Identification of Drought-Tolerance Genes in the Germination Stage of Soybean
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Methods
Optimum PEG-6000 Concentration Screening
2.3. Phenotype Identification and Drought Tolerance Evaluation in the Germination Stage
2.4. Phenotypic Data Analysis
2.5. Genotype Identification and Analysis
Genotype Identification
2.6. Analysis of Gene Diversity, Linkage Disequilibrium, and Population Structure
2.7. Genome-Wide Association Analysis
3. Results
3.1. Selection of the Optimal Concentration of PEG-6000
3.2. Phenotype Analysis of Soybean Germplasm at the Germination Stage
3.2.1. Descriptive Analysis of Four Germination-Related Traits and Drought Tolerance Traits
3.2.2. Analysis of Drought Tolerance
3.3. Analysis of Soybean Genetic Diversity
3.3.1. Analysis of Genetic Diversity and Linkage Disequilibrium
3.3.2. Population Genetic Structural Analysis
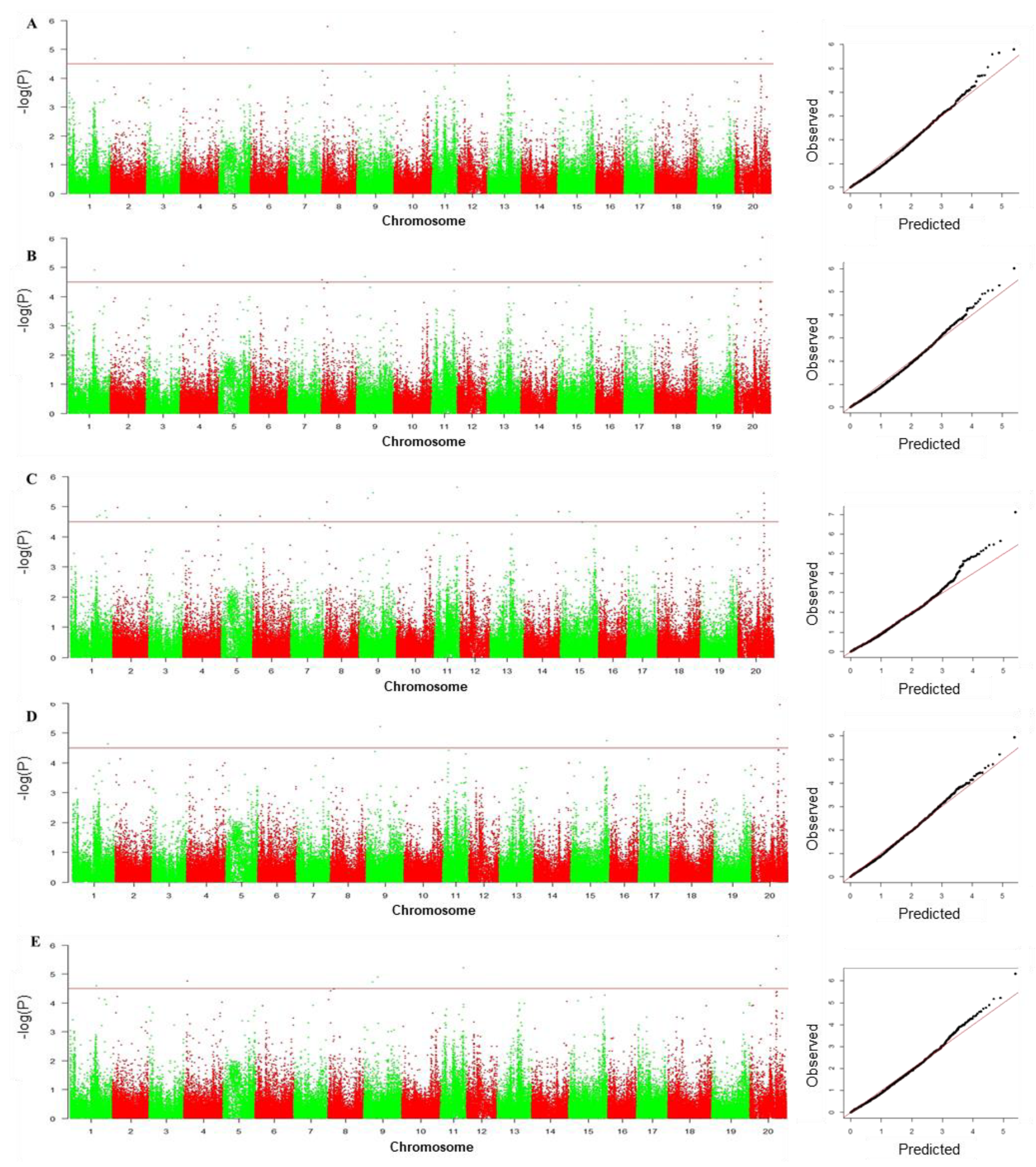
3.4. GWAS to Identify SNPs Associated with Drought Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Du, W.; Yu, D.; Fu, S. Detection of quantitative trait loci for yield and drought tolerance traits in soybean using a recombinant inbred line population. J. Integr. Plant Biol. 2009, 51, 868–878. [Google Scholar] [CrossRef] [PubMed]
- Vijay, R.; Ravichandran, V.; Boominathan, P. Assessment of soybean genotypes for PEG induced drought tolerance at germination and seedling level. Madras Agric. J. 2018, 105, 1. [Google Scholar] [CrossRef]
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Liu, B.; Piao, S.; Wang, X.; Lobell, D.B.; Huang, Y.; Huang, M.; Yan, Y.; Bassu, S.; Ciais, P.; et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc. Natl. Acad. Sci. USA 2017, 114, 9326–9331. [Google Scholar] [CrossRef] [Green Version]
- Ning, H.; Yuan, J.; Dong, Q.; Li, W.; Xue, H.; Wang, Y.; Li, W. Identification of QTLs related to the vertical distribution and seed-set of pod number in soybean [Glycine max L. Merr]. PLoS ONE 2018, 13, e0195830. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Oh, M.; Sakata, K.; Komatsu, S. Gel-free/label-free proteomic analysis of root tip of soybean over time under flooding and drought stresses. J. Proteom. 2016, 130, 42–55. [Google Scholar] [CrossRef]
- Hwang, S.; King, C.A.; Ray, J.D.; Cregan, P.B.; Chen, P.; Carter, T.E.; Li, Z.; Abdel-Haleem, H.; Matson, K.W.; Schapaugh, W.; et al. Confirmation of delayed canopy wilting QTLs from multiple soybean mapping populations. Theor. Appl. Genet. 2015, 128, 2047–2065. [Google Scholar] [CrossRef]
- Ye, H.; Roorkiwal, M.; Valliyodan, B.; Zhou, L.; Chen, P.; Varshney, R.K.; Nguyen, H.T. Genetic diversity of root system architecture in response to drought stress in grain legumes. J. Exp. Bot. 2018, 69, 3267–3277. [Google Scholar] [CrossRef] [Green Version]
- Condon, A.G.; Richards, R.A.; Rebetzks, G.J.; Farquhar, G.D. Breeding for high water-use efficiency. J. Exp. Bot. 2004, 55, 2447–2460. [Google Scholar] [CrossRef] [Green Version]
- Frederick, J.R.; Camp, C.R.; Bauer, P.J. Drought-stress effects on branch and mainstem seed yield and yield components of determinate soybean. Crop Sci. 2001, 41, 759–763. [Google Scholar] [CrossRef]
- Mishra, V.; Cherkauer, K.A. Retrospective droughts in the crop growing season: Implications to corn and soybean yield in the Midwestern United States. Agric. For. Meteorol. 2010, 150, 1030–1045. [Google Scholar] [CrossRef]
- Devi, J.M.; Sinclair, T.R.; Chen, P.; Carter, T.E. Evaluation of elite southern maturity soybean breeding lines for drought tolerant traits. Agron. J. 2014, 106, 1947–1954. [Google Scholar] [CrossRef]
- Zhang, W.B.; Qiu, P.C.; Jiang, H.W.; Liu, C.Y.; Li, C.D.; Hu, G.H.; Chen, Q.S. Dissection of genetic overlap of drought and low-temperature tolerance QTLs at the germination stage using backcross introgression lines in soybean. Mol. Biol. Rep. 2012, 39, 6087–6094. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.X.; Li, H.H.; Gou, Z.W.; Zhang, Y.J.; Wang, X.R.; Ren, H.L.; Wen, Z.X.; Kang, B.K.; Li, Y.H.; Yu, L.L.; et al. Genome-wide association study of soybean seed germination under drought stress. Mol. Genet. Genom. 2020, 295, 661–673. [Google Scholar] [CrossRef]
- Du, W.J.; Wang, M.; Fu, S.X.; Yu, D.Y. Mapping WTLs for seed yield and drought susceptiblity index in soybean (Glycine max L.) across different environments. J. Genet. Genom. 2009, 36, 721–731. [Google Scholar] [CrossRef]
- Ebdon, J.S.; Kopp, K.L. Relationships between water use efficiency, carbon isotope discrimination, and turf performance in genotypes of Kentucky bluegrass during drought. Crop Sci. 2004, 44, 1754–1762. [Google Scholar] [CrossRef]
- Sloane, R.J.; Patterson, R.P.; Carter, J.T.E. Field drought tolerance of a soybean plant introduction. Crop Sci. 1990, 30, 118–123. [Google Scholar] [CrossRef]
- Mian, M.A.R.; Bailey, M.A.; Ashley, D.A.; Wells, R.; Carter, T.E.; Parrott, W.A.; Boerma, H.R. Molecular markers associated with water use efficiency and leaf ash in soybean. Crop Sci. 1996, 36, 1252–1257. [Google Scholar] [CrossRef] [Green Version]
- Hufstetler, E.V.; Boerma, H.R.; Carter, T.E.; Earl, H.J. Genotypic variation for three physiological traits affecting drought tolerance in soybean. Crop Sci. 2007, 47, 25–35. [Google Scholar] [CrossRef]
- Abdel-Haleem, H.; Carter, T.E.; Purcell, L.C.; King, C.A.; Ries, L.L.; Chen, P.; Schapaugh, W.; Sinclair, T.R.; Boerma, H.R. Mapping of quantitative trait loci for canopy-wilting trait in soybean (Glycine max L. Merr). Theor. Appl. Genet. 2012, 5, 837–846. [Google Scholar] [CrossRef]
- Oya, T.; Nepomuceno, A.L.; Neumaier, N.; Farias, J.R.B.; Tobita, S.; Ito, O. Drought tolerance characteristics of Brazilian soybean [Glycine max] cultivars: Evaluation and characterization of drought tolerance of various Brazilian soybean cultivars in the field. Plant Prod. Sci. 2004, 7, 129–137. [Google Scholar] [CrossRef] [Green Version]
- Dantas, S.A.G.; Silva, F.C.S.; Silva, L.J.F.; Silva, L. Strategy for selection of soybean genotypes tolerant to drought during germination. Genet. Mol. Res. 2017, 16, 4–8. [Google Scholar] [CrossRef] [PubMed]
- Dhanapal, A.P.; Ray, J.D.; Singh, S.K.; Hoyos-Villegas, V.; Smith, J.R.; Purcell, L.C.; King, C.A.; Cregan, P.B.; Song, Q.; Fritschi, F.B. Genome-wide association study (GWAS) of carbon isotope ratio (δ 13 C) in diverse soybean [Glycine max L. Merr] genotypes. Theor. Appl. Genet. 2015, 128, 73–91. [Google Scholar] [CrossRef] [PubMed]
- Kaler, A.S.; Dhanapal, A.P.; Ray, J.D.; King, C.A.; Fritschi, F.B.; Purcell, L.C. Genome-wide association mapping of carbon isotope and oxygen isotope ratios in diverse soybean genotypes. Crop Sci. 2017, 57, 3085–3100. [Google Scholar] [CrossRef]
- Specht, J.E.; Chase, K.; Macrander, M.; Graef, G.L.; Chung, J.; Markwell, J.P.; Germann, M.; Orf, J.H.; Lark, K.G. Soybean response to water. Crop Sci. 2001, 41, 493–509. [Google Scholar] [CrossRef]
- Flintgarcia, S.A.; Thornsberry, J.M.; Bucklerive, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ku, Y.S.; Au-Yeung, W.K.; Yung, Y.L.; Li, M.W.; Wen, C.Q.; Liu, X.; Lam, H.M. Drought stress and tolerance in soybean. In A Comprehensive Survey of Internaitonal Soybean Research-Genetics, Physiology, Agronomy and Nitrogen Relationships; InTech: Rijeka, Croatia, 2013; pp. 209–237. [Google Scholar]
- Thabet, S.G.; Moursi, Y.S.; Karam, M.A.; Graner, A.; Alqudah, A.M. Genetic basis of drought tolerance during seed germination in barley. PLoS ONE 2018, 13, e0206682. [Google Scholar] [CrossRef]
- Liu, C.; Yang, Z.; Hu, Y.G. Drought resistance of wheat alien chromosome addition lines evaluated by membership function value based on multiple traits and drought resistance index of grain yield. Field Crops Res. 2015, 179, 103–112. [Google Scholar] [CrossRef]
- Holland, J.B.; Nyquist, W.E.; Cervantesmartinez, C.T. Estimating and interpreting heritability for plant breeding: An update. Plant Breed. Rev. 2010, 22, 9–112. [Google Scholar]
- Kisha, T.J.; Sneller, C.H.; Diers, B.W. Relationship between genetic distance among parents and genetic variance in populations of soybean. Crop Sci. 1997, 37, 1317–1325. [Google Scholar] [CrossRef]
- Mahajan, A.; Sim, X.; Ng, H.J.; Manning, A.; Rivas, M.A.; Highland, H.M.; Flannick, J. Identification and functional characterization of G6PC2 coding variants influencing glycemic traits define an effector transcript at the G6PC2-ABCB11 locus. PLoS Genet. 2015, 11, e1004876. [Google Scholar] [CrossRef]
- Zhao, S.; Jing, W.; Samuels, D.C.; Sheng, Q.; Shyr, Y.; Guo, Y. Strategies for processing and quality control of Illumina genotyping arrays. Brief. Bioinform. 2018, 19, 765–775. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.J.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.H.; Li, D.L.; Jiao, Y.Q.; Schnable, J.C.; Li, Y.F.; Li, H.H.; Chen, H.Z.; Hong, H.L.; Zhang, T.; Liu, B.; et al. Identification of loci controlling adaptation in Chinese soya bean landraces via a combination of conventional and bioclimatic GWAS. Plant Biotechnol. J. 2020, 18, 389–401. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeng, A.; Chen, P.; Korth, K.; Hancock, F.; Pereira, A.; Brye, K.; Wu, C.; Shi, A. Genome-wide association study (GWAS) of salt tolerance in worldwide soybean germplasm lines. Mol. Breed. 2017, 37, 30. [Google Scholar] [CrossRef]
- Wen, Z.; Tan, R.; Yuan, J.; Bales, C.; Du, W.; Zhang, S.; Chilvers, M.I.; Schmidt, C.; Song, Q.; Cregan, P.B.; et al. Genome-wide association mapping of quantitative resistance to sudden death syndrome in soybean. BMC Genom. 2014, 15, 809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farnir, F.; Coppieters, W.; Arranz, J.J.; Berzi, P.; Cambisano, N.; Grisart, B.; Karim, L.; Marcq, F.; Moreau, L.; Mni, M.; et al. Extensive genome-wide linkage disequilibrium in cattle. Genome Res. 2000, 10, 220–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Holland, J.B.; McMullen, M.D.; Buckler, E.S. Genetic design and statistical power of nested association mapping in maize. Genetics 2008, 178, 539–551. [Google Scholar] [CrossRef] [Green Version]
- Panthee, D.R.; Pantalone, V.R.; West, D.R.; Saxton, A.M.; Sams, C.E. Quantitative trait loci for seed protein and oil concen-tration, and seed size in soybean. Crop Sci. 2005, 45, 2015–2022. [Google Scholar] [CrossRef]
- Han, Y.; Li, D.; Zhu, D.; Li, H.; Li, X.; Teng, W.; Li, W. QTL analysis of soybean seed weight across multi-genetic backgrounds and environments. Theor. Appl. Genet. 2012, 125, 671–683. [Google Scholar] [CrossRef]
- Guo, G.Y.; Sun, R.; Hou, M.; Guo, Y.X.; Xin, D.W.; Jiang, H.W.; Hu, G.H.; Chen, Q.S. Quantitative trait locus (QTL) analysis of pod related traits in different environments in soybean. Afr. J. Biotechnol. 2011, 10, 11848–11854. [Google Scholar]
- Wu, J.J.; Xu, P.F.; Liu, L.J.; Zhang, S.; Wang, J.S.; Lin, W.G.; Li, W. Mapping QTLs for phosphorus-deficiency tolerance in soybean at seedling stage. In Proceedings of the International Conference on Biomedical Engineering and Biotechnology, Macao, China, 28–30 May 2012; pp. 370–378. [Google Scholar]
- Li, D.; Sun, M.; Han, Y.; Teng, W.; Li, W. Identification of QTL underlying soluble pigment content in soybean stems related to resistance to soybean white mold (Sclerotinia sclerotiorum). Euphytica 2010, 172, 49–57. [Google Scholar] [CrossRef]
- Stombaugh, S.K.; Orf, J.H.; Jung, H.G.; Chase, K.; Lark, K.G.; Somers, D.A. Quantitative trait loci associated with cell wall polysaccharides in soybean seed. Crop Sci. 2004, 44, 2101–2106. [Google Scholar] [CrossRef] [Green Version]
- Moongkanna, J.; Nakasathien, S.; Novitzky, W.P.; Kwanyuen, P.; Sinchaisri, P.; Srinives, P. SSR markers linking to seed traits and total oil content in soybean. Thai J. Agric. Sci. 2011, 44, 233–241. [Google Scholar]
- Vieira, A.J.D.; Oliveira, D.A.D.; Soares, T.C.B.; Schuster, I.; Piovesan, N.D.; Martínez, C.A.; Moreira, M.A. Use of the QTL approach to the study of soybean trait relationships in two populations of recombinant inbred lines at the F7 and F8 generations. Braz. J. Plant Physiol. 2006, 18, 281–290. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Kim, M.Y.; Van, K.; Lee, Y.H.; Li, H.; Liu, X.; Lee, S.H. QTL identification of yield-related traits and their association with flowering and maturity in soybean. J. Crop Sci. Biotechnol. 2011, 14, 65–70. [Google Scholar] [CrossRef]
- Teng, W.; Han, Y.; Du, Y.; Sun, D.; Zhang, Z.; Qiu, L.; Sun, G.; Li, W. QTL analyses of seed weight during the development of soybean (Glycine max L. Merr). Heredity 2009, 102, 372–380. [Google Scholar] [CrossRef] [Green Version]
- Manavalan, L.P.; Prince, S.J.; Musket, T.A.; Chaky, J.; Deshmukh, R.; Vuong, T.D.; Song, L.; Creqan, P.B.; Nelson, J.C.; Shannon, J.G.; et al. Identification of novel QTL governing root architectural traits in an interspecific soybean population. PLoS ONE 2015, 10, e0120490. [Google Scholar] [CrossRef] [Green Version]
- Kabelka, E.A.; Diers, B.W.; Fehr, W.R.; LeRoy, A.R.; Baianu, I.C.; You, T.; Neece, D.J.; Nelson, R.L. Putative alleles for in-creased yield from soybean plant introductions. Crop Sci. 2004, 44, 784–791. [Google Scholar] [CrossRef] [Green Version]
- Guzman, P.S.; Diers, B.W.; Neece, D.J.; Martin, S.K.; LeRoy, A.R.; Grau, C.R.; Hughes, T.J.; Nelson, R.L. QTL associated with yield in three backcross-derived populations of soybean. Crop Sci. 2007, 47, 111–122. [Google Scholar] [CrossRef]
- Yuan, J.; Njiti, V.N.; Meksem, K.; Iqbal, M.J.; Triwitayakorn, K.; Kassem, M.A.; Davis, G.T.; Schmidt, M.E.; Lightfoot, D.A. Quantitative trait loci in two soybean recombinant inbred line populations segregating for yield and disease resistance. Crop Sci. 2002, 42, 271–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, H.; Yu, Y.; Yang, H.; Xu, L.; Dong, W.; Du, H.; Cui, W.; Zhang, H. Inheritance and QTL mapping of related root traits in soybean at the seedling stage. Theor. Appl. Genet. 2014, 127, 2127–2137. [Google Scholar] [CrossRef] [PubMed]
- Kosturkova, G.; Todorova, R.; Sakthivelu, G.; Akitha-Devi, M.K.; Giridhar, P.; Rajasekaran, T.; Ravishankar, G.A. Response of Bulgarian and Indian soybean genotypes to drought and water deficiency in field and laboratory conditions. Gen. Appl. Plant Physiol. 2008, 34, 239–250. [Google Scholar]
- Hyten, D.L.; Pantalone, V.R.; Sams, C.E.; Saxton, A.M.; Landau-Ellis, D.; Stefaniak, T.R. Seed quality QTL in a prominent soybean population. Theor. Appl. Genet. 2004, 109, 552–561. [Google Scholar] [CrossRef] [PubMed]
- Palomeque, L.; Liu, L.; Li, W.; Hedges, B.; Cober, E.; Smid, M.; Lukens, L.; Rajcan, I. Validation of mega-environment universal and specific QTL associated with seed yield and agronomic traits in soybeans. Theor. Appl. Genet. 2010, 120, 997–1003. [Google Scholar] [CrossRef]
- Pan, S. Analysis of Fixed SNP Reveals Insight of Morphology Differences between Wild and Cultivated Soybeans. Ph.D. Thesis, Chinese University of Hong Kong, Hong Kong, China, 2013. [Google Scholar]
- Mirzaei, S.; Batley, J.; Ferguson, B.J.; Gresshoff, P.M. Transcriptome profiling of the shoot and root tips of S562L, a soybean GmCLAVATA1A mutant. Atlas J. Biol. 2014, 3, 183–205. [Google Scholar] [CrossRef]
- Wang, X.; Komatsu, S. Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean. J. Proteom. 2018, 172, 201–215. [Google Scholar] [CrossRef]

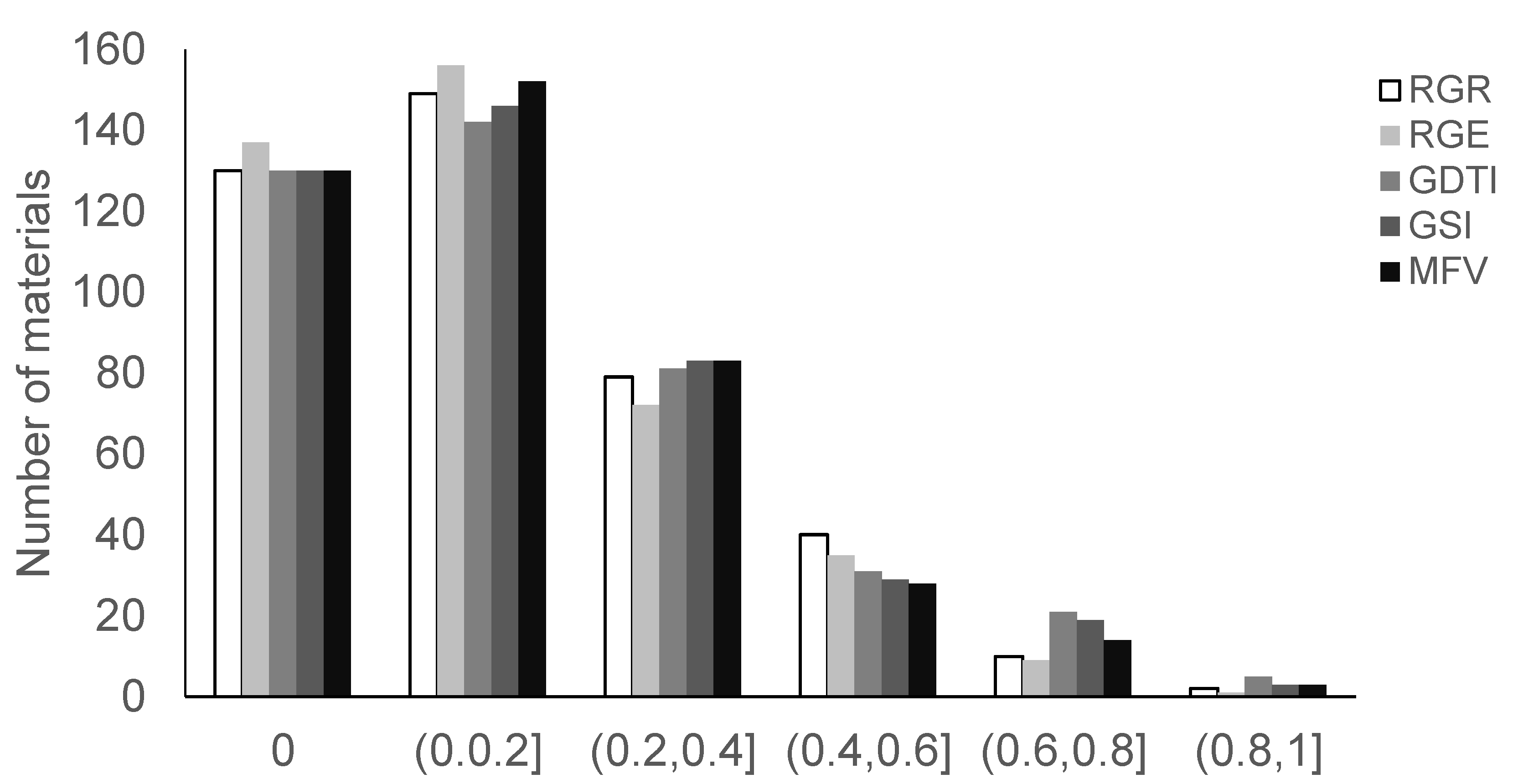
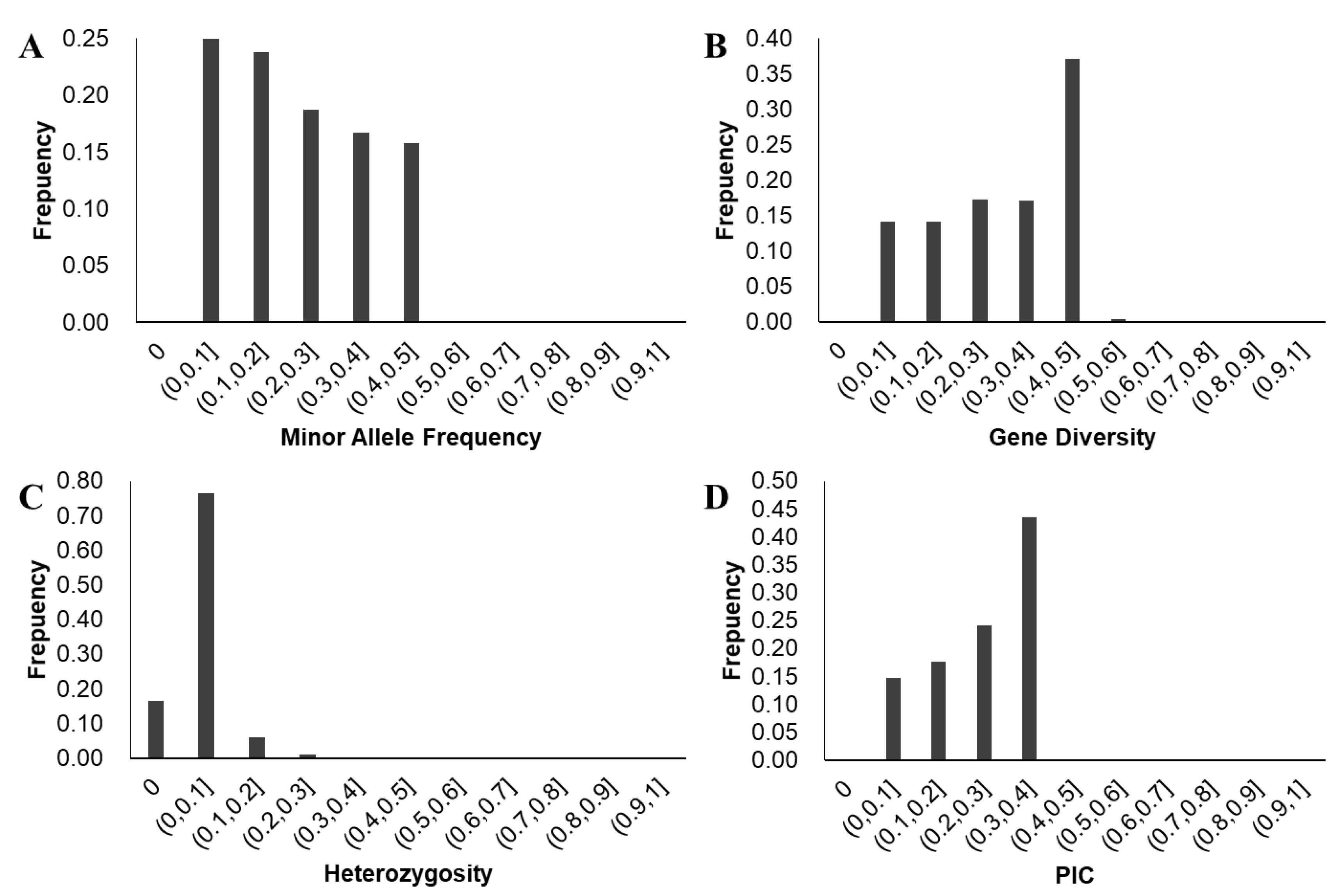
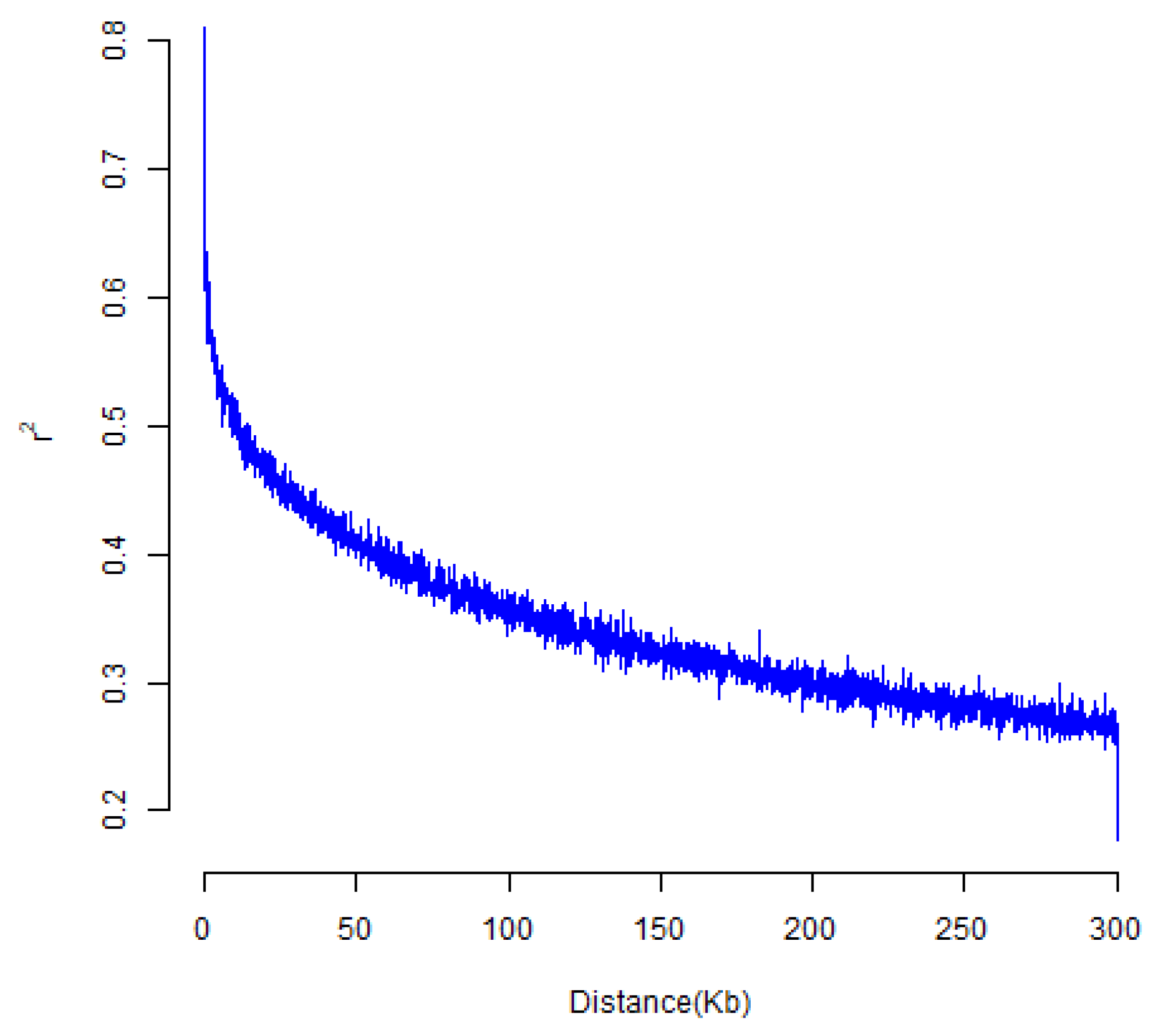
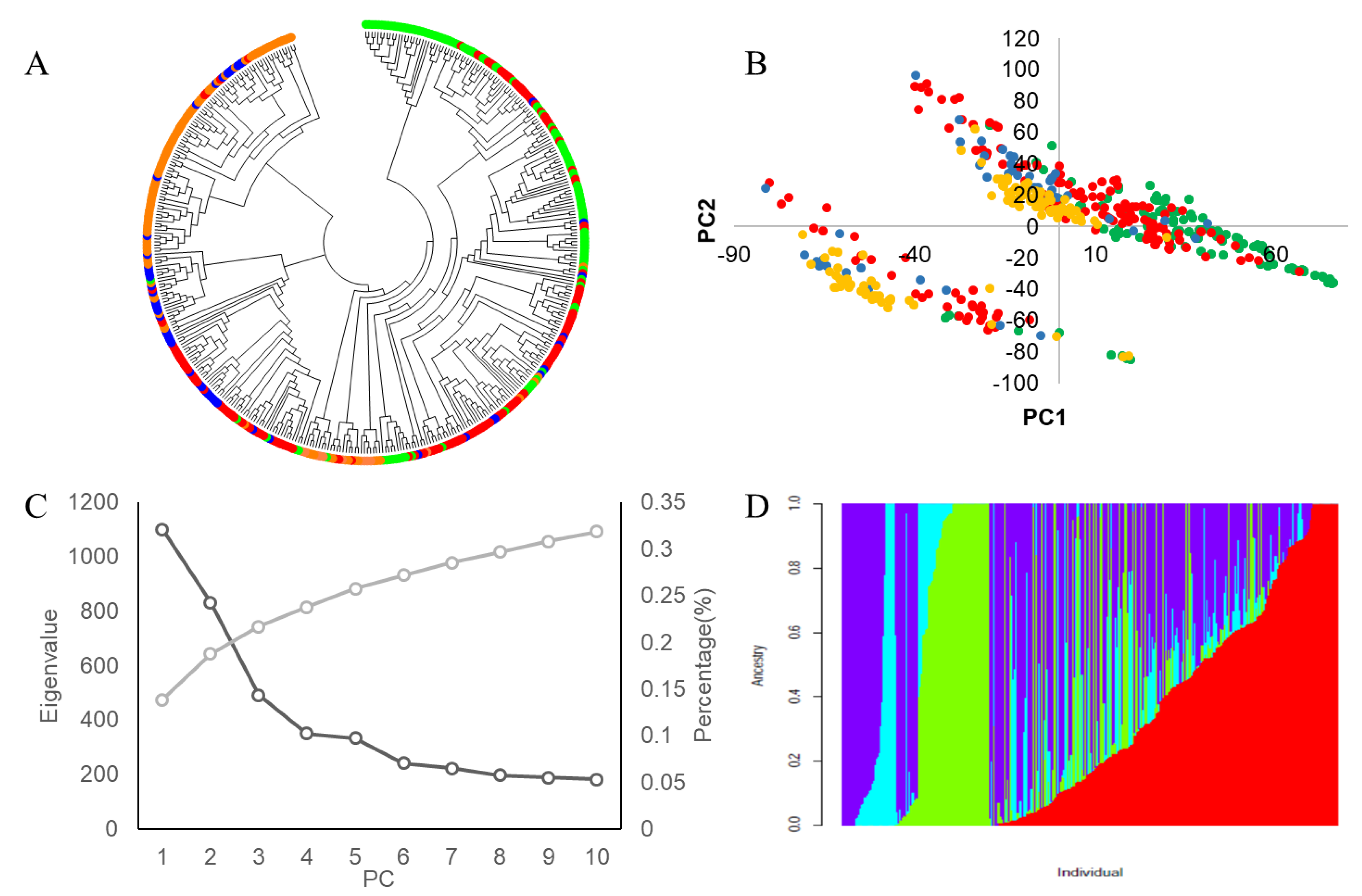
| Trait | Source | DF | Sum of Square | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|---|
| GR | Geno | 409 | 251,929.70 | 615.97 | 7.29 | <0.0001 |
| Treatment | 1 | 437,4687.00 | 4,374,687.00 | 51,778.10 | <0.0001 | |
| Block/Treat | 2 | 357.14 | 178.57 | 2.11 | 0.1211 | |
| Geno×Treat | 409 | 246,113.60 | 601.75 | 7.12 | <0.0001 | |
| GE | Geno | 409 | 236,174.00 | 577.44 | 7.74 | <0.0001 |
| Treatment | 1 | 4,470,355.00 | 4,470,355.00 | 59,943.60 | <0.0001 | |
| Block/Treat | 2 | 359.92 | 179.96 | 2.41 | 0.0898 | |
| Geno×Treat | 409 | 226,055.60 | 552.70 | 7.41 | <0.0001 | |
| GDI | Geno | 409 | 1,658,797.00 | 4055.74 | 10.23 | <0.0001 |
| Treatment | 1 | 19,290,378.00 | 19,290,378.00 | 48,679.80 | <0.0001 | |
| Block/Treat | 2 | 2222.70 | 1111.35 | 2.80 | 0.0608 | |
| Geno×Treat | 409 | 613,890.90 | 1500.96 | 3.79 | <0.0001 | |
| GI | Geno | 409 | 3440.47 | 8.41 | 8.82 | <0.0001 |
| Treatment | 1 | 30,333.58 | 30,333.58 | 31,790.80 | <0.0001 | |
| Block/Treat | 2 | 9.94 | 4.97 | 5.21 | 0.0055 | |
| Geno×Treat | 409 | 1756.14 | 4.29 | 4.50 | <0.0001 |
| Traits | Mean | SD | Skewness | Kurtosis | Range | CV |
|---|---|---|---|---|---|---|
| RGR | 0.16 | 0.20 | 1.55 | 2.03 | 0~1 | 129.88 |
| RGE | 0.14 | 0.19 | 1.63 | 2.40 | 0~1 | 136.51 |
| GDTI | 0.09 | 0.12 | 1.60 | 2.21 | 0~0.57 | 135.45 |
| GSI | 0.08 | 0.11 | 1.43 | 1.44 | 0~0.48 | 129.00 |
| MFV | 0.15 | 0.19 | 1.41 | 1.60 | 0~1 | 121.81 |
| Trait | Source | DF | Sum of Square | Mean Square | F Value | Pr > F |
|---|---|---|---|---|---|---|
| RGR | Geno | 409 | 44.94 | 0.11 | 11.67 | <0.0001 |
| Block | 2 | 0.01 | 0.00 | 0.28 | 0.75 | |
| RGE | Geno | 409 | 39.22 | 0.10 | 10.94 | <0.0001 |
| Block | 2 | 0.01 | 0.00 | 0.47 | 0.63 | |
| GDTI | Geno | 409 | 13.88 | 0.03 | 9.01 | <0.0001 |
| Block | 2 | 0.00 | 0.00 | 0.28 | 0.75 | |
| GSI | Geno | 409 | 12.41 | 0.03 | 8.19 | <0.0001 |
| Block | 2 | 0.00 | 0.00 | 0.16 | 0.86 |
| Trait | RGR | RGE | GDTI | GSI |
|---|---|---|---|---|
| RGE | 0.9840 *** | |||
| GDTI | 0.9640 *** | 0.9766 *** | ||
| GSI | 0.9490 *** | 0.9524 *** | 0.9729 *** | |
| MFV | 0.9862 *** | 0.9902 *** | 0.9909 *** | 0.9819 *** |
| Marker | Chr | Position | Associated Traits (R2) | Reported QTLs/Genes |
|---|---|---|---|---|
| Gm01_35877607 | 1 | 35877607 | RGR(7.20), RGE(7.53), GDTI(6.93), MFV(6.95) | Seed set [5]; seed weight [40,41] |
| Gm01_38948188 | 1 | 38948188 | GDTI(6.73) | Pod wall weight [42] |
| Gm01_47042336 | 1 | 47042336 | GSI(6.31), GDTI(6.51) | Drought index [1]; root area [43]; root length [43] |
| Gm01_48619013 | 1 | 48619013 | GDTI(6.16) | Drought index [1] |
| Gm02_6357585 | 2 | 6357585 | GDTI(7.11) | Canopy wilt [7] |
| Gm03_39037 | 3 | 39037 | GDTI(6.08) | |
| Gm04_4484515 | 4 | 4484515 | RGR(6.84), RGE(7.22), GDTI(6.99), MFV(6.74) | Canopy wilt [7]; seed set [5]; seed weight [44] |
| Gm04_50945875 | 4 | 50945875 | GDTI(6.69) | Seed number [44]; WUE [23] |
| Gm05_38540838 | 5 | 38540838 | RGR(7.60) | Cellwall polysacch composition [45] |
| Gm06_9791913 | 6 | 9791913 | GDTI(6.17) | Seed weight [46]; shoot weight [47] |
| Gm07_24735482 | 7 | 24735482 | GDTI(6.59) | |
| Gm08_1438457 | 8 | 1438457 | RGE(6.08) | Seed weight per plant [48] |
| Gm08_4052111 | 8 | 4052111 | GDTI(6.91) | Canopy wilt [7]; seed weight [49] |
| Gm08_7972856 | 8 | 7972856 | RGR(8.03) | Root density, lateral [50]; seed set [5] |
| Gm09_11414508 | 9 | 11414508 | RGE(5.19), GDTI(6.00), MFV(5.32) | Seed yield [51,52] |
| Gm09_18023730 | 9 | 18023730 | GSI(6.46), GDTI(6.77), MFV(6.03) | Seed yield [52,53] |
| Gm11_30280479 | 11 | 30280479 | RGR(8.33), RGE(7.04), GDTI(8.08), MFV(7.53) | Seed set [5] |
| Gm13_35517964 | 13 | 35517964 | GDTI(6.55) | |
| Gm14_46603856 | 14 | 46603856 | GDTI(6.19) | Canopy wilt [20] |
| Gm15_11950665 | 15 | 11950665 | GDTI(6.52) | Seed weight [48] |
| Gm15_47429024 | 15 | 47429024 | GSI(6.45) | |
| Gm19_49449499 | 19 | 49449499 | GDTI(6.46) | Canopy wilt [7]; drought tolerance [13] |
| Gm20_4618170 | 20 | 4618170 | GDTI(6.28) | |
| Gm20_13921498 | 20 | 13921498 | RGR(6.66), RGE(7.26), GDTI(7.01), MFV(6.64) | Seed weight [41] |
| Gm20_34956219 | 20 | 34956219 | RGR(6.88), RGE(7.87), GSI(7.22), GDTI(8.33), MFV(7.81) | Canopy wilt [20]; root density, lateral [54]; seed set [41]; WUE [24] |
| Gm20_36902659 | 20 | 36902659 | RGR(7.69), RGE(8.03), GSI(8.19), GDTI(9.66), MFV(8.57) | Root density, lateral [54] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhao, X.; Liu, Z.; Li, H.; Zhang, Y.; Yu, L.; Qi, X.; Gao, H.; Li, Y.; Qiu, L. Identification of Drought-Tolerance Genes in the Germination Stage of Soybean. Biology 2022, 11, 1812. https://doi.org/10.3390/biology11121812
Zhao X, Liu Z, Li H, Zhang Y, Yu L, Qi X, Gao H, Li Y, Qiu L. Identification of Drought-Tolerance Genes in the Germination Stage of Soybean. Biology. 2022; 11(12):1812. https://doi.org/10.3390/biology11121812
Chicago/Turabian StyleZhao, Xingzhen, Zhangxiong Liu, Huihui Li, Yanjun Zhang, Lili Yu, Xusheng Qi, Huawei Gao, Yinghui Li, and Lijuan Qiu. 2022. "Identification of Drought-Tolerance Genes in the Germination Stage of Soybean" Biology 11, no. 12: 1812. https://doi.org/10.3390/biology11121812
APA StyleZhao, X., Liu, Z., Li, H., Zhang, Y., Yu, L., Qi, X., Gao, H., Li, Y., & Qiu, L. (2022). Identification of Drought-Tolerance Genes in the Germination Stage of Soybean. Biology, 11(12), 1812. https://doi.org/10.3390/biology11121812







