High-Throughput Sequencing-Based Analysis of Changes in the Vaginal Microbiome during the Disease Course of Patients with Bacterial Vaginosis: A Case–Control Study
Abstract
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Sample Collection
2.3. DNA Extraction and PCR Amplification
2.4. Illumina MiSeq Sequencing
2.5. Processing of Sequencing Data
2.6. Statistical Analysis
3. Results
3.1. Characteristics of the Study Population
3.2. Sequencing Results
3.3. Species Composition
3.4. Community Composition
3.5. Vaginal Microbiome Richness and Diversity
3.6. Typing Analysis at Phylum Level
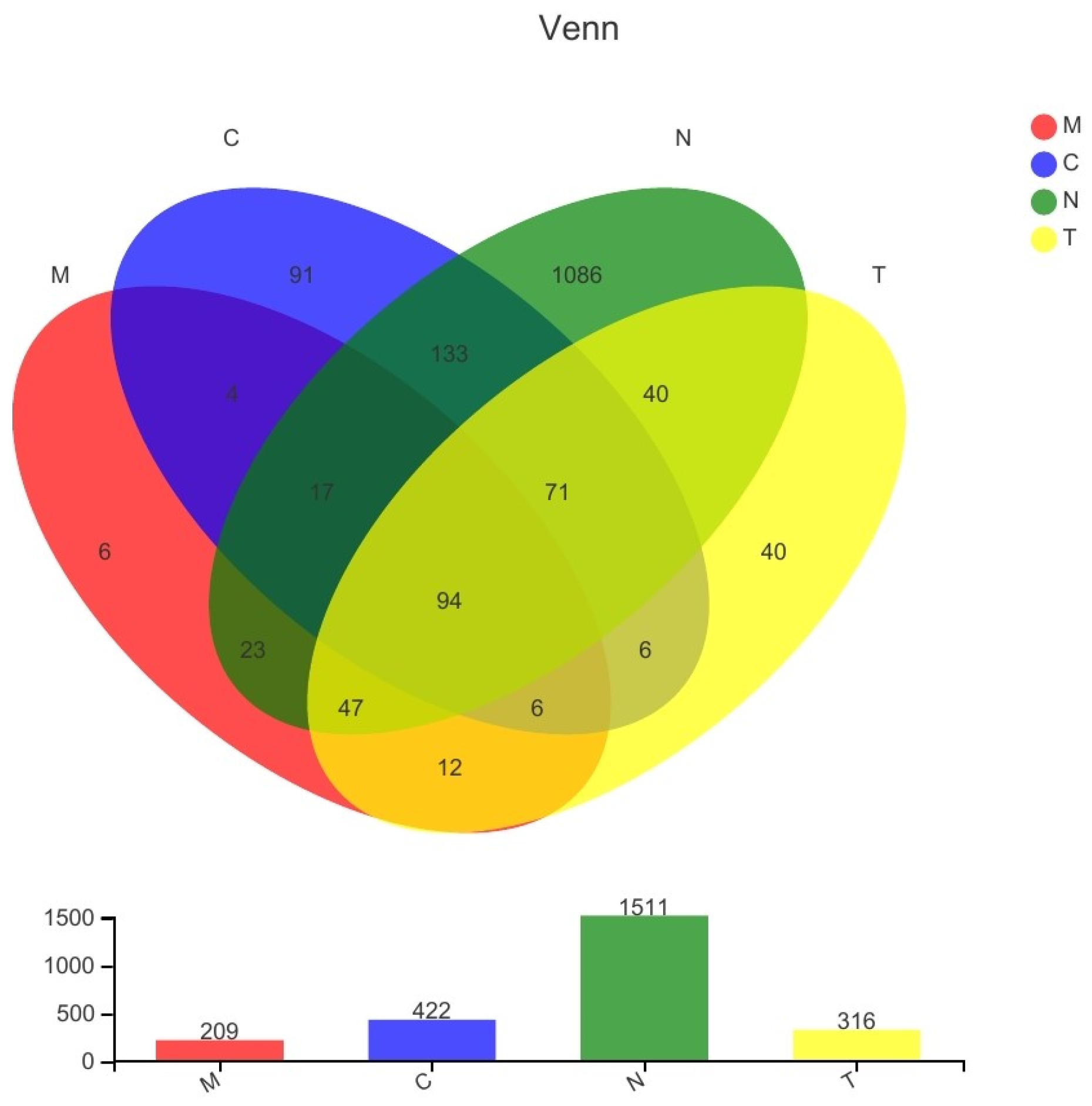
3.7. Differences in Bacterial Communities among Different Groups
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kairys, N.; Garg, M. Bacterial Vaginosis; StatPearls Publishing LLC.: Treasure Island, FL, USA, 2021. [Google Scholar]
- Lin, Y.P.; Chen, W.C.; Cheng, C.M.; Shen, C.J. Vaginal pH value for clinical diagnosis and treatment of common vaginitis. Diagnostics 2021, 11, 1996. [Google Scholar] [CrossRef] [PubMed]
- Morrill, S.; Gilbert, N.M.; Lewis, A.L. Gardnerella vaginalis as a cause of bacterial vaginosis: Appraisal of the evidence from In Vivo models. Front. Cell. Infect. Microbiol. 2020, 10, 168. [Google Scholar] [CrossRef] [PubMed]
- Vodstrcil, L.A.; Muzny, C.A.; Plummer, E.L.; Sobel, J.D.; Bradshaw, C.S. Bacterial vaginosis: Drivers of recurrence and challenges and opportunities in partner treatment. BMC Med. 2021, 19, 194. [Google Scholar] [CrossRef] [PubMed]
- Ravel, J.; Moreno, I.; Simón, C. Bacterial vaginosis and its association with infertility, endometritis, and pelvic inflammatory disease. Am. J. Obstet. Gynecol. 2021, 224, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lu, Y.; Chen, T.; Li, R. The female vaginal microbiome in health and bacterial vaginosis. Front. Cell. Infect. Microbiol. 2021, 11, 631972. [Google Scholar] [CrossRef] [PubMed]
- Coudray, M.S.; Madhivanan, P. Bacterial vaginosis—A brief synopsis of the literature. Eur. J. Obstet. Gynecol. Reprod. Biol. 2020, 245, 143–148. [Google Scholar] [CrossRef]
- Superti, F.; De Seta, F. Warding off recurrent yeast and bacterial vaginal infections: Lactoferrin and lactobacilli. Microorganisms 2020, 8, 130. [Google Scholar] [CrossRef]
- Wang, Z.; He, Y.; Zheng, Y. Probiotics for the treatment of bacterial vaginosis: A meta-analysis. Int. J. Environ. Res. Public Health 2019, 16, 3859. [Google Scholar] [CrossRef]
- Han, Y.; Ren, Q.L. Does probiotics work for bacterial vaginosis and vulvovaginal candidiasis. Curr. Opin. Pharmacol. 2021, 61, 83–90. [Google Scholar] [CrossRef]
- Zheng, N.; Guo, R.; Wang, J.; Zhou, W.; Ling, Z. Contribution of Lactobacillus iners to vaginal health and diseases: A systematic review. Front. Cell. Infect. Microbiol. 2021, 11, 792787. [Google Scholar] [CrossRef]
- Almeida, M.O.; Viana, M.V.C.; Cerqueira, J.C.; Aburjaile, F.F.; Junior, A.A.Z.; Azevedo, V.; Carvalho, R.D.O. Novel insights in bacterial vaginosis etiology through genomic approaches. An. Da Acad. Bras. De Ciências 2021, 93, e20200945. [Google Scholar] [CrossRef] [PubMed]
- Grewal, K.; MacIntyre, D.A.; Bennett, P.R. The reproductive tract microbiota in pregnancy. Biosci. Rep. 2021, 41, BSR20203908. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Chopra, C.; Mehta, M.; Sharma, V.; Mallubhotla, S.; Sistla, S.; Sistla, J.C.; Bhushan, I. An insight into vaginal microbiome techniques. Life 2021, 11, 1229. [Google Scholar] [CrossRef] [PubMed]
- Lev-Sagie, A.; De Seta, F.; Verstraelen, H.; Ventolini, G.; Lonnee-Hoffmann, R.; Vieira-Baptista, P. The vaginal microbiome: II. Vaginal dysbiotic conditions. J. Low. Genit. Tract Dis. 2022, 26, 79–84. [Google Scholar] [CrossRef] [PubMed]
- Usyk, M.; Schlecht, N.F.; Pickering, S.; Williams, L.; Sollecito, C.C.; Gradissimo, A.; Porras, C.; Safaeian, M.; Pinto, L.; Herrero, R.; et al. molBV reveals immune landscape of bacterial vaginosis and predicts human papillomavirus infection natural history. Nat. Commun. 2022, 13, 233. [Google Scholar] [PubMed]
- Chacra, L.A.; Fenollar, F.; Diop, K. Bacterial vaginosis: What do we currently know? Front. Cell. Infect. Microbiol. 2021, 11, 672429. [Google Scholar] [CrossRef] [PubMed]
- Redelinghuys, M.J.; Geldenhuys, J.; Jung, H.; Kock, M.M. Bacterial vaginosis: Current diagnostic avenues and future opportunities. Front. Cell. Infect. Microbiol. 2020, 10, 354. [Google Scholar] [CrossRef] [PubMed]
- Muzny, C.A.; Łaniewski, P.; Schwebke, J.R.; Herbst-Kralovetz, M.M. Host-vaginal microbiota interactions in the pathogenesis of bacterial vaginosis. Curr. Opin. Infect. Dis. 2020, 33, 59–65. [Google Scholar] [CrossRef]
- Muzny, C.A.; Taylor, C.M.; Swords, W.E.; Tamhane, A.; Chattopadhyay, D.; Cerca, N.; Schwebke, J.R. An updated conceptual model on the pathogenesis of bacterial vaginosis. J. Infect. Dis. 2019, 220, 1399–1405. [Google Scholar] [CrossRef]
- Bagga, R.; Arora, P. Genital micro-organisms in pregnancy. Front. Public Health 2020, 8, 225. [Google Scholar] [CrossRef]
- Zhao, F.; Chen, Y.; Gao, J.; Wu, M.; Li, C.; Wang, Z.; Huang, N.; Cui, L.; Du, M.; Ying, C. Characterization of vaginal microbiota in women with recurrent spontaneous abortion that can be modified by drug treatment. Front. Cell. Infect. Microbiol. 2021, 11, 680643. [Google Scholar] [CrossRef] [PubMed]
- Theis, K.R.; Florova, V.; Romero, R.; Borisov, A.B.; Winters, A.D.; Galaz, J.; Gomez-Lopez, N. Sneathia: An emerging pathogen in female reproductive disease and adverse perinatal outcomes. Crit. Rev. Microbiol. 2021, 47, 517–542. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.M. The immune response to Prevotella bacteria in chronic inflammatory disease. Immunology 2017, 151, 363–374. [Google Scholar] [CrossRef] [PubMed]
- Muzny, C.A.; Kardas, P. A narrative review of current challenges in the diagnosis and management of bacterial vaginosis. Sex. Transm. Dis. 2020, 47, 441–446. [Google Scholar] [CrossRef]
- Vodstrcil, L.A.; Plummer, E.L.; Doyle, M.; Fairley, C.K.; McGuiness, C.; Bateson, D.; Hocking, J.S.; Law, M.G.; Petoumenos, K.; Donovan, B.; et al. Treating male partners of women with bacterial law vaginosis (StepUp): A protocol for a randomised controlled trial to assess the clinical effectiveness of male partner treatment for reducing the risk of BV recurrence. BMC Infect. Dis. 2020, 20, 834. [Google Scholar]
- Plummer, E.L.; Vodstrcil, L.A.; Doyle, M.; Danielewski, J.A.; Murray, G.L.; Fehler, G.; Fairley, C.K.; Bulach, D.M.; Garland, S.M.; Chow, E.P.F.; et al. A prospective, open-label pilot study of concurrent male partner treatment for bacterial vaginosis. mBio 2021, 12, e0232321. [Google Scholar] [CrossRef]
- Zhang, Y.; Lyu, J.; Ge, L.; Huang, L.; Peng, Z.; Liang, Y.; Zhang, X.; Fan, S. Probiotic Lacticaseibacillus rhamnosus GR-1 and Limosilactobacillus reuteri RC-14 as an adjunctive treatment for bacterial vaginosis do not increase the cure rate in a Chinese cohort: A prospective, parallel-group, randomized, controlled study. Front. Cell. Infect. Microbiol. 2021, 11, 669901. [Google Scholar] [CrossRef]
- Gottschick, C.; Deng, Z.L.; Vital, M.; Masur, C.; Abels, C.; Pieper, D.H.; Rohde, M.; Mendling, W.; Wagner-Döbler, I. Treatment of biofilms in bacterial vaginosis by an amphoteric tenside pessary-clinical study and microbiota analysis. Microbiome 2017, 5, 119. [Google Scholar] [CrossRef]
- Caselli, E.; D′Accolti, M.; Santi, E.; Soffritti, I.; Conzadori, S.; Mazzacane, S.; Greco, P.; Contini, C.; Bonaccorsi, G. Vaginal microbiota and cytokine microenvironment in HPV clearance/persistence in women surgically treated for cervical intraepithelial neoplasia: An observational prospective study. Front. Cell. Infect. Microbiol. 2020, 10, 540900. [Google Scholar] [CrossRef] [PubMed]
- Happel, A.U.; Kullin, B.; Gamieldien, H.; Wentzel, N.; Zauchenberger, C.Z.; Jaspan, H.B.; Dabee, S.; Barnabas, S.L.; Jaumdally, S.Z.; Dietrich, J.; et al. Exploring potential of vaginal Lactobacillus isolates from South African women for enhancing treatment for bacterial vaginosis. PLoS Pathog. 2020, 16, e1008559. [Google Scholar] [CrossRef]
- Cohen, C.R.; Wierzbicki, M.R.; French, A.L.; Morris, S.; Newmann, S.; Reno, H.; Green, L.; Miller, S.; Powell, J.; Parks, T.; et al. Randomized trial of lactin-V to prevent recurrence of bacterial vaginosis. N. Engl. J. Med. 2020, 382, 1906–1915. [Google Scholar] [CrossRef] [PubMed]
- Latham-Cork, H.C.; Walker, K.F.; Thornton, J.G.; Gunnarsson, O.S.; Säfholm, A.; Cardell, M.; Strevens, H. A novel non-antimicrobial treatment of bacterial vaginosis: An open label two-private centre study. Eur. J. Obstet. Gynecol. Reprod. Biol. 2021, 256, 419–424. [Google Scholar] [CrossRef] [PubMed]




| Characteristic | N (n = 12) | C (n = 11) | M (n = 9) | T (n = 8) | Total (n = 40) | p-Value | |
|---|---|---|---|---|---|---|---|
| Age, years (mean ± SD) | 37.50 ± 8.81 | 34.91 ± 6.24 | 42.67 ± 10.50 | 38.13 ± 8.29 | 38.10 ± 8.63 | 0.260 | |
| HPV status, n/N (%) | HPV (−) | 12/12(100.00) | 10/11(90.91) | 7/9(77.78) | 7/8(87.50) | 36/40(90.00) | 0.408 |
| HPV (+) | 0/12(0.00) | 1/11(9.09) | 2/9(22.22) | 1/8(12.50) | 4/40(10.00) | ||
| Pathological examination results | NILM | 12/12(100.00) | 8/11(72.72) | 5/9(55.56) | 5/8(62.50) | 30/40(75.00) | 0.271 |
| LSIL | 0/12(0.00) | 1/11(9.09) | 3/9(33.33) | 2/8(25.00) | 6/40(15.00) | ||
| HSIL | 0/12(0.00) | 2/11(18.18) | 1/9(11.11) | 1/8(12.50) | 4/40(10.00) | ||
| Nugent score | 0.58 ± 0.51 | 0.64 ± 0.67 | 7.56 ± 0.53 | 2.25 ± 3.06 | 2.50 ± 3.15 | 0.000 | |
| Lactobacillus | 3.42 ± 0.51 | 3.55 ± 0.52 | 0.33 ± 0.50 | 3.00 ± 1.41 | 2.68 ± 1.49 | 0.000 | |
| Gardnerella vaginalis and Bacteroides | 0.00 ± 0.00 | 0.18 ± 0.60 | 3.89 ± 0.33 | 1.25 ± 1.83 | 1.18 ± 1.77 | 0.000 | |
| Mobiluncus | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | 0.00 ± 0.00 | -- | |
| Domain | Kingdom | Phylum | Class | Order | Family | Genus | Species | OTUs * |
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | 25 | 51 | 137 | 244 | 535 | 869 | 1674 |
| Estimators | C-Mean | C-SD | M-Mean | M-SD | T-Mean | T-SD | N-Mean | N-SD | p-Value | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C-M | C-T | C-N | M-T | M-N | N-T | |||||||||
| Sobs | 70.455 | 54.764 | 58.778 | 20.614 | 61.75 | 43.824 | 211.08 | 191.18 | 0.554 | 0.715 | 0.029 * | 0.857 | 0.029 * | 0.045 * |
| ACE | 101.9 | 63.542 | 102.07 | 48.078 | 86.702 | 41.282 | 234.46 | 182.67 | 0.994 | 0.563 | 0.033 * | 0.493 | 0.048 * | 0.038 * |
| Chao | 92.789 | 65.227 | 83.894 | 28.826 | 81.244 | 45.249 | 229.5 | 183.93 | 0.709 | 0.672 | 0.029 * | 0.886 | 0.030 * | 0.039 * |
| Shannon | 0.410 | 0.282 | 1.827 | 0.702 | 0.836 | 0.845 | 2.264 | 1.877 | 0.000 ** | 0.135 | 0.004 ** | 0.018 * | 0.517 | 0.060 |
| Simpson | 0.812 | 0.156 | 0.264 | 0.142 | 0.682 | 0.332 | 0.387 | 0.361 | 0.000 ** | 0.268 | 0.001 ** | 0.003 ** | 0.351 | 0.081 |
| Coverage | 0.999 | 0.001 | 0.999 | 0.000 | 0.999 | 0.000 | 0.999 | 0.000 | 0.155 | 0.069 | 0.714 | 0.470 | 0.416 | 0.259 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; Peng, Y.; Jiang, N.; Shi, Y.; Ying, C. High-Throughput Sequencing-Based Analysis of Changes in the Vaginal Microbiome during the Disease Course of Patients with Bacterial Vaginosis: A Case–Control Study. Biology 2022, 11, 1797. https://doi.org/10.3390/biology11121797
Gao J, Peng Y, Jiang N, Shi Y, Ying C. High-Throughput Sequencing-Based Analysis of Changes in the Vaginal Microbiome during the Disease Course of Patients with Bacterial Vaginosis: A Case–Control Study. Biology. 2022; 11(12):1797. https://doi.org/10.3390/biology11121797
Chicago/Turabian StyleGao, Jing, Yiqian Peng, Nanyan Jiang, Youhao Shi, and Chunmei Ying. 2022. "High-Throughput Sequencing-Based Analysis of Changes in the Vaginal Microbiome during the Disease Course of Patients with Bacterial Vaginosis: A Case–Control Study" Biology 11, no. 12: 1797. https://doi.org/10.3390/biology11121797
APA StyleGao, J., Peng, Y., Jiang, N., Shi, Y., & Ying, C. (2022). High-Throughput Sequencing-Based Analysis of Changes in the Vaginal Microbiome during the Disease Course of Patients with Bacterial Vaginosis: A Case–Control Study. Biology, 11(12), 1797. https://doi.org/10.3390/biology11121797





