RETRACTED: Identify Biomarkers and Design Effective Multi-Target Drugs in Ovarian Cancer: Hit Network-Target Sets Model Optimizing
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Detection of Differentially Expressed Genes
2.3. Gene Regulatory Network Construction
2.4. Core Nodes/Genes Identification and Identification of Driver Nodes
2.5. Modular Screening and Stability
2.6. Core Module Identification
2.7. Performance Assessment of the OHNS
- (a)
- Characteristic path length (L) [41] based on Equation (3);
- (b)
- Giant component (GC): The giant component is the most significant connected component in each network. The fraction was calculated by dividing the number of nodes in the giant component by the total number of nodes in each network [10];
- (c)
- Calculation of the F-measure: To assess the F-measure, taking into account the precision and recall of the predicted HNS using the following formula, the key cancer genes are annotated in the list of drug targets and biomarker genes (Supplementary Table S1) for ovarian cancer (Comparative Toxicogenomics Database, http://ctdbase.org/ (accessed on 23 May 2022)) were chosen:
- (d)
- Perturbation effects: Cancer Dependency Map’s genome-scale CRISPR-Cas9 knockout data were used (https://depmap.org/portal/ (accessed on 23 May 2022)). In CM, CN, and DN, the required genes for perturbing 178 cancer cell lines were gathered, respectively. A lower Chronos score suggests a higher probability that the target gene is crucial in a particular cell line. A gene with a score of 0 is not considered influential; a score of −1 is comparable to the median of all genes considered necessary.
2.8. Gene Ontology and Functional Enrichment Analysis
2.9. Prognostic Risk Assessment
2.10. Tissue-Specific Enrichment and Correlation Analyses of Hub Genes
2.11. Replication and Validation Analyses
2.12. Samples Collection
2.13. Sample Preparation
2.14. Validation of RNA-Seq Results Using qRT-PCR
2.15. Western Blotting
2.16. Immunohistochemistry
2.17. Statistical Analysis
3. Results
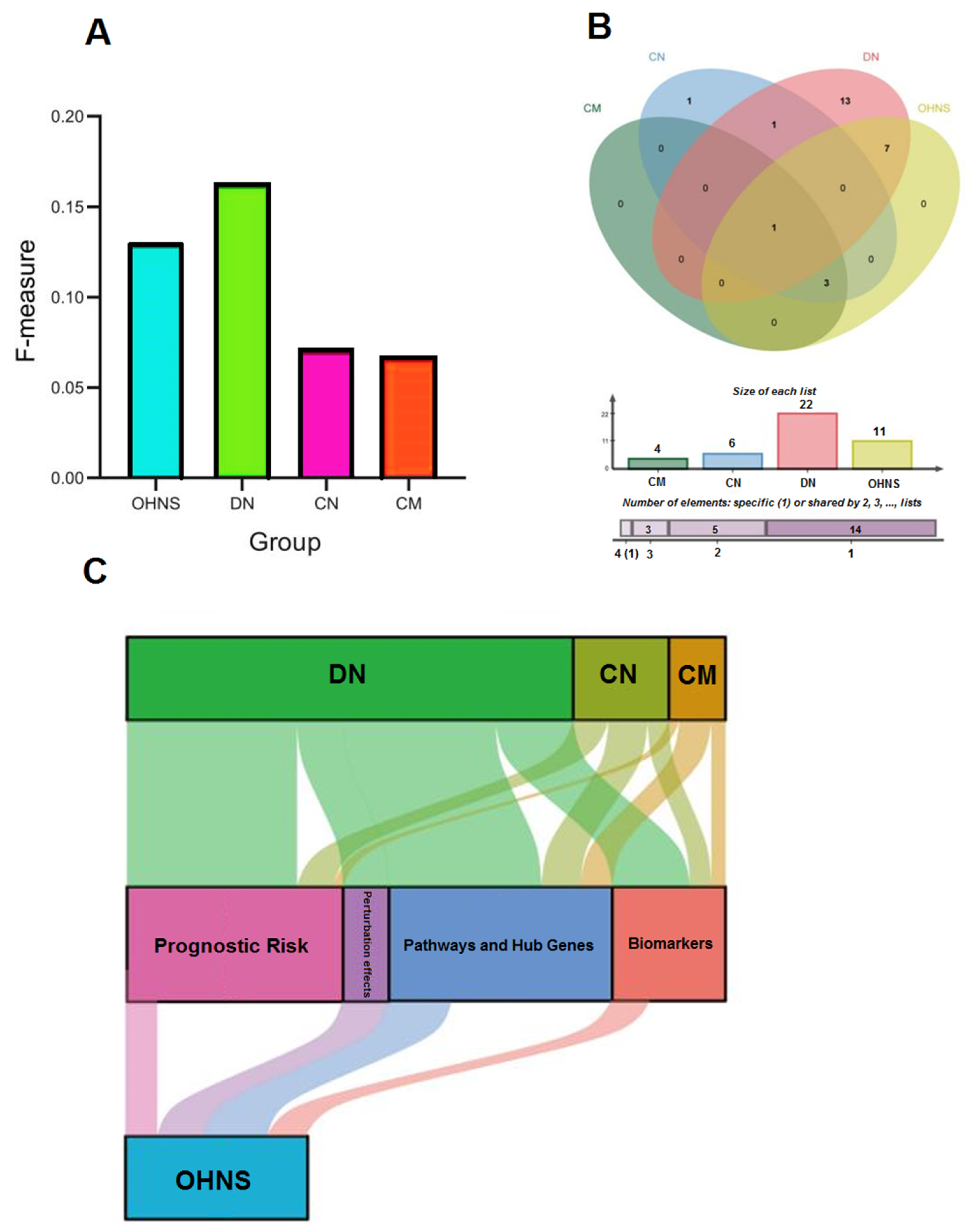
3.1. Hit Network-Target Sets Identification
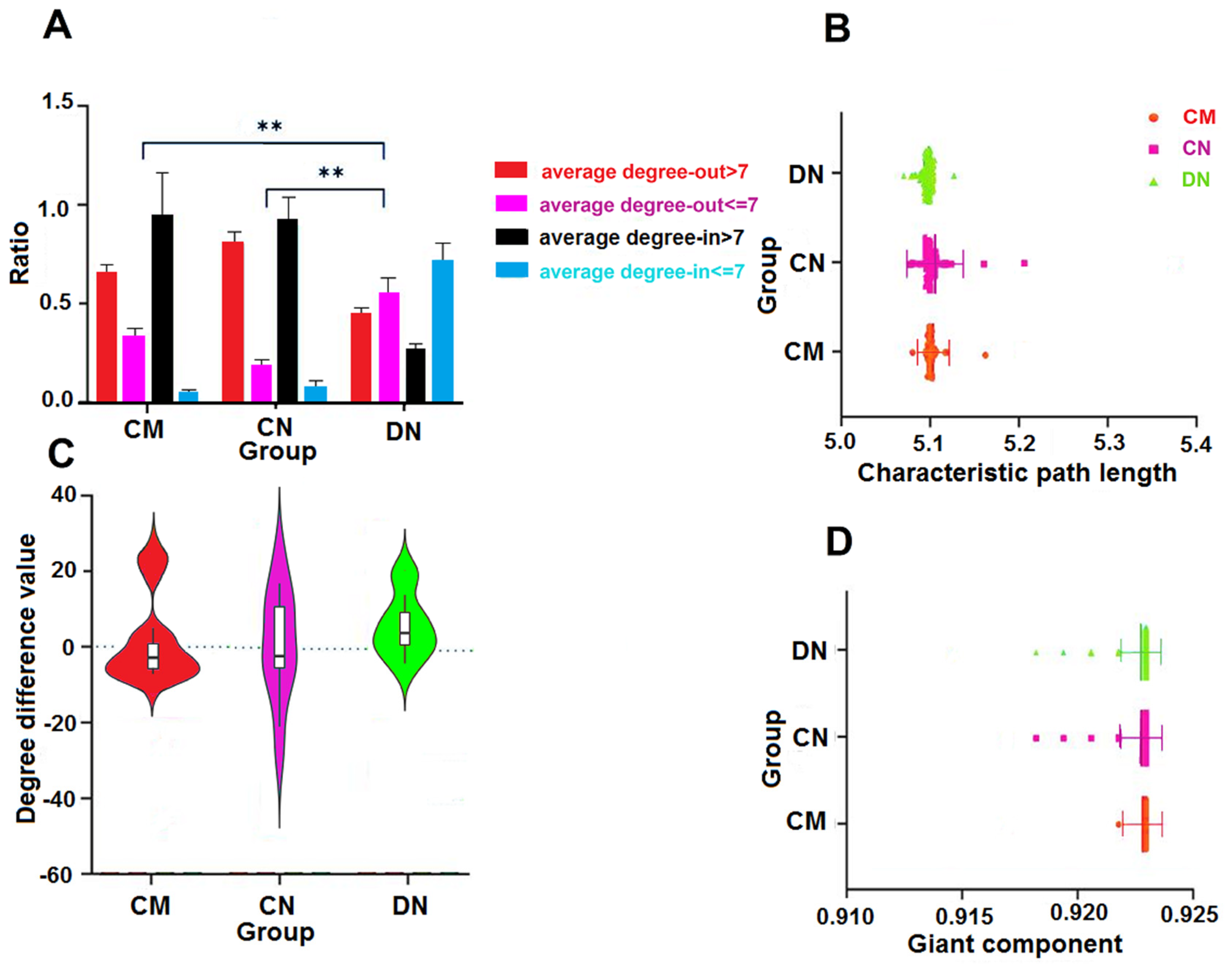
3.2. Characteristics of Clustering and Scattering of the Network Distribution
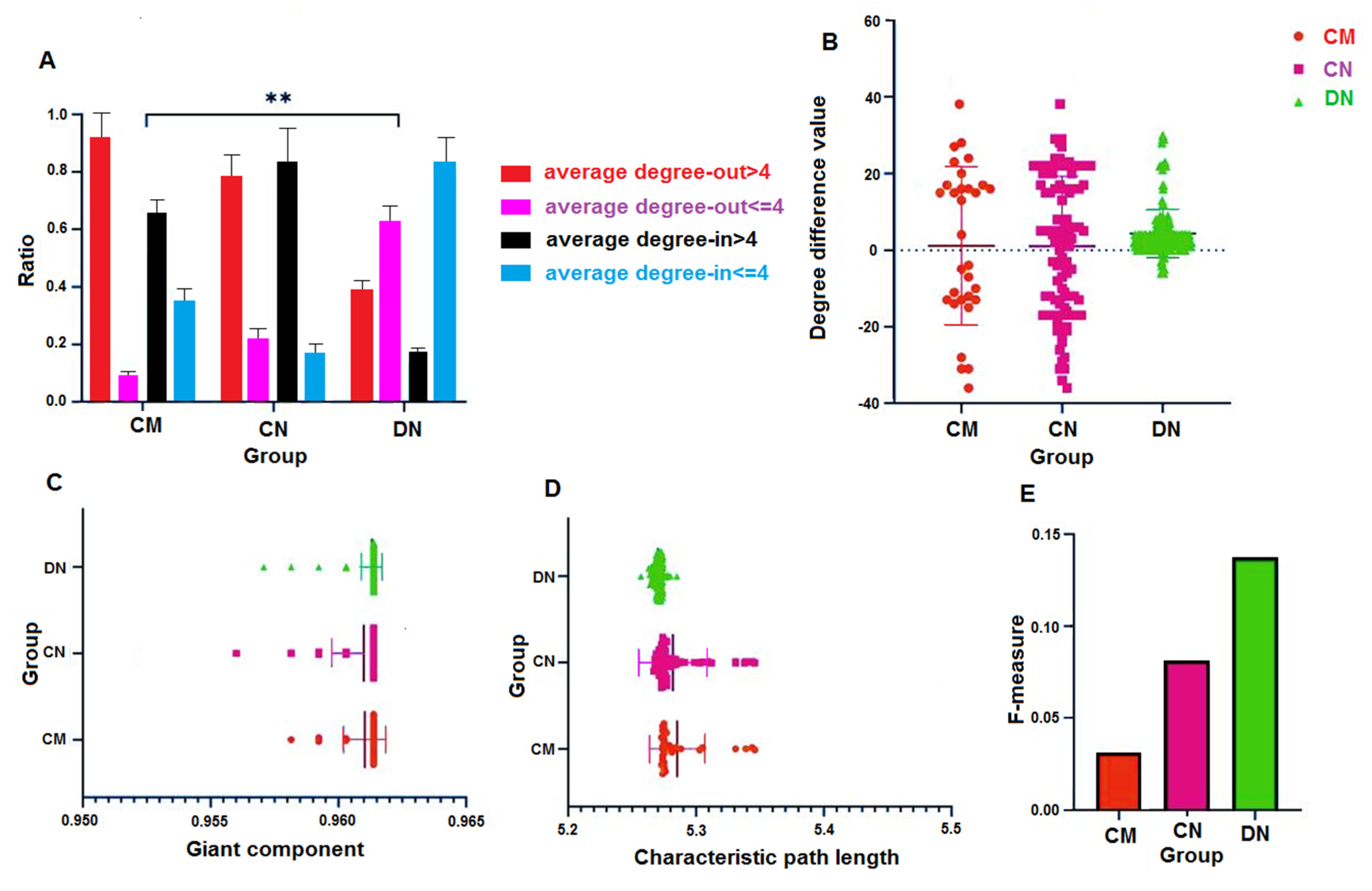
3.3. Out-Degree-Dominant Characteristics of Driver Nodes
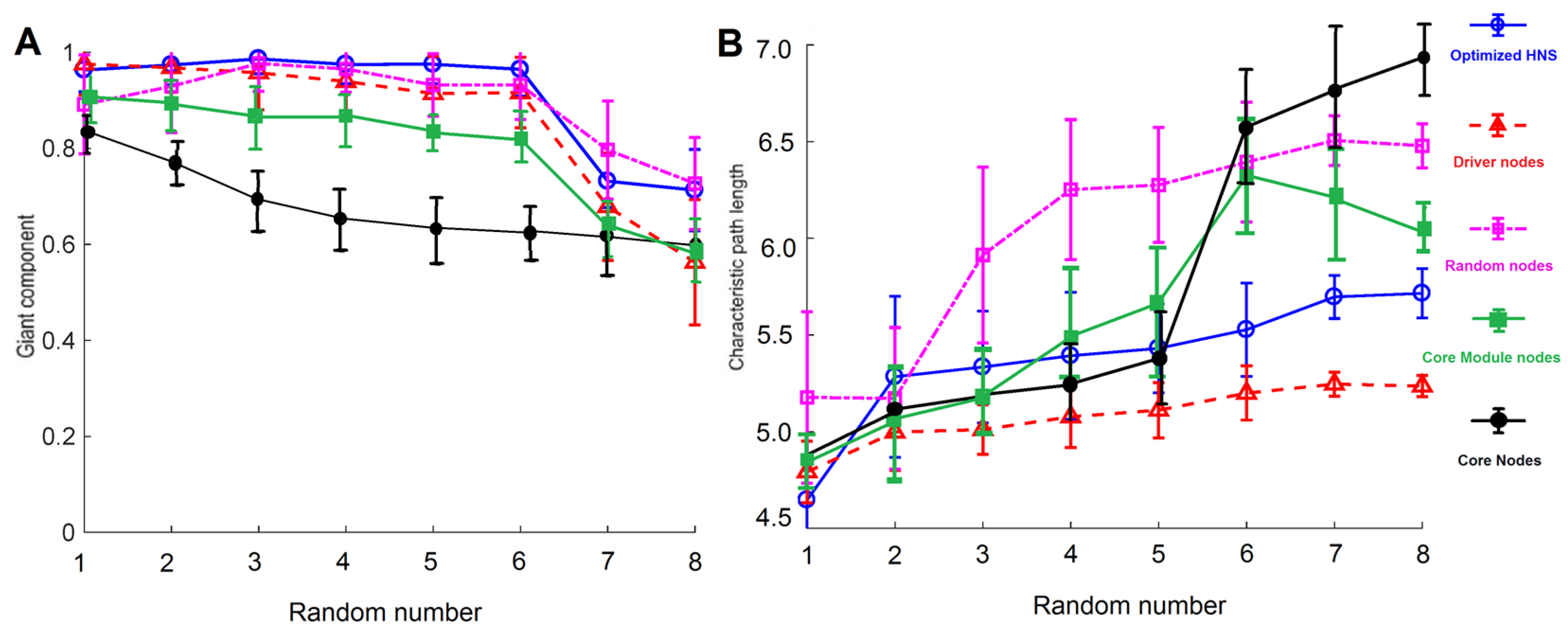
3.4. Characteristic Path Lengths and Giant Components of HNSs
3.5. Characteristic Path Lengths and Giant Components
3.6. F-Measure and Perturbation Effect
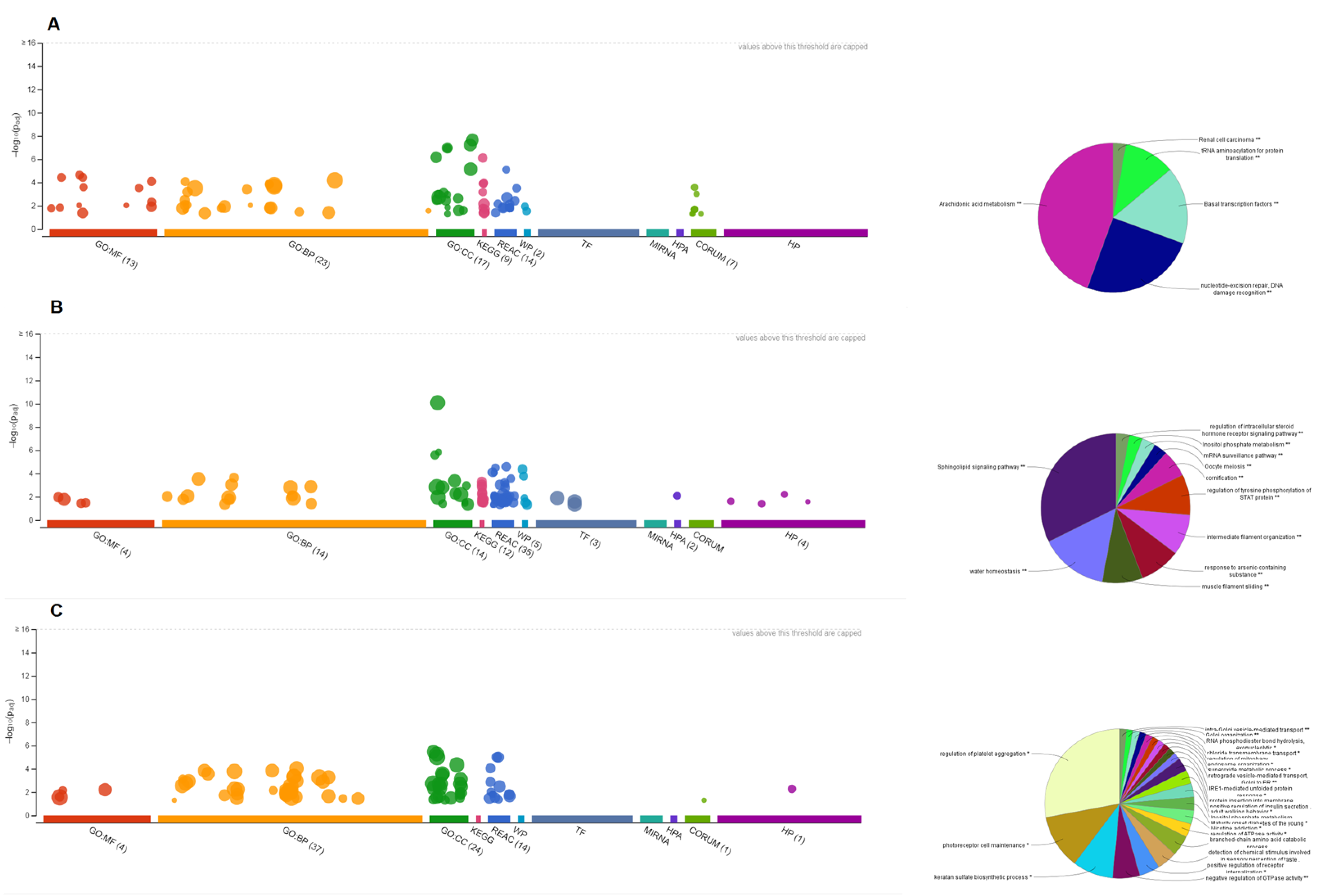
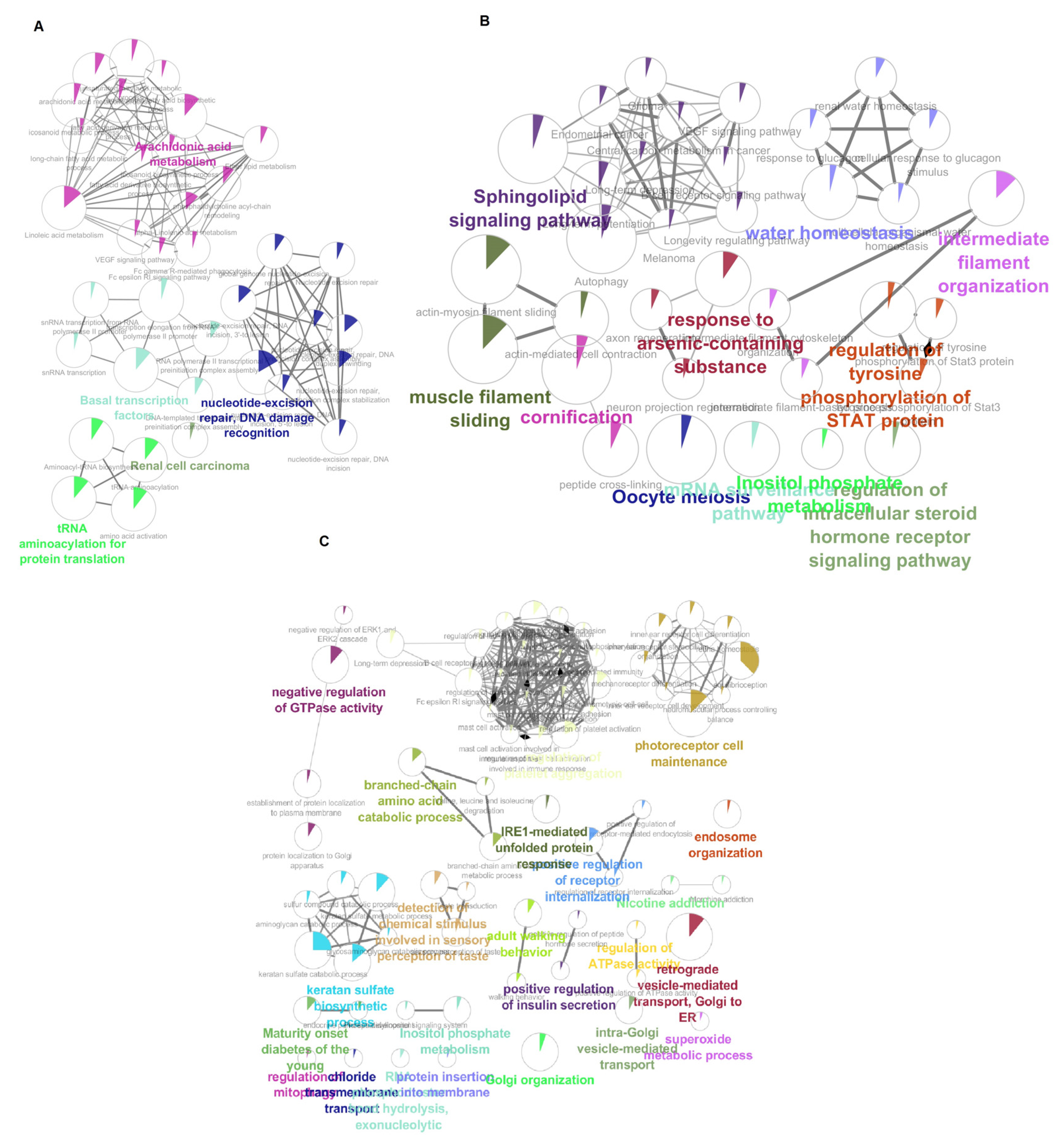
3.7. Pathway Enrichment Analysis of HNSs
3.8. Survival Analysis of Perturbed Genes
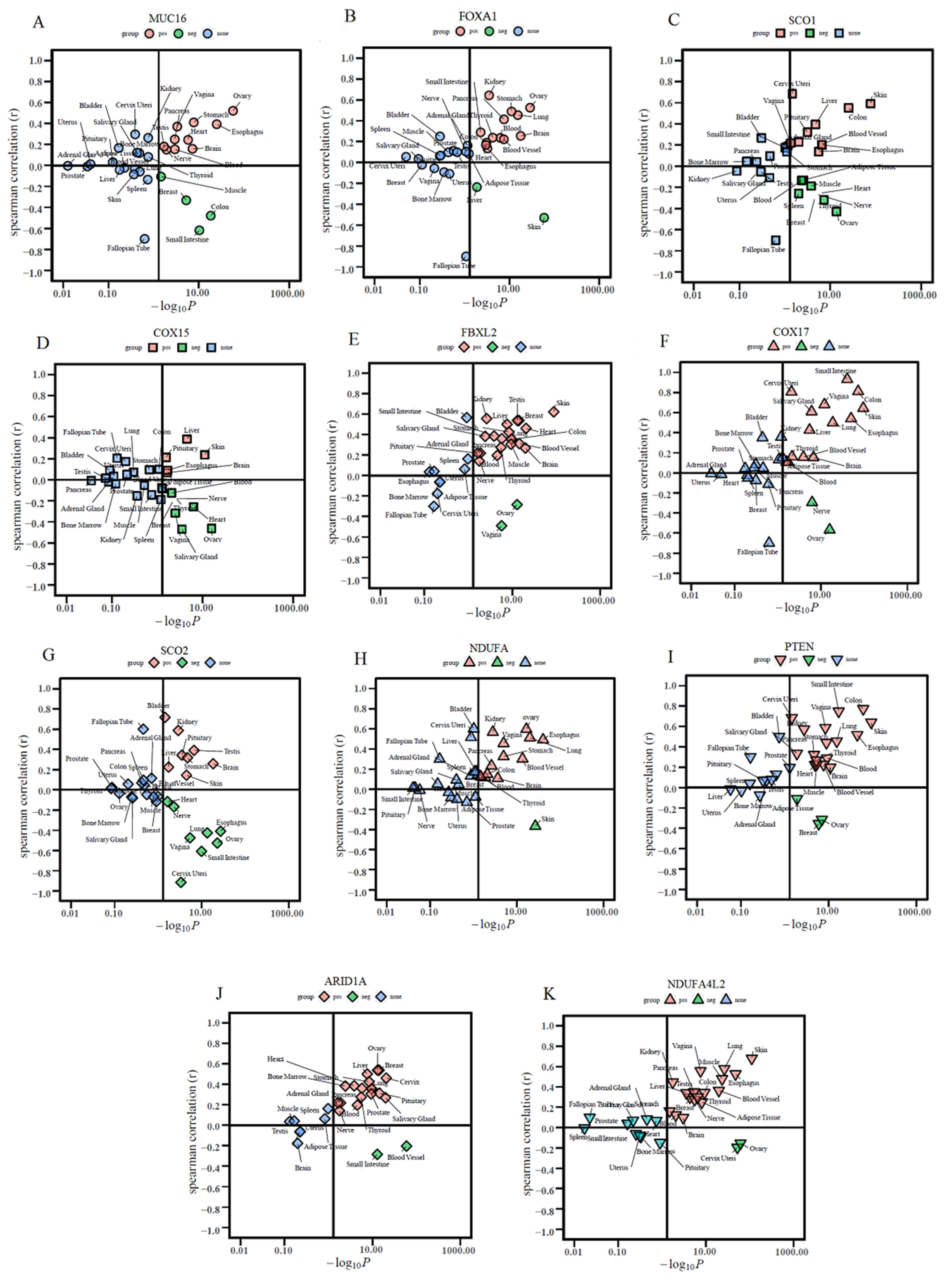
3.9. Tissue-Specific Enrichment and Correlation Analyses of Hub Genes
3.10. Replication and Validation Analyses
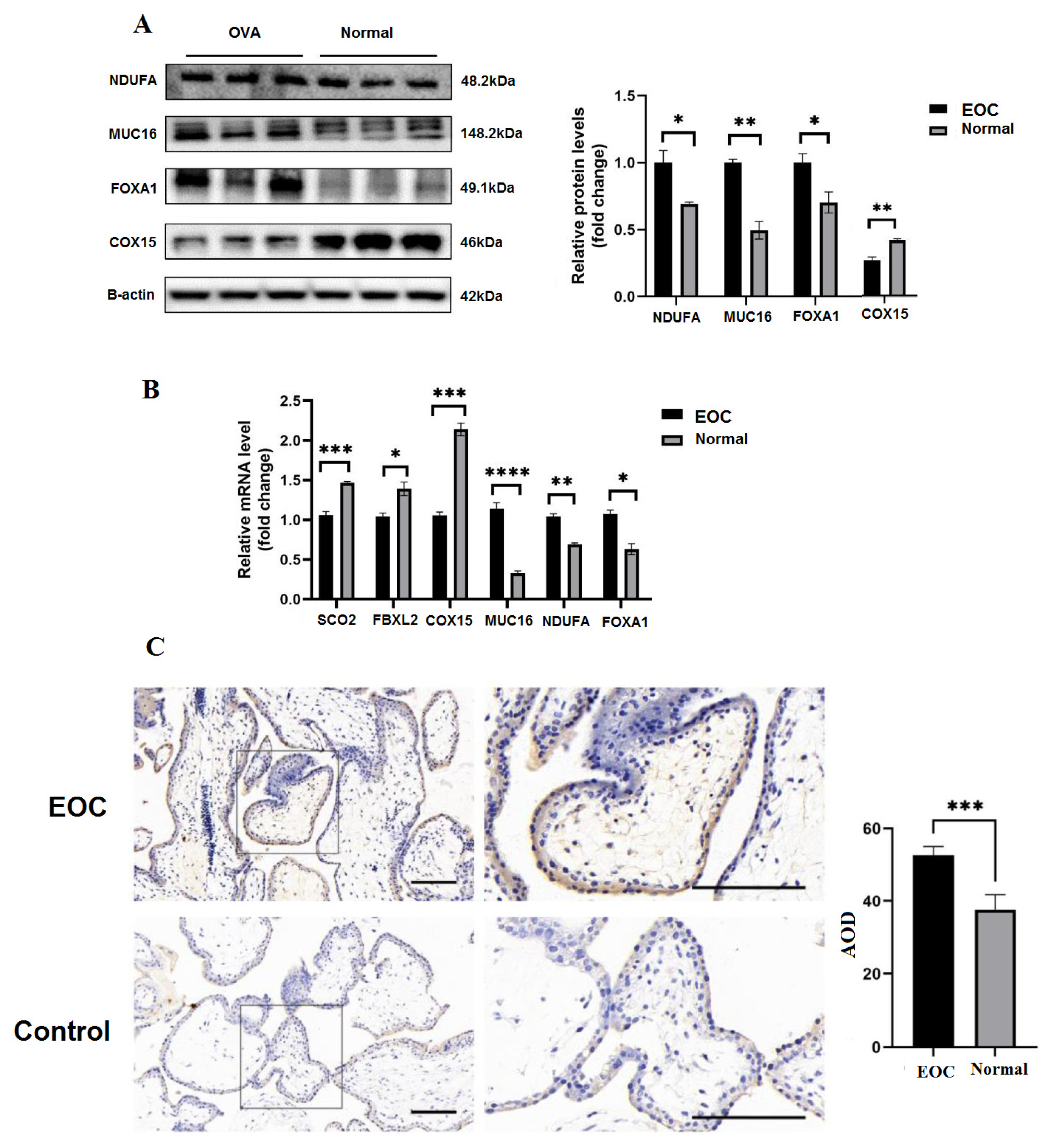
3.11. The Deficiency of Proteins Expression and Biological Functions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dai, Y.F.; Zhao, X.M. A survey on the computational approaches to identify drug targets in the postgenomic era. BioMed Res. Int. 2015, 2015, 239654. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.Y.; Chen, D.B.; Ren, X.L.; Zhang, Q.M.; Zhang, Y.C.; Zhou, T. Vital nodes identification in complex networks. Phys. Rep.-Rev. Sect. Phys. Lett. 2016, 650, 1–63. [Google Scholar]
- Kim, S.S.; Dai, C.; Hormozdiari, F.; van de Geijn, B.; Gazal, S.; Park, Y.; O’Connor, L.; Amariuta, T.; Loh, P.R.; Finucane, H.; et al. Genes with high network connectivity are enriched for disease heritability. Am. J. Hum. Genet. 2019, 104, 896–913. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.; Mariani, M.S.; Medo, M.; Zhang, Y.C.; Zhou, M.Y. Ranking in evolving complex networks. Phys. Rep.-Rev. Sect. Phys. Lett. 2017, 689, 1–54. [Google Scholar] [CrossRef]
- Amberger, J.; Bocchini, C.; Scott, A.; Hamosh, A. OMIM.org: Leveraging knowledge across phenotype-gene relationships. Nucleic Acids Res. 2019, 47, 1038–1043. [Google Scholar] [CrossRef]
- Yu, G.C.; Wang, L.G.; Han, Y.Y.; He, Q.Y. clusterProfiler: An R Package for Comparing Biological Themes Among Gene Clusters. Omics-A J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Jang, H.S.; Shah, N.M.; Du, A.Y.; Dailey, Z.Z.; Pehrsson, E.C.; Godoy, P.M.; Zhang, D.; Li, D.; Xing, X.; Kim, S.; et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat. Genet. 2019, 51, 611–617. [Google Scholar] [CrossRef]
- Luo, P.; Ding, Y.; Lei, X.; Wu, F.-X. deepDriver: Predicting Cancer Driver Genes Based on Somatic Mutations Using Deep Convolutional Neural Networks. Front. Genet. 2019, 10, 13. [Google Scholar] [CrossRef]
- Guo, W.-F.; Zhang, S.-W.; Liu, L.-L.; Liu, F.; Shi, Q.-Q.; Zhang, L.; Tang, Y.; Zeng, T.; Chen, L. Discovering personalized driver mutation profiles of single samples in cancer by network control strategy. Bioinformatics 2018, 34, 1893–1903. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, C.H.; Ding, Y.Y.; Wen, X.; Wang, B.; Gao, L.; Tan, K. Optimal control nodes in disease-perturbed networks as targets for combination therapy. Nat. Commun. 2019, 10, 2180. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhao, Z.; Yu, Y.; Liu, J.; Wang, P.; Li, B.; Zhang, X.; Chen, Y.; Wang, Z. Mining the Synergistic Core Allosteric Modules Variation and Sequencing Pharmacological Module Drivers in a Preclinical Model of Ischemia. Cpt-Pharmacomet. Syst. Pharmacol. 2018, 7, 269–280. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Y.; Slotine, J.J.; Barabasi, A.L. Controllability of complex networks. Nature 2011, 473, 167–173. [Google Scholar] [CrossRef] [PubMed]
- Chin, C.H.; Chen, S.H.; Wu, H.H.; Ho, C.W.; Ko, M.T.; Lin, C.Y. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst. Biol. 2014, 8, S11. [Google Scholar] [CrossRef] [PubMed]
- Schwartz, J.M.; Otokuni, H.; Akutsu, T.; Nacher, J.C. Probabilistic controllability approach to metabolic fluxes in normal and cancer tissues. Nat. Commun. 2019, 10, 2725. [Google Scholar] [CrossRef] [PubMed]
- Jeong, H.; Mason, S.P.; Barabasi, A.L.; Oltvai, Z.N. Lethality and centrality in protein networks. Nature 2001, 411, 41–42. [Google Scholar] [CrossRef]
- Liu, X.; Pan, L. Detection of driver metabolites in the human liver metabolic network using structural controllability analysis. BMC Syst. Biol. 2014, 8, 51. [Google Scholar] [CrossRef]
- Grondin, C.J.; Davis, A.P.; Wiegers, J.A.; Wiegers, T.C.; Sciaky, D.; Johnson, R.J.; Mattingly, C.J. Predicting Molecular Mechanisms, Pathways, and Health Outcomes Induced by Juul E-Cigarette Aerosol Chemicals Using the Comparative Toxicogenomics Database. Curr. Res. Toxicol. 2021, 2, 272–281. [Google Scholar] [CrossRef]
- Stelzer, G.; Rosen, N.; Plaschkes, I.; Zimmerman, S.; Twik, M.; Fishilevich, S.; Stein, T.I.; Nudel, R.; Lieder, I.; Mazor, Y.; et al. The Genecards Suite: From Gene Data Mining to Disease Genome Sequence Analyses. Curr. Protoc. Bioinform. 2016, 54, 1–30. [Google Scholar] [CrossRef]
- Brown, G.R.; Hem, V.; Katz, K.S.; Ovetsky, M.; Wallin, C.; Ermolaeva, O.; Tolstoy, I.; Tatusova, T.; Pruitt, K.; Maglott, D.R.; et al. Gene: A Gene-Centered Information Resource at Ncbi. Nucleic Acids Res. 2015, 43, 36–42. [Google Scholar] [CrossRef]
- Watts, D.J.; Strogatz, S.H. Collective dynamics of ‘small-world’ networks. Nature 1998, 393, 440–442. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, S.; Li, F.; Zhou, Y.; Zhang, Y.; Wang, Z.; Zhang, R.; Zhu, J.; Ren, Y.; Tan, Y.; et al. Therapeutic Target Database 2020: Enriched Resource for Facilitating Research and Early Development of Targeted Therapeutics. Nucleic Acids Res. 2020, 48, D1031–D1041. [Google Scholar] [CrossRef]
- Kim, S.; Chen, J.; Cheng, T.; Gindulyte, A.; He, J.; He, S.; Li, Q.; Shoemaker, B.A.; Thiessen, P.A.; Yu, B.; et al. Pubchem in 2021: New Data Content and Improved Web Interfaces. Nucleic Acids Res. 2021, 49, 1388-d95. [Google Scholar] [CrossRef] [PubMed]
- Piñero, J.; Ramírez-Anguita, J.M.; Saüch-Pitarch, J.; Ronzano, F.; Centeno, E.; Sanz, F.; Furlong, L.I. The Disgenet Knowledge Platform for Disease Genomics: 2019 Update. Nucleic Acids Res. 2020, 48, 845-d55. [Google Scholar] [CrossRef] [PubMed]
- Leek, J.T.; Johnson, W.E.; Parker, H.S.; Jaffe, A.E.; Storey, J.D. The Sva Package for Removing Batch Effects and Other Unwanted Variation in High-Throughput Experiments. Bioinformatics 2012, 28, 882–883. [Google Scholar] [CrossRef] [PubMed]
- Blankenberg, D.; Gordon, A.; Von Kuster, G.; Coraor, N.; Taylor, J.; Nekrutenko, A.; Galaxy Team. Manipulation of FASTQ data with Galaxy. Bioinformatics 2010, 26, 1783–1785. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Bader, G.D.; Donaldson, I.; Wolting, C.; Ouellette, B.F.; Pawson, T.; Hogue, C.W. BIND—The biomolecular interaction network database. Nucleic Acids Res. 2001, 29, 242–245. [Google Scholar] [CrossRef]
- Xenarios, I.; Salwinski, L.; Duan, X.J.; Higney, P.; Kim, S.M.; Eisenberg, D. DIP, the Database of Interacting Proteins: A research tool for studying cellular networks of protein interactions. Nucleic Acids Res. 2002, 30, 303–305. [Google Scholar] [CrossRef]
- Chatr-Aryamontri, A.; Breitkreutz, B.J.; Heinicke, S.; Boucher, L.; Winter, A.; Stark, C.; Nixon, J.; Ramage, L.; Kolas, N.; O’Donnell, L.; et al. The BioGRID interaction database: 2013 update. Nucleic Acids Res. 2012, 41, 816–823. [Google Scholar] [CrossRef] [PubMed]
- Pagel, P.; Kovac, S.; Oesterheld, M.; Brauner, B.; Dunger-Kaltenbach, I.; Frishman, G.; Montrone, C.; Mark, P.; Stümpflen, V.; Mewes, H.W.; et al. The MIPS mammalian protein–protein interaction database. Bioinformatics 2005, 21, 832–834. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Lyon, D.; Junge, A.; Wyder, S.; Huerta-Cepas, J.; Simonovic, M.; Doncheva, N.T.; Morris, J.H.; Bork, P.; et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019, 47, 607–613. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Mulligan, M.K.; Mozhui, K.; Prins, P.; Williams, R.W. Genenetwork: A Toolbox for Systems Genetics. Methods Mol. Biol. 2017, 1488, 75–120. [Google Scholar]
- Hopcroft, J.E.; Karp, R.M. A n^5/2 Algorithm for Maximum Matchings in Bipartite Graphs. Annu. Symp. Switch. Autom. Theory 1973, 2, 225–231. [Google Scholar] [CrossRef]
- Lu, C.; Hu, X.; Wang, G.; Leach, L.J.; Luo, Z.W. Why do essential proteins tend to be clustered in the yeast interactome network? Mol. BioSyst. 2010, 6, 871–877. [Google Scholar] [CrossRef]
- Repana, D.; Nulsen, J.; Dressler, L.; Bortolomeazzi, M.; Venkata, S.K.; Tourna, A.; Yakovleva, A.; Palmieri, T.; Ciccarelli, F.D. The Network of Cancer Genes (NCG): A comprehensive catalogue of known and candidate cancer genes from cancer sequencing screens. Genome Biol. 2019, 20, 1. [Google Scholar] [CrossRef]
- Lotia, S.; Montojo, J.; Dong, Y.; Bader, G.D.; Pico, A.R. Cytoscape app store. Bioinformatics 2013, 29, 1350–1351. [Google Scholar] [CrossRef]
- Przulj, N.; Wigle, D.A.; Jurisica, I. Functional topology in a network of protein interactions. Bioinformatics 2004, 20, 340–348. [Google Scholar] [CrossRef]
- Morris, J.H.; Apeltsin, L.; Newman, A.M.; Baumbach, J.; Wittkop, T.; Su, G.; Bader, G.D.; Ferrin, T.E. clusterMaker: A multi-algorithm clustering plugin for Cytoscape. BMC Bioinform. 2011, 12, 436. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Huang, D.W.; Tan, Q.; Guo, Y.; Bour, S.; Liu, D.; Stephens, R.; Baseler, M.W.; Lane, H.C.; Lempicki, R.A. DAVID Knowledgebase: A gene-centered database integrating heterogeneous gene annotation resources to facilitate high-throughput gene functional analysis. BMC Bioinform. 2007, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J.G. Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.-H.; Pagès, F.; Trajanoski, Z.; Galon, J. ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef]
- Warde-Farley, D.; Donaldson, S.L.; Comes, O.; Zuberi, K.; Badrawi, R.; Chao, P.; Franz, M.; Grouios, C.; Kazi, F.; Lopes, C.T.; et al. The Genemania Prediction Server: Biological Network Integration for Gene Prioritization and Predicting Gene Function. Nucleic Acids Res. 2010, 38, 214–220. [Google Scholar] [CrossRef]
- GTEx Consortium. The Genotype-Tissue Expression (Gtex) Project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Yang, S.Y.C.; Lheureux, S.; Karakasis, K.; Burnier, J.V.; Bruce, J.P.; Clouthier, D.L.; Danesh, A.; Quevedo, R.; Dowar, M.; Hanna, Y.; et al. Landscape of genomic alterations in high-grade serous ovarian cancer from exceptional long- and short-term survivors. Genome Med. 2018, 10, 81. [Google Scholar] [CrossRef]
- Albert, R.; Jeong, H.; Barabasi, A.L. Error and attack tolerance of complex networks. Nature 2000, 406, 378–382. [Google Scholar] [CrossRef]
- Nishikawa, T.; Motter, A.E.; Lai, Y.C.; Hoppensteadt, F.C. Heterogeneity in oscillator networks: Are smaller worlds easier to synchronize? Phys. Rev. Lett. 2003, 91, 014101. [Google Scholar] [CrossRef]
- Wang, W.; Slotine, J.J. On partial contraction analysis for coupled nonlinear oscillators. Biol. Cybern. 2005, 92, 38–53. [Google Scholar] [CrossRef]
- Giuntoli, R.L.; Rodriguez, G.C.; Whitaker, R.S.; Dodge, R.; Voynow, J.A. Mucin gene expression in ovarian cancers. Cancer Res. 1998, 58, 5546–5550. [Google Scholar] [PubMed]
- Singh, A.P.; Senapati, S.; Ponnusamy, M.P.; Jain, M.; Lele, S.M.; Davis, J.S.; Remmenga, S.; Batra, S.K. Clinical potential of mucins in diagnosis, prognosis, and therapy of ovarian cancer. Lancet Oncol. 2008, 9, 1076–1085. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Guan, C.; Fang, C.; Jin, X.; Yu, J.; Zhang, Y.; Zheng, L. Clinical significance and prognostic value of Forkhead box A1 expression in human epithelial ovarian cancer. Oncol. Lett. 2018, 15, 4457–4462. [Google Scholar] [CrossRef] [PubMed]
- Heo, J.; Eki, R.; Abbas, T. Deregulation of F-box proteins and its consequence on cancer development, progression and metastasis. Semin. Cancer Biol. 2016, 36, 33–51. [Google Scholar] [CrossRef] [PubMed]
- Won, K.Y.; Lim, S.-J.; Kim, G.Y.; Kim, Y.W.; Han, S.-A.; Song, J.Y.; Lee, D.-K. Regulatory role of p53 in cancer metabolism via SCO2 and TIGAR in human breast cancer. Hum. Pathol. 2012, 43, 221–228. [Google Scholar] [CrossRef] [PubMed]
- Mabuchi, S.; Kuroda, H.; Takahashi, R.; Sasano, T. The PI3K/AKT/mTOR pathway as a therapeutic target in ovarian cancer. Gynecol. Oncol. 2015, 137, 173–179. [Google Scholar] [CrossRef]
- Abdel-Fatah, T.M.; Arora, A.; Moseley, P.; Coveney, C.; Perry, C.; Johnson, K.; Kent, C.; Ball, G.; Chan, S.; Madhusudan, S. ATM, ATR and DNA-PKcs expressions correlate to adverse clinical outcomes in epithelial ovarian cancers. BBA Clin. 2014, 2, 10–17. [Google Scholar] [CrossRef]
- Hager, K.M.; Gu, W. Understanding the non-canonical pathways involved in p53-mediated tumor suppression. Carcinogenesis 2014, 35, 740–746. [Google Scholar] [CrossRef]
- Minton, D.R.; Fu, L.; Mongan, N.P.; Shevchuk, M.M.; Nanus, D.M.; Gudas, L.J. Role of NADH Dehydrogenase (Ubiquinone) 1 Alpha Subcomplex 4-Like 2 in Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. 2016, 22, 2791–2801. [Google Scholar] [CrossRef]
- Mamelak, A.J.; Kowalski, J.; Murphy, K.; Yadava, N.; Zahurak, M.; Kouba, D.J.; Howell, B.G.; Tzu, J.; Cummins, D.L.; Liegeois, N.J.; et al. Downregulation of NDUFA1 and other oxidative phosphorylation-related genes is a consistent feature of basal cell carcinoma. Exp. Dermatol. 2005, 14, 336–348. [Google Scholar] [CrossRef]
- Phan, L.M.; Yeung, S.C.J.; Lee, M.H. Cancer metabolic reprogramming: Importance, main features, and potentials for precise targeted anti-cancer therapies. Cancer Biopsy. Med. 2014, 11, 1–19. [Google Scholar]
- Hay, N. Reprogramming glucose metabolism in cancer: Can it be exploited for cancer therapy? Nat. Rev. Cancer 2016, 16, 635–649. [Google Scholar] [CrossRef] [PubMed]
- Turgeon, M.O.; Perry, N.J.S.; Poulgiannis, G. DNA damage, repair and cancer metabolism. Front. Oncol. 2018, 8, 5955. [Google Scholar] [CrossRef] [PubMed]
- Pandey, N.; Lanke, V.; Vinod, P.K. Network-based metabolic characterization of renal cell carcinoma. Sci. Rep. 2020, 10, 5955. [Google Scholar] [CrossRef]
- Fuh, K.C.; Java, J.J.; Chan, J.K.; Kapp, D.S.; Monk, B.J.; Burger, R.A.; Young, R.C.; Alberts, D.S.; McGuire, W.P.; Markman, M.; et al. Differences in presentation and survival of Asians compared to Caucasians with ovarian cancer: An NRG Oncology/GOG Ancillary study of 7914 patients. Gynecol. Oncol. 2019, 154, 420–425. [Google Scholar] [CrossRef]
- Cheng, F.; Kovacs, I.A.; Barabasi, A.L. Network-based prediction of drug combinations. Nat. Commun. 2019, 10, 1197. [Google Scholar] [CrossRef]
- Gates, A.J.; Correia, R.B.; Wang, X.; Rocha, L.M. The effective graph reveals redundancy, canalization, and control pathways in biochemical regulation and signaling. Proc. Natl. Acad. Sci. USA 2021, 118, e2022598118. [Google Scholar] [CrossRef]
- Wang, Y.; Yang, H.; Chen, L.; Jafari, M.; Tang, J. Network-based modeling of herb combinations in traditional Chinese medicine. Brief. Bioinform. 2021, 22, 106. [Google Scholar] [CrossRef]

| Characteristic | Correlation | CM-OUT | CM-IN | CN-OUT | CN-IN | DN-OUT | DN-IN |
|---|---|---|---|---|---|---|---|
| GC | Pearson correlation coefficient | 0.045 | 0.121 | 0.065 | 0.131 | 0.024 | 0.084 |
| Significance | 0.801 | 0.334 | 0.499 | 0.177 | 0.562 | 0.271 | |
| L | Pearson correlation coefficient | 0.035 | 0.211 | 0.041 | 0.112 | 0.341 ** | 0.503 ** |
| Significance | 0.821 | 0.062 | 0.694 | 0.082 | 0.00 | 0.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Esmaeilzadeh, A.A.; Kashian, M.; Salman, H.M.; Alsaffar, M.F.; Jaber, M.M.; Soltani, S.; Ilhan, A.; Bahrami, A. RETRACTED: Identify Biomarkers and Design Effective Multi-Target Drugs in Ovarian Cancer: Hit Network-Target Sets Model Optimizing. Biology 2022, 11, 1851. https://doi.org/10.3390/biology11121851
Esmaeilzadeh AA, Kashian M, Salman HM, Alsaffar MF, Jaber MM, Soltani S, Ilhan A, Bahrami A. RETRACTED: Identify Biomarkers and Design Effective Multi-Target Drugs in Ovarian Cancer: Hit Network-Target Sets Model Optimizing. Biology. 2022; 11(12):1851. https://doi.org/10.3390/biology11121851
Chicago/Turabian StyleEsmaeilzadeh, Amir Abbas, Mahdis Kashian, Hayder Mahmood Salman, Marwa Fadhil Alsaffar, Mustafa Musa Jaber, Siamak Soltani, Ahmet Ilhan, and Abolfazl Bahrami. 2022. "RETRACTED: Identify Biomarkers and Design Effective Multi-Target Drugs in Ovarian Cancer: Hit Network-Target Sets Model Optimizing" Biology 11, no. 12: 1851. https://doi.org/10.3390/biology11121851
APA StyleEsmaeilzadeh, A. A., Kashian, M., Salman, H. M., Alsaffar, M. F., Jaber, M. M., Soltani, S., Ilhan, A., & Bahrami, A. (2022). RETRACTED: Identify Biomarkers and Design Effective Multi-Target Drugs in Ovarian Cancer: Hit Network-Target Sets Model Optimizing. Biology, 11(12), 1851. https://doi.org/10.3390/biology11121851






