Genomics, Origin and Selection Signals of Loudi Cattle in Central Hunan
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Sample Collection and Sequencing
2.3. Read Mapping and Variant Calling
2.4. Genetic Diversity Analysis
2.5. Population Structure Analysis
2.6. Admixture Event Ancestry Inference
2.7. Detection of Selection Signatures
2.8. Enrichment Analyses and Visualization
2.9. Data Availability
3. Results
3.1. Genome Resequencing, SNP Detection and Diversity
3.2. Population Structure and Demography
3.3. Origin and Ancestry Inference
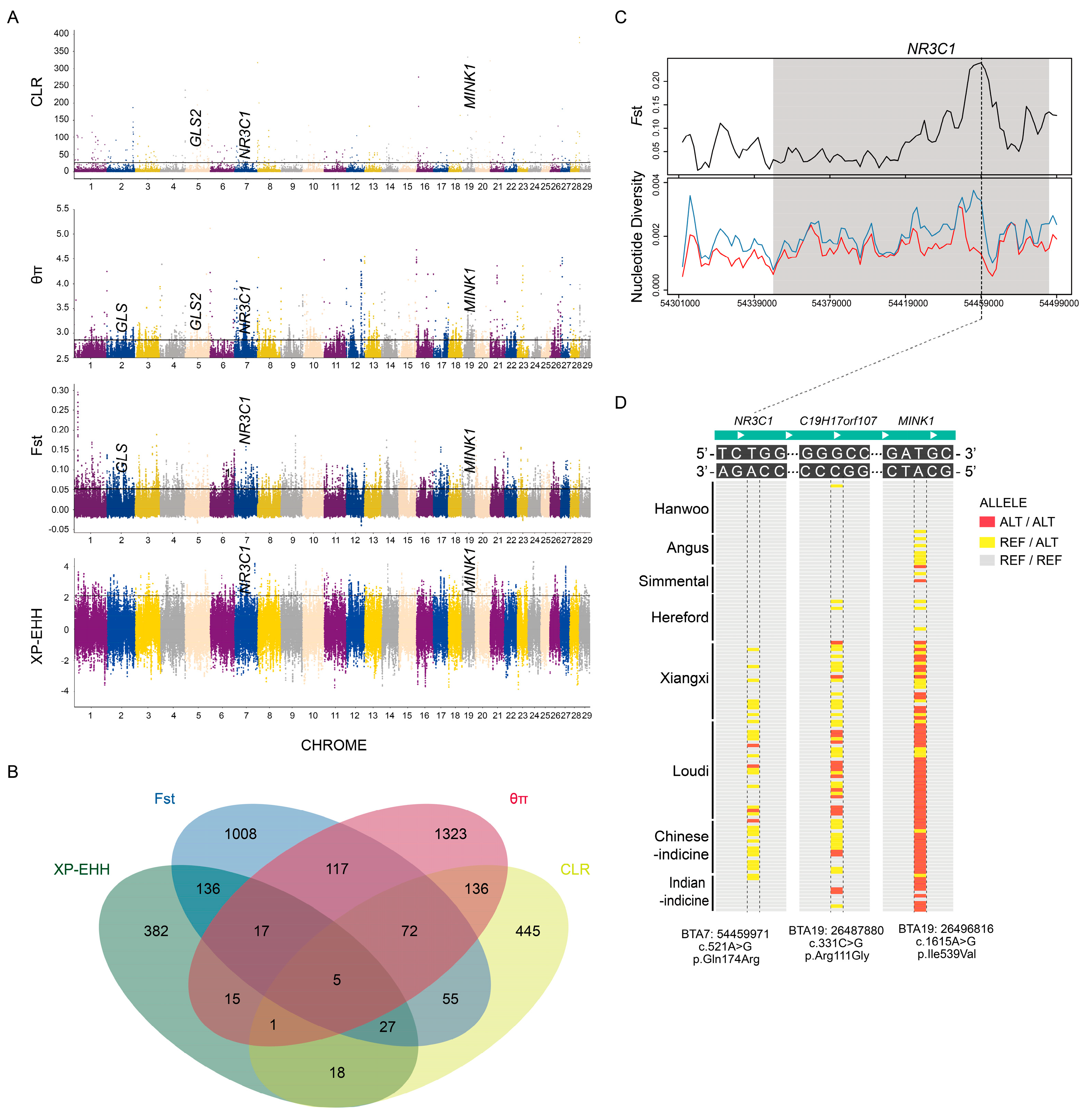
3.4. Patterns of Selection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Decker, J.E.; McKay, S.D.; Rolf, M.M.; Kim, J.; Molina Alcala, A.; Sonstegard, T.S.; Hanotte, O.; Gotherstrom, A.; Seabury, C.M.; Praharani, L.; et al. Worldwide patterns of ancestry, divergence, and admixture in domesticated cattle. PLoS Genet. 2014, 10, e1004254. [Google Scholar] [CrossRef] [PubMed]
- Bradley, D.G.; MacHugh, D.E.; Cunningham, P.; Loftus, R.T. Mitochondrial diversity and the origins of African and European cattle. Proc. Natl. Acad. Sci. USA 1996, 93, 5131–5135. [Google Scholar] [CrossRef] [PubMed]
- MacHugh, D.E.; Shriver, M.D.; Loftus, R.T.; Cunningham, P.; Bradley, D.G. Microsatellite DNA variation and the evolution, domestication and phylogeography of taurine and zebu cattle (Bos taurus and Bos indicus). Genetics 1997, 146, 1071–1086. [Google Scholar] [CrossRef]
- Chen, N.; Cai, Y.; Chen, Q.; Li, R.; Wang, K.; Huang, Y.; Hu, S.; Huang, S.; Zhang, H.; Zheng, Z.; et al. Whole-genome resequencing reveals world-wide ancestry and adaptive introgression events of domesticated cattle in East Asia. Nat. Commun. 2018, 9, 2337. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.D.; Ding, X.D.; Wang, S.; Wojcik, J.M.; Zhang, Y.; Tokarska, M.; Li, Y.; Wang, M.S.; Faruque, O.; Nielsen, R.; et al. Pervasive introgression facilitated domestication and adaptation in the Bos species complex. Nat. Ecol. Evol. 2018, 2, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- Barendse, W. Climate Adaptation of Tropical Cattle. Annu. Rev. Anim. Biosci. 2017, 5, 133–150. [Google Scholar] [CrossRef] [PubMed]
- Li, X. Fireside chat about Loudi cattle (in Chinese). In Loudi Daily; Publicity Department of Loudi Municipal Committee: Loudi, China, 2017; p. 1. [Google Scholar]
- Jin, L.; Qu, K.; Hanif, Q.; Zhang, J.; Liu, J.; Chen, N.; Suolang, Q.; Lei, C.; Huang, B. Whole-Genome Sequencing of Endangered Dengchuan Cattle Reveals Its Genomic Diversity and Selection Signatures. Front. Genet. 2022, 13, 833475. [Google Scholar] [CrossRef]
- Sun, L.; Qu, K.; Ma, X.; Hanif, Q.; Zhang, J.; Liu, J.; Chen, N.; Suolang, Q.; Lei, C.; Huang, B. Whole-Genome Analyses Reveal Genomic Characteristics and Selection Signatures of Lincang Humped Cattle at the China-Myanmar Border. Front. Genet. 2022, 13, 833503. [Google Scholar] [CrossRef]
- Luo, X.; Li, J.; Xiao, C.; Sun, L.; Xiang, W.; Chen, N.; Lei, C.; Lei, H.; Long, Y.; Long, T.; et al. Whole-Genome Resequencing of Xiangxi Cattle Identifies Genomic Diversity and Selection Signatures. Front. Genet. 2022, 13, 816379. [Google Scholar] [CrossRef]
- Sterky, F.H.; Trotter, J.H.; Lee, S.J.; Recktenwald, C.V.; Du, X.; Zhou, B.; Zhou, P.; Schwenk, J.; Fakler, B.; Sudhofa, T.C. Carbonic anhydrase-related protein CA10 is an evolutionarily conserved pan-neurexin ligand (vol 114, pg E1253, 2017). Proc. Natl. Acad. Sci. USA 2017, 114, E2984. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R.; Genome Project Data Processing, S. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Wang, K.; Li, M.; Hakonarson, H. ANNOVAR: Functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010, 38, e164. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Bhati, M.; Kadri, N.K.; Crysnanto, D.; Pausch, H. Assessing genomic diversity and signatures of selection in Original Braunvieh cattle using whole-genome sequencing data. BMC Genom. 2020, 21, 27. [Google Scholar] [CrossRef]
- Kirin, M.; McQuillan, R.; Franklin, C.S.; Campbell, H.; McKeigue, P.M.; Wilson, J.F. Genomic runs of homozygosity record population history and consanguinity. PLoS ONE 2010, 5, e13996. [Google Scholar] [CrossRef]
- Lorenzana, G.P.; Figueiro, H.V.; Kaelin, C.B.; Barsh, G.S.; Johnson, J.; Karlsson, E.; Morato, R.G.; Sana, D.A.; Cullen, L.; May, J.A., Jr.; et al. Whole-genome sequences shed light on the demographic history and contemporary genetic erosion of free-ranging jaguar (Panthera onca) populations. J. Genet. Genom. 2022, 49, 77–80. [Google Scholar] [CrossRef]
- Keller, M.C.; Visscher, P.M.; Goddard, M.E. Quantification of Inbreeding Due to Distant Ancestors and Its Detection Using Dense Single Nucleotide Polymorphism Data (vol 189, pg 237, 2011). Genetics 2012, 190, 283. [Google Scholar]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Zhang, C.; Dong, S.S.; Xu, J.Y.; He, W.M.; Yang, T.L. PopLDdecay: A fast and effective tool for linkage disequilibrium decay analysis based on variant call format files. Bioinformatics 2019, 35, 1786–1788. [Google Scholar] [CrossRef] [PubMed]
- Patterson, N.; Price, A.L.; Reich, D. Population structure and eigenanalysis. PLoS Genet. 2006, 2, e190. [Google Scholar] [CrossRef] [PubMed]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Maples, B.K.; Gravel, S.; Kenny, E.E.; Bustamante, C.D. RFMix: A discriminative modeling approach for rapid and robust local-ancestry inference. Am. J. Hum. Genet. 2013, 93, 278–288. [Google Scholar] [CrossRef]
- Nielsen, R.; Williamson, S.; Kim, Y.; Hubisz, M.J.; Clark, A.G.; Bustamante, C. Genomic scans for selective sweeps using SNP data. Genome Res. 2005, 15, 1566–1575. [Google Scholar] [CrossRef]
- DeGiorgio, M.; Huber, C.D.; Hubisz, M.J.; Hellmann, I.; Nielsen, R. SweepFinder2: Increased sensitivity, robustness and flexibility. Bioinformatics 2016, 32, 1895–1897. [Google Scholar] [CrossRef]
- Sabeti, P.C.; Varilly, P.; Fry, B.; Lohmueller, J.; Hostetter, E.; Cotsapas, C.; Xie, X.; Byrne, E.H.; McCarroll, S.A.; Gaudet, R.; et al. Genome-wide detection and characterization of positive selection in human populations. Nature 2007, 449, 913–918. [Google Scholar] [CrossRef] [PubMed]
- Bu, D.C.; Luo, H.T.; Huo, P.P.; Wang, Z.H.; Zhang, S.; He, Z.H.; Wu, Y.; Zhao, L.H.; Liu, J.J.; Guo, J.C.; et al. KOBAS-i: Intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 2021, 49, W317–W325. [Google Scholar] [CrossRef]
- Leinonen, R.; Sugawara, H.; Shumway, M.; International Nucleotide Sequence Database, C. The sequence read archive. Nucleic Acids Res. 2011, 39, D19–D21. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Chen, X.; Zhang, S.; Zhu, J.; Tang, B.; Wang, A.; Dong, L.; Zhang, Z.; Yu, C.; Sun, Y.; et al. The Genome Sequence Archive Family: Toward Explosive Data Growth and Diverse Data Types. Genom. Proteom. Bioinform. 2021, 19, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Members, C.-N.; Partners. Database Resources of the National Genomics Data Center, China National Center for Bioinformation in 2022. Nucleic Acids Res. 2022, 50, D27–D38. [Google Scholar] [CrossRef]
- Trizzino, M.; Zucco, A.; Deliard, S.; Wang, F.; Barbieri, E.; Veglia, F.; Gabrilovich, D.; Gardini, A. EGR1 is a gatekeeper of inflammatory enhancers in human macrophages. Sci. Adv. 2021, 7, eaaz8836. [Google Scholar] [CrossRef]
- Qi, X.W.; Wang, H.G.; Xia, L.C.; Lin, R.W.; Li, T.; Guan, C.N.; Liu, T.T. miR-30b-5p releases HMGB1 via UBE2D2/KAT2B/HMGB1 pathway to promote pro-inflammatory polarization and recruitment of macrophages. Atherosclerosis 2021, 324, 38–45. [Google Scholar] [CrossRef]
- Larabi, A.; Barnich, N.; Nguyen, H.T.T. New insights into the interplay between autophagy, gut microbiota and inflammatory responses in IBD. Autophagy 2020, 16, 38–51. [Google Scholar] [CrossRef]
- Zheng, P.L.; Chen, Q.Z.; Tian, X.Y.; Qian, N.N.; Chai, P.Y.; Liu, B.; Hu, J.J.; Blackstone, C.; Zhu, D.S.; Teng, J.L.; et al. DNA damage triggers tubular endoplasmic reticulum extension to promote apoptosis by facilitating ER-mitochondria signaling. Cell Res. 2018, 28, 833–854. [Google Scholar] [CrossRef]
- Olahova, M.; Peter, B.; Szilagyi, Z.; Diaz-Maldonado, H.; Singh, M.; Sommerville, E.W.; Blakely, E.L.; Collier, J.J.; Hoberg, E.; Stranecky, V.; et al. POLRMT mutations impair mitochondrial transcription causing neurological disease. Nat. Commun. 2021, 12, 1135. [Google Scholar] [CrossRef]
- Yan, L.; Shamir, A.; Skirzewski, M.; Leiva-Salcedo, E.; Kwon, O.B.; Karavanova, I.; Paredes, D.; Malkesman, O.; Bailey, K.R.; Vullhorst, D.; et al. Neuregulin-2 ablation results in dopamine dysregulation and severe behavioral phenotypes relevant to psychiatric disorders. Mol. Psychiatry 2018, 23, 1233–1243. [Google Scholar] [CrossRef] [PubMed]
- Labreche, K.; Simeonova, I.; Kamoun, A.; Gleize, V.; Chubb, D.; Letouze, E.; Riazalhosseini, Y.; Dobbins, S.E.; Elarouci, N.; Ducray, F.; et al. TCF12 is mutated in anaplastic oligodendroglioma. Nat. Commun. 2015, 6, 7207. [Google Scholar] [CrossRef] [PubMed]
- Deb, S.; Felix, D.A.; Koch, P.; Deb, M.K.; Szafranski, K.; Buder, K.; Sannai, M.; Groth, M.; Kirkpatrick, J.; Pietsch, S.; et al. Tnfaip2/exoc3-driven lipid metabolism is essential for stem cell differentiation and organ homeostasis. Embo Rep. 2021, 22, e49328. [Google Scholar] [CrossRef] [PubMed]
- Jing, X.; Ren, D.; Gao, F.; Chen, Y.; Wu, X.; Han, Y.; Han, Q.; Li, L.; Wang, X.; Tang, W.; et al. Gene deficiency or pharmacological inhibition of PDCD4-mediated FGR signaling protects against acute kidney injury. Acta Pharm. Sin. B 2021, 11, 394–405. [Google Scholar] [CrossRef]
- Sveidahl Johansen, O.; Ma, T.; Hansen, J.B.; Markussen, L.K.; Schreiber, R.; Reverte-Salisa, L.; Dong, H.; Christensen, D.P.; Sun, W.; Gnad, T.; et al. Lipolysis drives expression of the constitutively active receptor GPR3 to induce adipose thermogenesis. Cell 2021, 184, 3502–3518.e3533. [Google Scholar] [CrossRef]
- Mejhert, N.; Kuruvilla, L.; Gabriel, K.R.; Elliott, S.D.; Guie, M.A.; Wang, H.; Lai, Z.W.; Lane, E.A.; Christiano, R.; Danial, N.N.; et al. Partitioning of MLX-Family Transcription Factors to Lipid Droplets Regulates Metabolic Gene Expression. Mol. Cell 2020, 77, 1251–1264.e1259. [Google Scholar] [CrossRef]
- Fuhrmann, D.C.; Brune, B. Mitochondrial composition and function under the control of hypoxia. Redox Biol. 2017, 12, 208–215. [Google Scholar] [CrossRef]
- Munier, C.C.; De Maria, L.; Edman, K.; Gunnarsson, A.; Longo, M.; MacKintosh, C.; Patel, S.; Snijder, A.; Wissler, L.; Brunsveld, L.; et al. Glucocorticoid receptor Thr524 phosphorylation by MINK1 induces interactions with 14-3-3 protein regulators. J. Biol. Chem. 2021, 296, 100551. [Google Scholar] [CrossRef]
- Chen, S.; Lin, B.Z.; Baig, M.; Mitra, B.; Lopes, R.J.; Santos, A.M.; Magee, D.A.; Azevedo, M.; Tarroso, P.; Sasazaki, S.; et al. Zebu cattle are an exclusive legacy of the South Asia neolithic. Mol. Biol. Evol. 2010, 27, 1–6. [Google Scholar] [CrossRef]
- Edelman, N.B.; Mallet, J. Prevalence and Adaptive Impact of Introgression. Annu. Rev. Genet. 2021, 55, 265–283. [Google Scholar] [CrossRef]
- Yue, M.; Luo, D.J.; Yu, S.S.; Liu, P.; Zhou, Q.; Hu, M.J.; Liu, Y.Y.; Wang, S.; Huang, Q.; Niu, Y.X.; et al. Misshapen/NIK-related kinase (MINK1) is involved in platelet function, hemostasis, and thrombus formation. Blood 2016, 127, 927–937. [Google Scholar] [CrossRef] [PubMed]
- Zhu, K.; Jin, X.; Chi, Z.; Chen, S.; Wu, S.; Sloan, R.D.; Lin, X.; Neculai, D.; Wang, D.; Hu, H.; et al. Priming of NLRP3 inflammasome activation by Msn kinase MINK1 in macrophages. Cell Mol. Immunol. 2021, 18, 2372–2382. [Google Scholar] [CrossRef] [PubMed]
- Witze, A. Extreme heatwaves: Surprising lessons from the record warmth. Nature 2022, 608, 464–465. [Google Scholar] [CrossRef] [PubMed]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jin, L.; Zhang, B.; Luo, J.; Li, J.; Liang, J.; Wu, W.; Xie, Y.; Li, F.; Lei, C.; Yi, K. Genomics, Origin and Selection Signals of Loudi Cattle in Central Hunan. Biology 2022, 11, 1775. https://doi.org/10.3390/biology11121775
Jin L, Zhang B, Luo J, Li J, Liang J, Wu W, Xie Y, Li F, Lei C, Yi K. Genomics, Origin and Selection Signals of Loudi Cattle in Central Hunan. Biology. 2022; 11(12):1775. https://doi.org/10.3390/biology11121775
Chicago/Turabian StyleJin, Liangliang, Baizhong Zhang, Jing Luo, Jianbo Li, Juyong Liang, Wanghe Wu, Yongzhong Xie, Fuqiang Li, Chuzhao Lei, and Kangle Yi. 2022. "Genomics, Origin and Selection Signals of Loudi Cattle in Central Hunan" Biology 11, no. 12: 1775. https://doi.org/10.3390/biology11121775
APA StyleJin, L., Zhang, B., Luo, J., Li, J., Liang, J., Wu, W., Xie, Y., Li, F., Lei, C., & Yi, K. (2022). Genomics, Origin and Selection Signals of Loudi Cattle in Central Hunan. Biology, 11(12), 1775. https://doi.org/10.3390/biology11121775





