allergen: A tool for diagnosis and therapy of fish allergy. J. Immunol. 2002, 168, 4576–4584. [Google Scholar] [CrossRef]
- Bugajska-Schretter, A.; Elfman, L.; Fuchs, T.; Kapiotis, S.; Rumpold, H.; Valenta, R.; Spitzauer, S. Parvalbumin, a cross-reactive fish allergen, contains IgE-binding epitopes sensitive to periodate treatment and Ca2+ depletion. J. Allergy Clin. Immunol. 1998, 101, 67–74. [Google Scholar] [CrossRef]
- Kondo, Y.; Komatsubara, R.; Nakajima, Y.; Yasuda, T.; Kakami, M.; Tsuge, I.; Urisu, A. Parvalbumin is not responsible for cross-reactivity between tuna and marlin: A case report. J. Allergy Clin. Immunol. 2006, 118, 1382–1383. [Google Scholar] [CrossRef]
- Koyama, H.; Kakami, M.; Kawamura, M.; Tokuda, R.; Kondo, Y.; Tsuge, I.; Yamada, K.; Yasuda, T.; Urisu, A. Grades of 43 fish species in Japan based on IgE-binding activity. Allergol. Int. 2006, 55, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Kuehn, A.; Codreanu-Morel, F.; Lehners-Weber, C.; Doyen, V.; Gomez-André, S.A.; Bienvenu, F.; Fischer, J.; Ballardini, N.; van Hage, M.; Perotin, J.M.; et al. Cross-reactivity to fish and chicken meat—A new clinical syndrome. Allergy 2016, 71, 1772–1781. [Google Scholar] [CrossRef]
- Ruethers, T.; Nugraha, R.; Taki, A.C.; O’Malley, A.; Karnaneedi, S.; Zhang, S.; Kapingidza, A.B.; Mehr, S.; Kamath, S.D.; Chruszcz, M.; et al. The first reptilian allergen and major allergen for fish-allergic patients: Crocodile β-parvalbumin. Pediatr. Allergy Immunol. 2022, 33, e13781. [Google Scholar] [CrossRef]
- Hilger, C.; Thill, L.; Grigioni, F.; Lehners, C.; Falagiani, P.; Ferrara, A.; Romano, C.; Stevens, W.; Hentges, F. IgE antibodies of fish allergic patients cross-react with frog parvalbumin. Allergy 2004, 59, 653–660. [Google Scholar] [CrossRef] [PubMed]
- Kalic, T.; Morel-Codreanu, F.; Radauer, C.; Ruethers, T.; Taki, A.C.; Swoboda, I.; Hilger, C.; Hoffmann-Sommergruber, K.; Ollert, M.; Hafner, C.; et al. Patients Allergic to Fish Tolerate Ray Based on the Low Allergenicity of Its Parvalbumin. J. Allergy Clin. Immunol. Pract. 2019, 7, 500–508.e11. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ahn, J.; Komatsubara, R.; Terada, A.; Yasuda, T.; Tsuge, I.; Urisu, A. Comparison of allergenic properties of salmon (Oncorhynchus nerka) between landlocked and anadromous species. Allergol. Int. 2009, 58, 295–299. [Google Scholar] [CrossRef] [Green Version]
- Dijkstra, J.M.; Grimholt, U.; Leong, J.; Koop, B.F.; Hashimoto, K. Comprehensive analysis of MHC class II genes in teleost fish genomes reveals dispensability of the peptide-loading DM system in a large part of vertebrates. BMC Evol. Biol. 2013, 13, 260. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dijkstra, J.M. TH2 and Treg candidate genes in elephant shark. Nature 2014, 511, E7–E9. [Google Scholar] [CrossRef]
- Dijkstra, J.M. A method for making alignments of related protein sequences that share very little similarity; shark interleukin 2 as an example. Immunogenetics 2021, 73, 35–51. [Google Scholar] [CrossRef] [PubMed]
- Nelson, J.S.; Grande, T.C.; Wilson, M.V.H. Fishes of the World, 5th ed.; John Wiley & Sons: Hoboken, NJ, USA, 2016. [Google Scholar]
- Jaillon, O.; Aury, J.M.; Brunet, F.; Petit, J.L.; Stange-Thomann, N.; Mauceli, E.; Bouneau, L.; Fischer, C.; Ozouf-Costaz, C.; Bernot, A.; et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature 2004, 431, 946–957. [Google Scholar] [CrossRef] [Green Version]
- Broughton, R.E.; Betancur-R, R.; Li, C.; Arratia, G.; Ortí, G. Multi-locus phylogenetic analysis reveals the pattern and tempo of bony fish evolution. PLoS Curr. 2013, 5. [Google Scholar] [CrossRef]
- Godzik, A.; Sander, C. Conservation of residue interactions in a family of Ca-binding proteins. Protein Eng. 1989, 2, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Philippe, H.; Brinkmann, H.; Lavrov, D.V.; Littlewood, D.T.; Manuel, M.; Wörheide, G.; Baurain, D. Resolving difficult phylogenetic questions: Why more sequences are not enough. PLoS Biol. 2011, 9, e1000602. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, Y.; Glazko, G.V.; Nei, M. Overcredibility of molecular phylogenies obtained by Bayesian phylogenetics. Proc. Natl. Acad. Sci. USA 2002, 99, 16138–16143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, Z.; Zhu, T. Bayesian selection of misspecified models is overconfident and may cause spurious posterior probabilities for phylogenetic trees. Proc. Natl. Acad. Sci. USA 2018, 115, 1854–1859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakatani, Y.; Takeda, H.; Kohara, Y.; Morishita, S. Reconstruction of the vertebrate ancestral genome reveals dynamic genome reorganization in early vertebrates. Genome Res. 2007, 17, 1254–1265. [Google Scholar] [CrossRef] [Green Version]
- Glasauer, S.M.; Neuhauss, S.C. Whole-genome duplication in teleost fishes and its evolutionary consequences. Mol. Genet. Genom. 2014, 289, 1045–1060. [Google Scholar] [CrossRef] [Green Version]
- Lapteva, Y.S.; Uversky, V.N.; Permyakov, S.E. Sequence microheterogeneity of parvalbumin; the major fish allergen. Biochim. Biophys. Acta 2013, 1834, 1607–1614. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Tavarez, R.; Carrera, M.; Pedrosa, M.; Quirce, S.; Rodríguez-Pérez, R.; Gasset, M. Reconstruction of fish allergenicity from the content and structural traits of the component β-parvalbumin isoforms. Sci. Rep. 2019, 9, 16298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bottoms, C.A.; Schuermann, J.P.; Agah, S.; Henzl, M.T.; Tanner, J.J. Crystal structure of rat alpha-parvalbumin at 1.05 Angstrom resolution. Protein Sci. 2004, 13, 1724–1734. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, T.C.; Corson, D.C.; Oikawa, K.; McCubbin, W.D.; Kay, C.M.; Sykes, B.D. 1H NMR spectroscopic studies of calcium-binding proteins. 3. Solution conformations of rat apo-alpha-parvalbumin and metal-bound rat alpha-parvalbumin. Biochemistry 1986, 25, 1835–1846. [Google Scholar] [CrossRef] [PubMed]
- Declercq, J.P.; Evrard, C.; Lamzin, V.; Parello, J. Crystal structure of the EF-hand parvalbumin at atomic resolution (0.91 A) and at low temperature (100 K). Evidence for conformational multistates within the hydrophobic core. Protein Sci. 1999, 8, 2194–2204. [Google Scholar] [CrossRef] [Green Version]
- Babini, E.; Bertini, I.; Capozzi, F.; Del Bianco, C.; Hollender, D.; Kiss, T.; Luchinat, C.; Quattrone, A. Solution structure of human beta-parvalbumin and structural comparison with its paralog alpha-parvalbumin and with their rat orthologs. Biochemistry 2004, 43, 16076–16085. [Google Scholar] [CrossRef]
- Kretsinger, R.H. Gene triplication deduced from the tertiary structure of a muscle calcium binding protein. Nat. New Biol. 1972, 240, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Barker, W.C.; Ketcham, L.K.; Dayhoff, M.O. A comprehensive examination of protein sequences for evidence of internal gene duplication. J. Mol. Evol. 1978, 10, 265–281. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, M.W.; Epstein, P.; Beaudet, A.L.; Payne, M.E.; Heizmann, C.W.; Means, A.R. Structural organization and chromosomal assignment of the parvalbumin gene. J. Biol. Chem. 1987, 262, 8696–8701. [Google Scholar] [CrossRef] [PubMed]
- Ahmed, F.R.; Rose, D.R.; Evans, S.V.; Pippy, M.E.; To, R. Refinement of recombinant oncomodulin at 1.30 A resolution. J. Mol. Biol. 1993, 230, 1216–1224. [Google Scholar] [CrossRef] [PubMed]
- Cates, M.S.; Berry, M.B.; Ho, E.L.; Li, Q.; Potter, J.D.; Phillips, G.N., Jr. Metal-ion affinity and specificity in EF-hand proteins: Coordination geometry and domain plasticity in parvalbumin. Structure 1999, 7, 1269–1278. [Google Scholar] [CrossRef] [PubMed]
- Hoh, F.; Cavé, A.; Strub, M.P.; Banères, J.L.; Padilla, A. Removing the invariant salt bridge of parvalbumin increases flexibility in the AB-loop structure. Acta Crystallogr. D Biol. Crystallogr. 2009, 65, 733–743. [Google Scholar] [CrossRef] [PubMed]
- Schuermann, J.P.; Tan, A.; Tanner, J.J.; Henzl, M.T. Structure of avian thymic hormone—A high-affinity avian beta-parvalbumin in the Ca2+-free and Ca2+-bound states. J. Mol. Biol. 2010, 397, 991–1002. [Google Scholar] [CrossRef] [Green Version]
- Mutus, B.; Palmer, E.J.; MacManus, J.P. Disulfide-linked dimer of oncomodulin: Comparison to calmodulin. Biochemistry 1988, 27, 5615–5622. [Google Scholar] [CrossRef]
- Werner, T.E.R.; Bernson, D.; Esbjörner, E.K.; Rocha, S.; Wittung-Stafshede, P. Amyloid formation of fish β-parvalbumin involves primary nucleation triggered by disulfide-bridged protein dimers. Proc. Natl. Acad. Sci. USA 2020, 117, 27997–28004. [Google Scholar] [CrossRef]
- Clayshulte, T.M.; Taylor, D.F.; Henzl, M.T. Reactivity of cysteine 18 in oncomodulin. J. Biol. Chem. 1990, 265, 1800–1805. [Google Scholar] [CrossRef]
- Vologzhannikova, A.A.; Khorn, P.A.; Shevelyova, M.P.; Kazakov, A.S.; Emelyanenko, V.I.; Permyakov, E.A.; Permyakov, S.E. The Highly Conservative Cysteine of Oncomodulin as a Feasible Redox Sensor. Biomolecules 2021, 11, 66. [Google Scholar] [CrossRef] [PubMed]
- Perez-Gordo, M.; Lin, J.; Bardina, L.; Pastor-Vargas, C.; Cases, B.; Vivanco, F.; Cuesta-Herranz, J.; Sampson, H.A. Epitope mapping of Atlantic salmon major allergen by peptide microarray immunoassay. Int. Arch. Allergy Immunol. 2012, 157, 31–40. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, A.; Tanaka, H.; Hamada, Y.; Ishizaki, S.; Nagashima, Y.; Shiomi, K. Comparison of allergenicity and allergens between fish white and dark muscles. Allergy 2006, 61, 357–363. [Google Scholar] [CrossRef]
- Griesmeier, U.; Bublin, M.; Radauer, C.; Vázquez-Cortés, S.; Ma, Y.; Fernández-Rivas, M.; Breiteneder, H. Physicochemical properties and thermal stability of Lep w 1, the major allergen of whiff. Mol. Nutr. Food Res. 2010, 54, 861–869. [Google Scholar] [CrossRef] [PubMed]
- Somkuti, J.; Bublin, M.; Breiteneder, H.; Smeller, L. Pressure-temperature stability, Ca2+ binding, and pressure-temperature phase diagram of cod parvalbumin: Gad m 1. Biochemistry 2012, 51, 5903–5911. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dijkstra, J.M.; Kondo, Y. Comprehensive Sequence Analysis of Parvalbumins in Fish and Their Comparison with Parvalbumins in Tetrapod Species. Biology 2022, 11, 1713. https://doi.org/10.3390/biology11121713
Dijkstra JM, Kondo Y. Comprehensive Sequence Analysis of Parvalbumins in Fish and Their Comparison with Parvalbumins in Tetrapod Species. Biology. 2022; 11(12):1713. https://doi.org/10.3390/biology11121713
Chicago/Turabian StyleDijkstra, Johannes M., and Yasuto Kondo. 2022. "Comprehensive Sequence Analysis of Parvalbumins in Fish and Their Comparison with Parvalbumins in Tetrapod Species" Biology 11, no. 12: 1713. https://doi.org/10.3390/biology11121713
APA StyleDijkstra, J. M., & Kondo, Y. (2022). Comprehensive Sequence Analysis of Parvalbumins in Fish and Their Comparison with Parvalbumins in Tetrapod Species. Biology, 11(12), 1713. https://doi.org/10.3390/biology11121713




