A De Novo Optimized Cell-Free System for the Expression of Soluble and Active Human Tumor Necrosis Factor-Alpha
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Vectors
2.2. Rare Codon Analysis of the DNA Coding Sequence for Human TNF-α
2.3. Codon Optimization of the DNA Coding Sequence for Human TNF-α
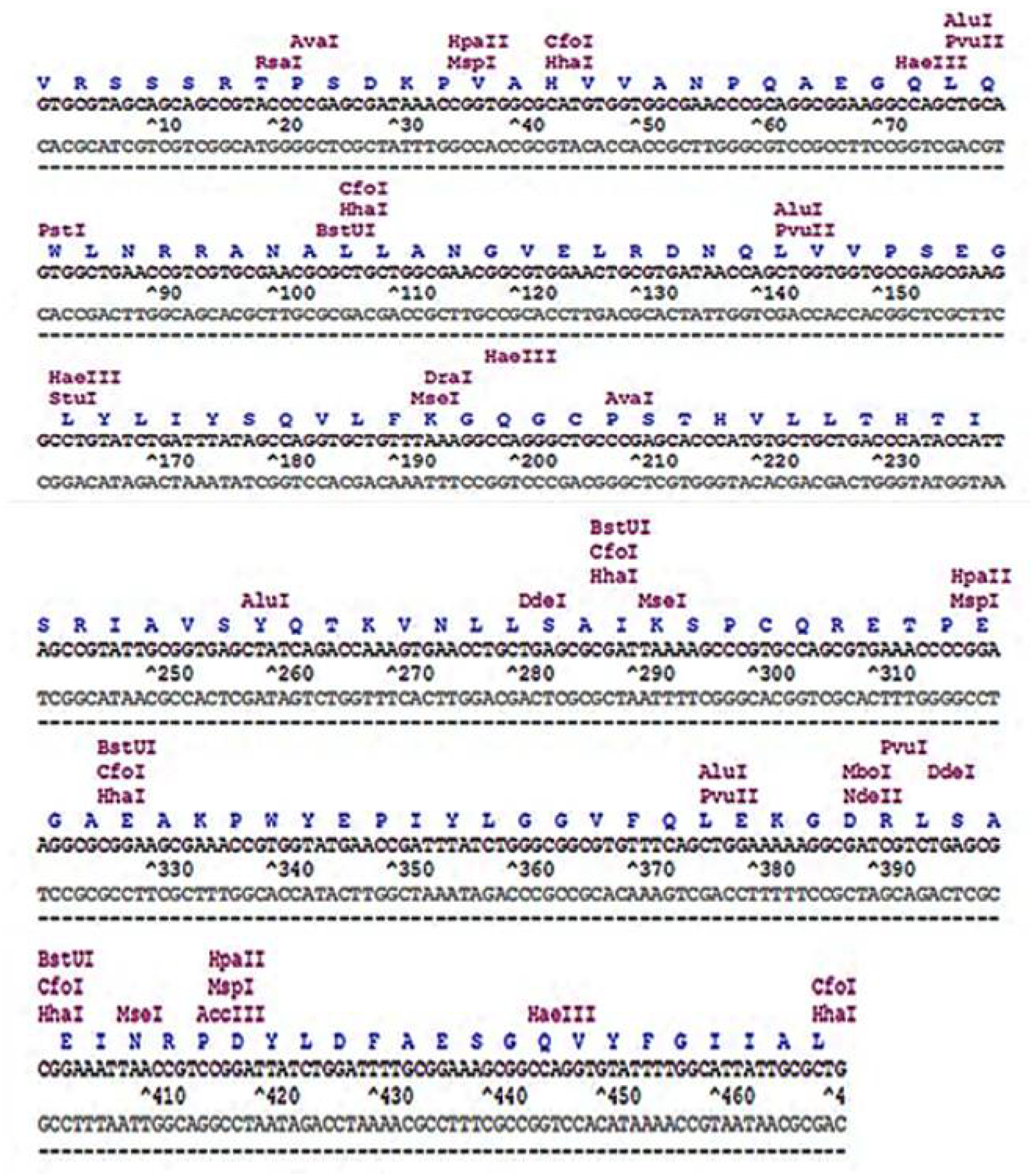
2.4. Restriction Map of Human TNF-α Gene
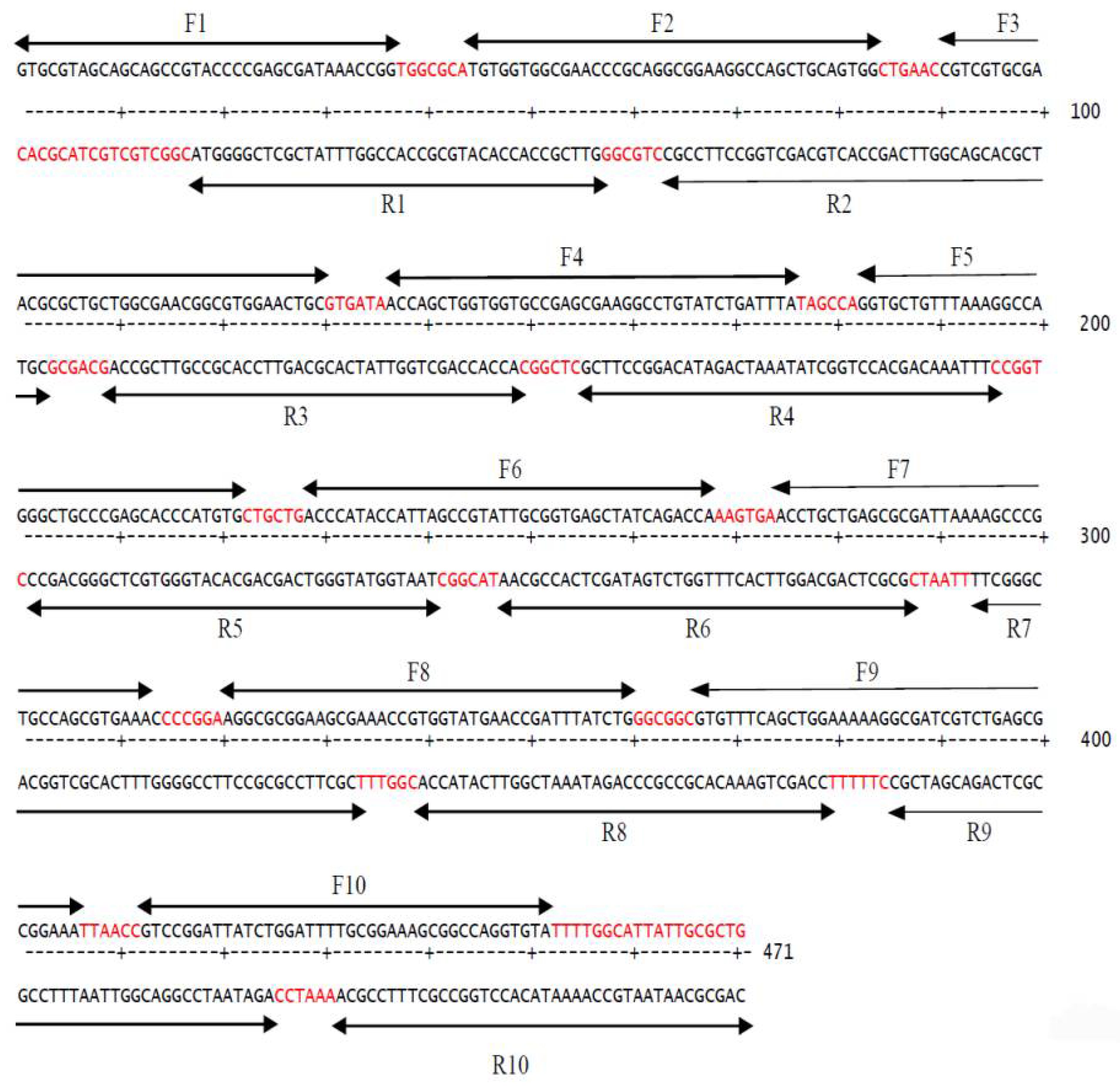
2.5. Primers Design for the Synthesis of Codon-Optimized Human TNF-α Gene
2.5.1. Oligonucleotide Primers for Gene Synthesis
2.5.2. Gene-Specific Primers for Amplification and Subsequent Cloning into Vectors
2.6. Two-Step PCR-Mediated Gene Construction
2.6.1. PCR Assembling Step
2.6.2. PCR Amplification Step
2.7. Cloning of Human TNF-α Gene
2.8. Construction of Expression Plasmid for CFPS
2.9. Cell-Free Synthesis of Recombinant Human TNF-α Protein
2.10. SDS-PAGE Analysis of Cell-Free Synthesized Human TNF-α Protein
2.11. Quantitative Estimation of Synthesized Human TNF-α by ELISA
2.12. Two-Step Chromatographic Purification of Cell-Free Synthesized Human TNF-α
2.13. RSM for the Optimization of Cell-Free Expression of Soluble Human TNF-α
2.13.1. Experimental Design
2.13.2. Statistical Analysis
2.14. Bioassays of Cell-Free Synthesized Human TNF-α Protein
2.14.1. Human Cancer Cell Lines
2.14.2. Cell Culture
2.14.3. Determination of the Cytotoxicity of Cell-Free Synthesized Human TNF-α against Normal Human Cells
2.14.4. Determination of the Cytotoxicity of Cell-Free Synthesized Human TNF-α against Human Cancer Cell Lines
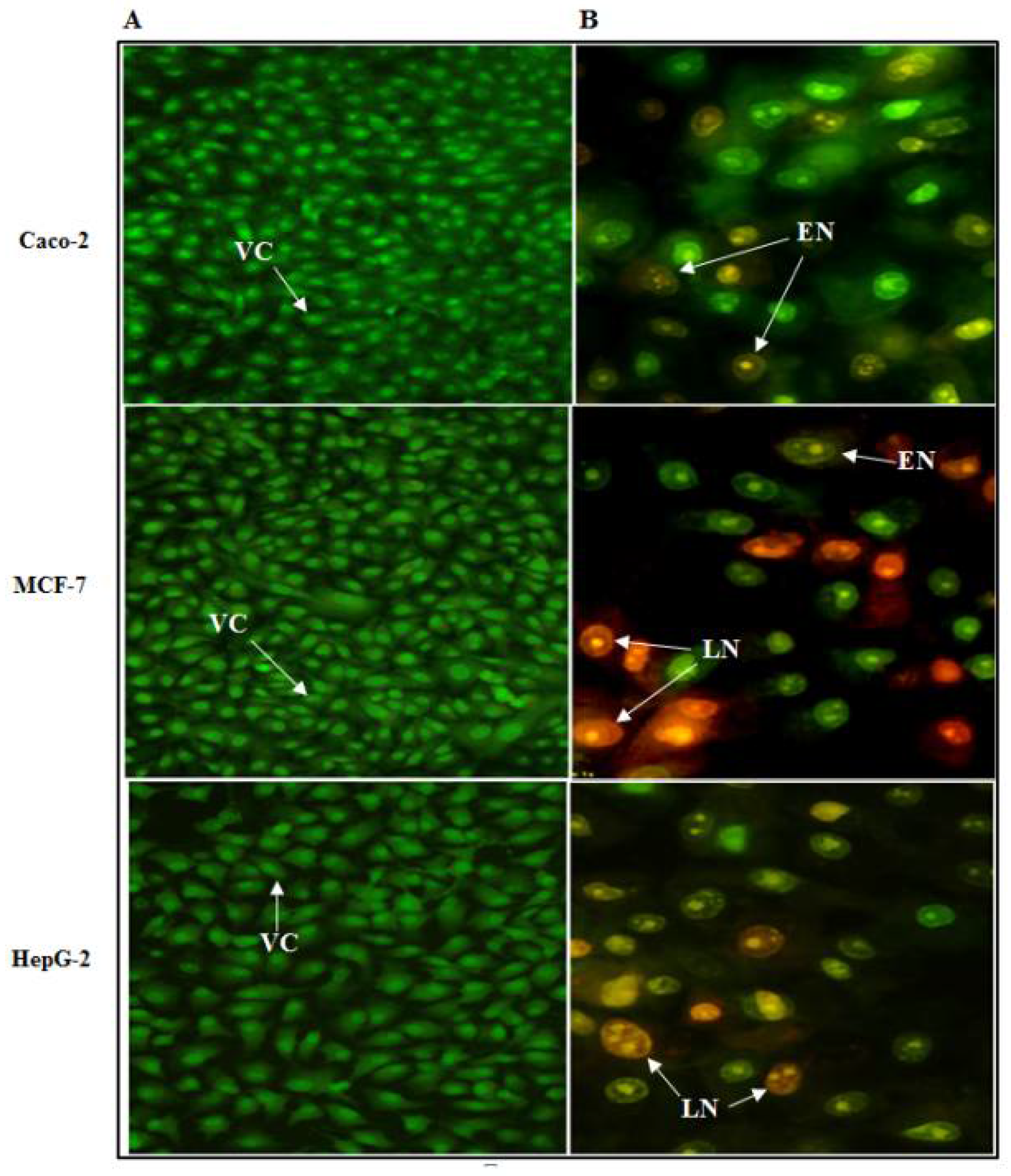
2.14.5. Assessment of Cancer Cell Death Using Nuclear Staining
3. Results
3.1. Codon Optimization of the Human TNF-α Gene
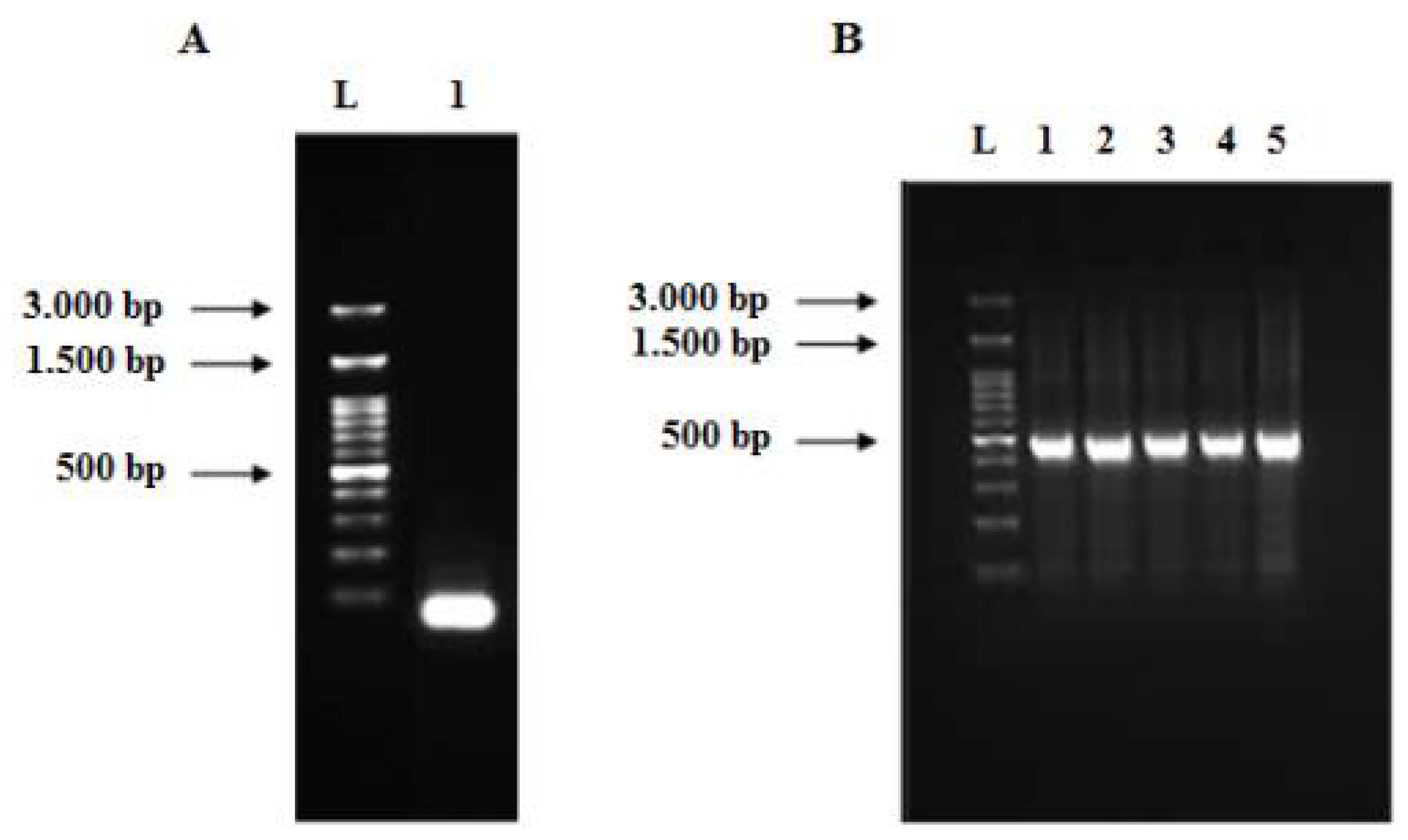
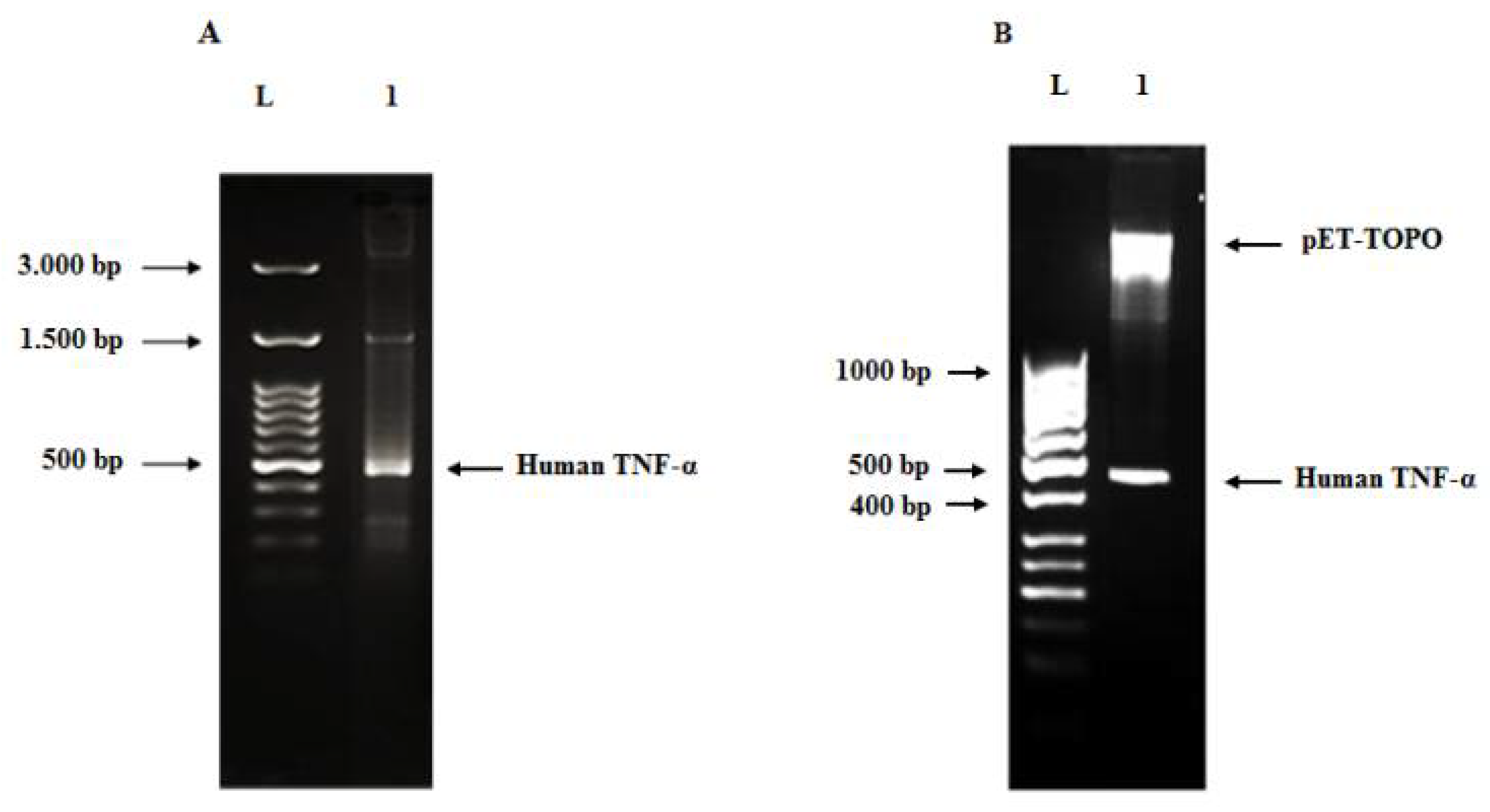
3.2. Synthesis of Full-Length of Codon-Optimized Human TNF-α Gene Using Assembly and Amplification PCR Strategy
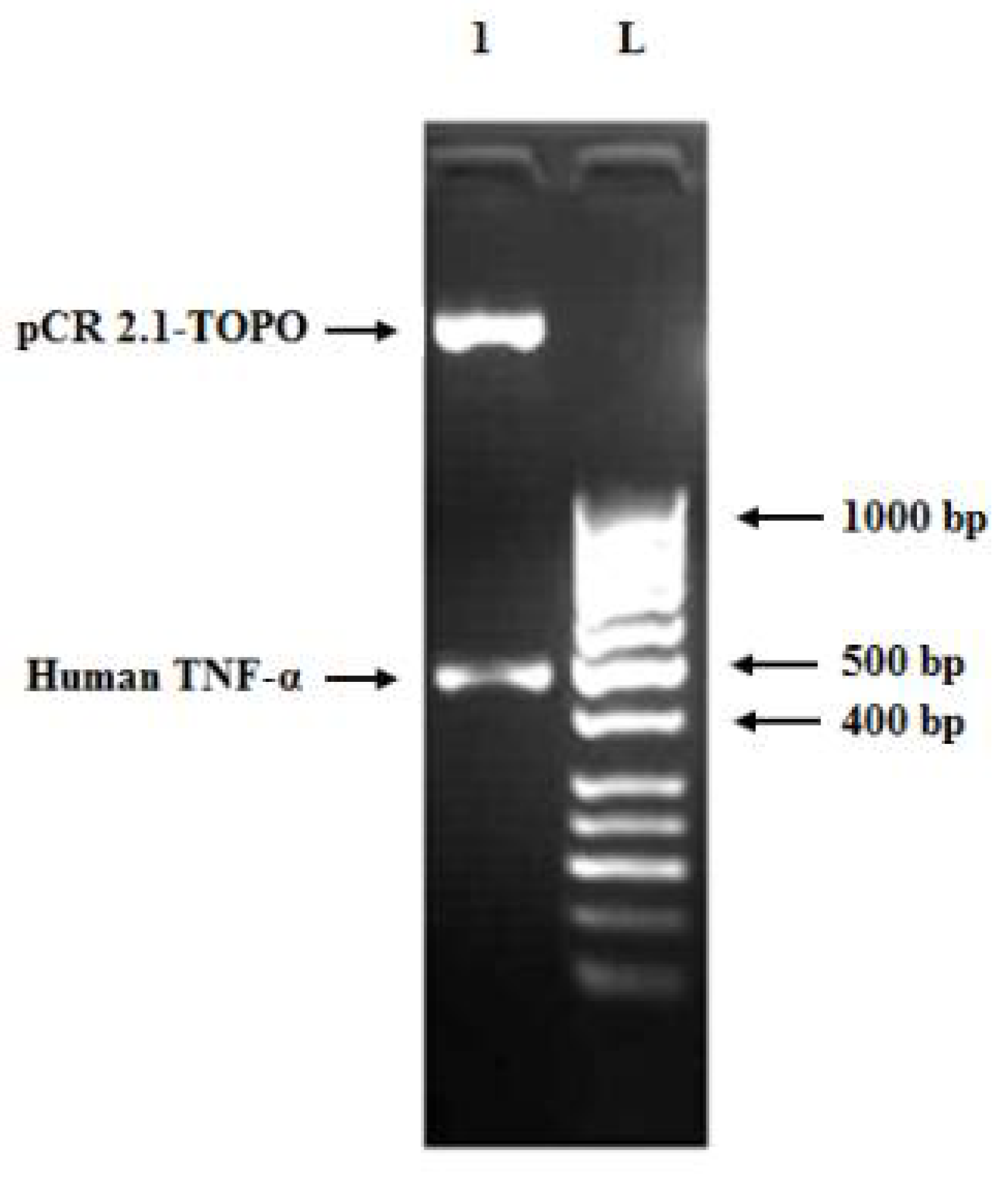
3.3. Cloning of Human TNF-α Gene into Cloning and Expression Vectors
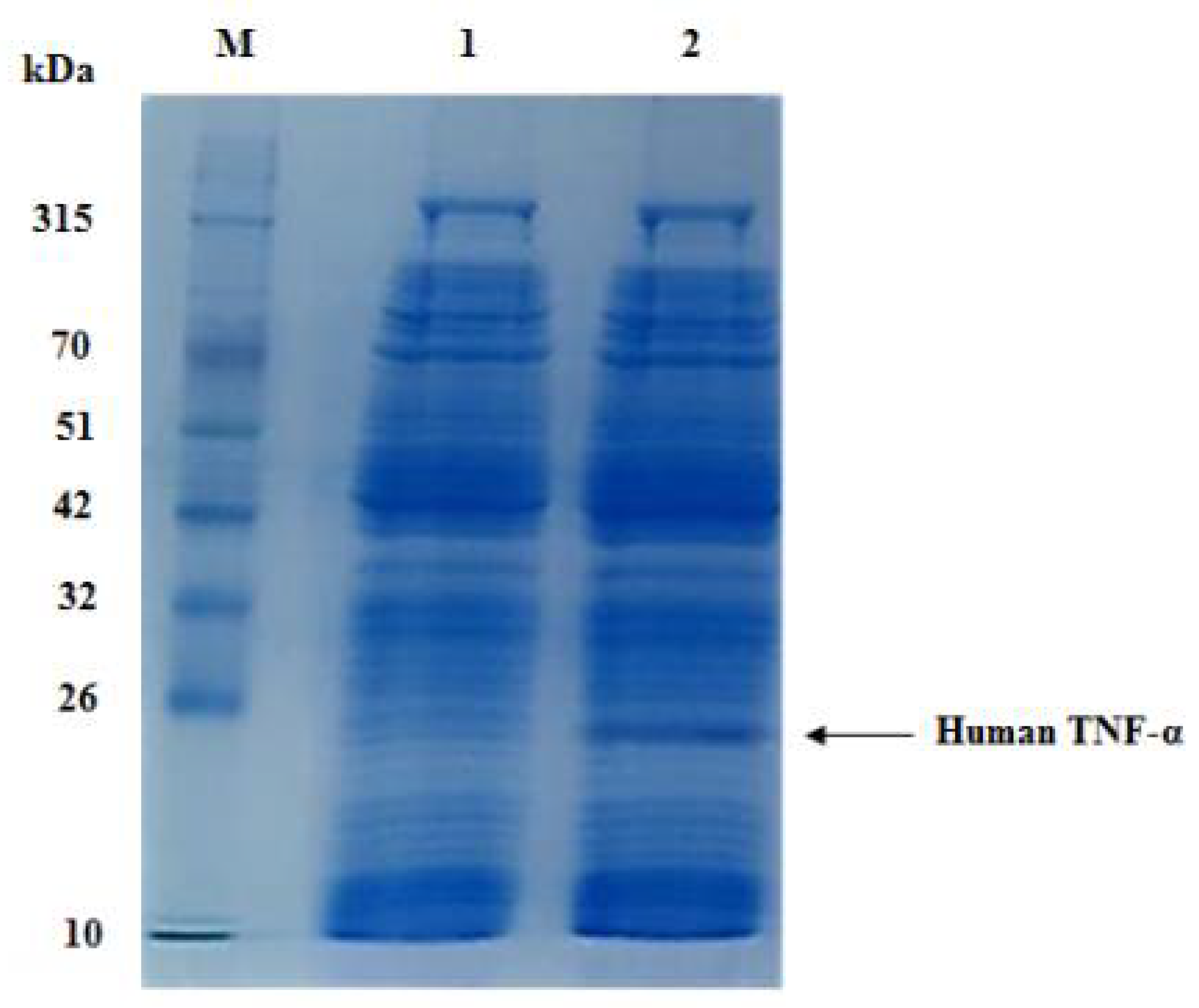
3.4. Cell-Free Synthesis of Recombinant Human TNF-α Protein
3.5. Two-Step Chromatographic Purification of Cell-Free Synthesized Human TNF-α Protein
3.6. RSM for the Optimization of Cell-Free Synthesis of Human TNF-α Protein
3.6.1. CCD Modelling
3.6.2. ANOVA
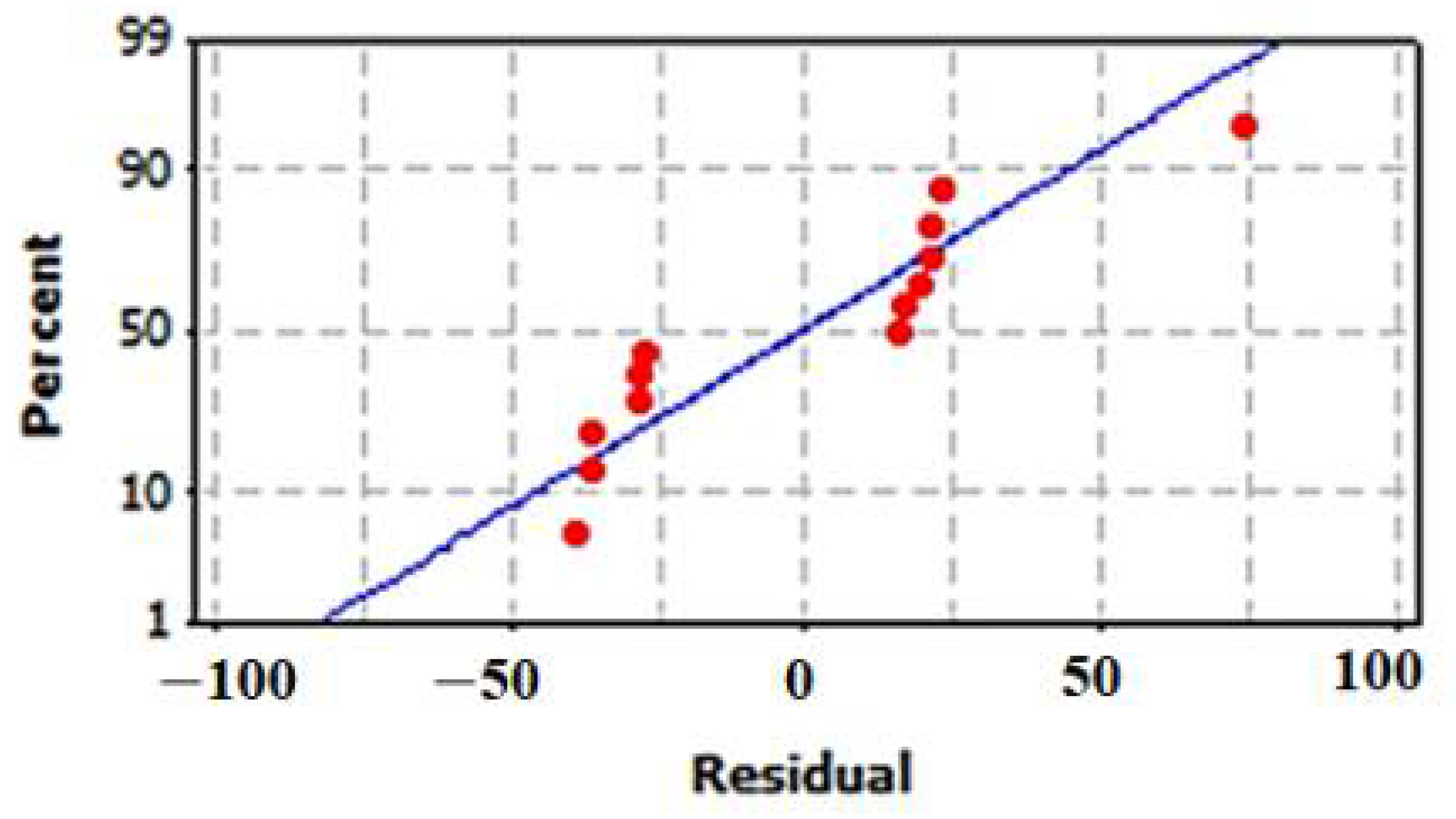
3.6.3. Model Validation Using Residuals
3.6.4. Interactive Effect of Independent Variables
3.6.5. Experimental Model Validation
3.7. Bioassays of Cell-Free Synthesized Human TNF-α Protein
3.7.1. Determination of the Cytotoxicity of Cell-Free Synthesized Human TNF-α against Normal Human Cells
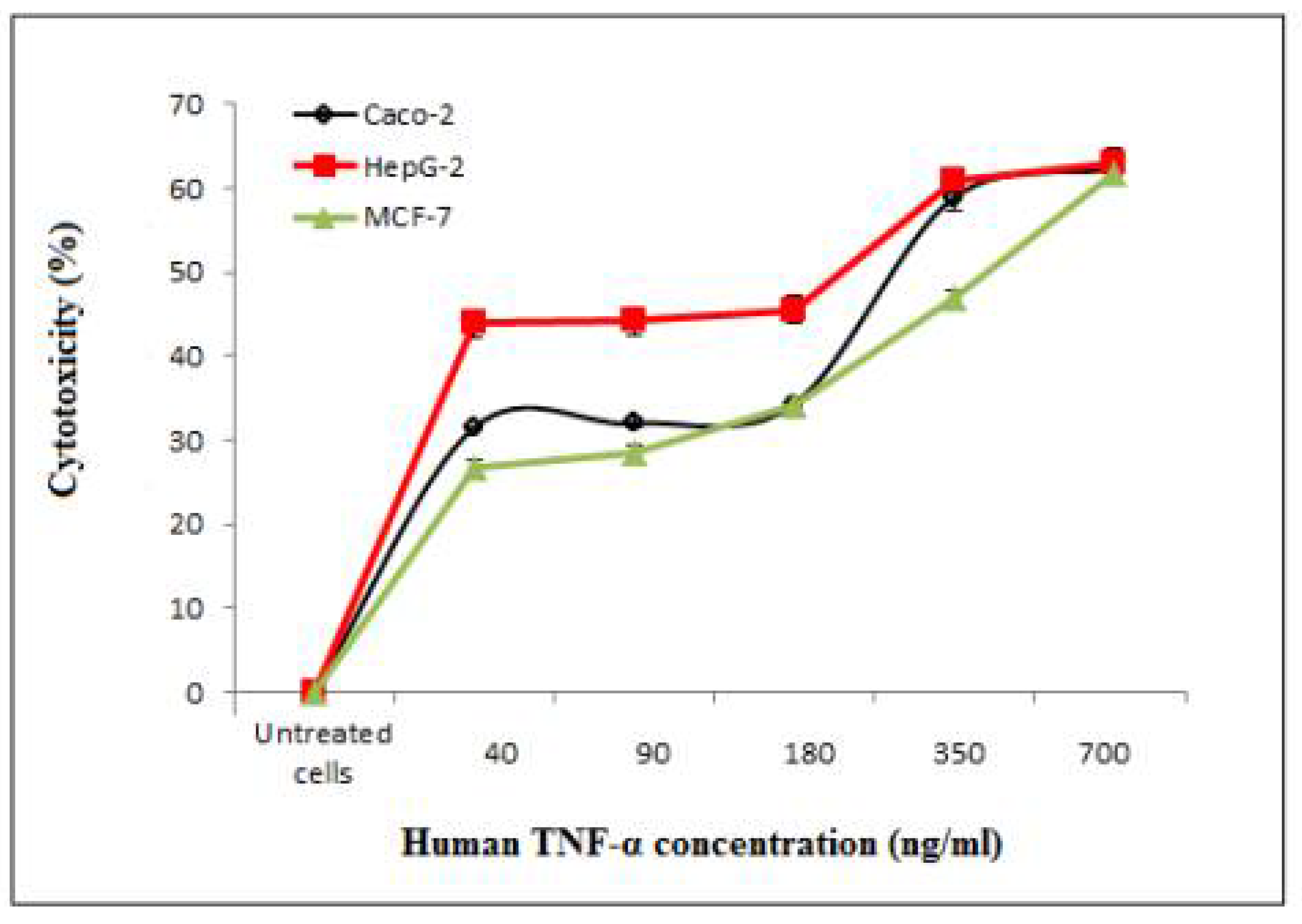
3.7.2. Determination of the Cytotoxicity of Cell-Free Synthesized Human TNF-α against Human Cancer Cell Lines
3.7.3. Assessment of Cancer Cell Death Using EtBr-AO Nuclear Staining
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, H.J.; Choi, B.Y.; Sohn, M.; Han, N.Y.; Kim, I.W.; Oh, J.M. Effects of Tumor Necrosis Factor-alpha Inhibitors on the Incidence of Tuberculosis. Korean J. Clin. Pharm. 2018, 28, 333–341. [Google Scholar] [CrossRef]
- Idress, M.; Milne, B.F.; Thompson, G.S.; Trembleau, L.; Jaspars, M.; Houssen, W.E. Structure-Based Design, Synthesis and Bioactivity of a New Anti-TNFα Cyclopeptide. Molecules 2020, 25, 922. [Google Scholar] [CrossRef] [Green Version]
- Chu, W.-M. Tumor necrosis factor. Cancer Lett. 2013, 328, 222–225. [Google Scholar] [CrossRef] [Green Version]
- Howerton, E.; Tarzami, S.T. Tumor necrosis factor-alpha and inflammation-mediated cardiac injury. J. Cell Sci. Ther. 2017, 8, 1–4. [Google Scholar] [CrossRef]
- Perse, M.; Unkovic, A. The Role of TNF in the Pathogenesis of Inflammatory Bowel Disease. In Biological Therapy for Inflammatory Bowel Disease; Leal, R.F., Torriani, T., Eds.; IntechOpen: London, UK, 2019; pp. 23–35. [Google Scholar] [CrossRef] [Green Version]
- Turner, M.D.; Nedjai, B.; Hurst, T.; Pennington, D.J. Cytokines and chemokines: At the crossroads of cell signalling and inflammatory disease. Biochim. Et Biophys. Acta (BBA)—Mol. Cell Res. 2014, 1843, 2563–2582. [Google Scholar] [CrossRef] [Green Version]
- Singha, T.K.; Gulati, P.; Kumar, S. Nonconventional induction strategies for production of recombinant human tumor necrosis factor-alpha in Escherichia coli. J. Appl. Biol. Biotechnol. 2018, 6, 23–27. [Google Scholar] [CrossRef] [Green Version]
- Ruder, B.; Atreya, R.; Becker, C. Tumour necrosis factor alpha in intestinal homeostasis and gut related diseases. Int. J. Mol. Sci. 2019, 20, 1887. [Google Scholar] [CrossRef] [Green Version]
- Uversky, V.N.; El-Baky, N.A.; El-Fakharany, E.M.; Sabry, A.; Mattar, E.H.; Uversky, A.V.; Redwan, E.M. Functionality of intrinsic disorder in tumor necrosis factor-α and its receptors. FEBS J. 2017, 284, 3589–3618. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharif, P.M.; Jabbari, P.; Razi, S.; Keshavarz-Fathi, M.; Rezaei, N. Importance of TNF-alpha and its alterations in the development of cancers. Cytokine 2020, 130, 155066. [Google Scholar] [CrossRef] [PubMed]
- Brenner, D.; Blaser, H.; Mak, T.W. Regulation of tumour necrosis factor signalling: Live or let die. Nat. Rev. Immunol. 2015, 15, 362–374. [Google Scholar] [CrossRef] [PubMed]
- Kalliolias, G.; Ivashkiv, L.B. TNF biology, pathogenic mechanisms and emerging therapeutic strategies. Nat. Rev. Rheumatol. 2016, 12, 49–62. [Google Scholar] [CrossRef]
- Singha, T.K.; Dagar, V.K.; Gulati, P.; Kumar, S. Kinetic study and optimization of recombinant human tumor necrosis factor-alpha (rhTNF-α) production in Escherichia coli. Prep. Biochem. Biotechnol. 2021, 51, 267–276. [Google Scholar] [CrossRef] [PubMed]
- Damough, S.; Sabzalinezhad, M.; Talebkhan, Y.; Nematollahi, L.; Bayat, E.; Torkashvand, F.; Adeli, A.; Jahandar, H.; Barkhordari, F.; Mahboudi, F. Optimization of culture conditions for high-level expression of soluble and active tumor necrosis factor-α in E. coli. Protein Expr. Purif. 2021, 179, 105805. [Google Scholar] [CrossRef] [PubMed]
- Ciacma, K.; Wieckiewicz, J.; Kędracka-Krok, S.; Kurtyka, M.; Stec, M.; Siedlar, M.; Baran, J. Secretion of tumoricidal human tumor necrosis factor-related apoptosis-inducing ligand (TRAIL) by recombinant Lactococcus lactis: Optimization of in vitro synthesis conditions. Microb. Cell Factories 2018, 17, 177. [Google Scholar] [CrossRef] [PubMed]
- Pozidis, C.; Lammertyn, E.; Politou, A.S.; Anné, J.; Tsiftsoglou, A.S.; Sianidis, G.; Economou, A. Protein secretion biotechnology using Streptomyces lividans: Large-scale production of functional trimeric tumor necrosis factor alpha. Biotechnol. Bioeng. 2001, 72, 611–619. [Google Scholar] [CrossRef]
- Zhang, X.-W.; Sun, T.; Zeng, X.-Y.; Liu, X.; Gu, D.-X. Expression of recombinant human tumor necrosis factor-α in a baculovirus expression system. Bioresour. Technol. 1999, 70, 299–301. [Google Scholar] [CrossRef]
- Ebihara, T.; Xu, J.; Tonooka, Y.; Nagasato, T.; Kakino, K.; Masuda, A.; Minamihata, K.; Kamiya, N.; Nakatake, H.; Chieda, Y.; et al. Active Human and Murine Tumor Necrosis Factor α Cytokines Produced from Silkworm Baculovirus Expression System. Insects 2021, 12, 517. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Zeng, L.; Wang, Y.-L.; Cui, S.-Q.; Hu, L.; Zheng, J.-M.; Huang, D.-N.; Hou, G. Construction and characterization of a transmembrane eukaryotic expression vector based on the membrane domain structure of TNF-α. Mol. Med. Rep. 2017, 16, 1021–1030. [Google Scholar] [CrossRef] [Green Version]
- Han, W.; Zhang, Y.; Yan, Z.; Shi, J. Construction of a new tumour necrosis factor fusion-protein expression vector for high-level expression of heterologous genes in Escherichia coli. Biotechnol. Appl. Biochem. 2003, 37, 109–113. [Google Scholar] [CrossRef]
- Alizadeh, A.; Hamzeh-Mivehroud, M.; Farajzadeh, M.; Moosavi-Movahedi, A.; Dastmalchi, S. A simple and rapid method for expression and purification of functional TNF-α using GST fusion system. Curr. Pharm. Biotechnol. 2015, 16, 707–715. [Google Scholar] [CrossRef]
- Castineiras, T.S.; Williams, S.G.; Hitchcock, A.; Cole, J.A.; Smith, D.C.; Overton, T.W. Optimizing host cell physiology and stress avoidance for the production of recombinant human tumour necrosis factor α in Escherichia coli. Microbiology 2018, 164, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Kajikawa, M.; Maenaka, K.; Park, E.Y. Silkworm expression system as a platform technology in life science. Appl. Microbiol. Biotechnol. 2009, 85, 459–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, B.; Jeon, C.O. High-throughput recombinant protein expression in Escherichia coli: Current status and future perspectives. Open Biol. 2016, 6, 160196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tripathi, N.K.; Shrivastava, A. Recent developments in bioprocessing of recombinant proteins: Expression hosts and process development. Front. Bioeng. Biotechnol. 2019, 7, 420. [Google Scholar] [CrossRef] [Green Version]
- Damiati, S.; Mhanna, R.; Kodzius, R.; Ehmoser, E.-K. Cell-free approaches in synthetic biology utilizing microfluidics. Genes 2018, 9, 144. [Google Scholar] [CrossRef] [Green Version]
- Dopp, J.L.; Tamiev, D.D.; Reuel, N.F. Cell-free supplement mixtures: Elucidating the history and biochemical utility of additives used to support in vitro protein synthesis in E. coli extract. Biotechnol. Adv. 2019, 37, 246–258. [Google Scholar] [CrossRef]
- Dondapati, S.K.; Stech, M.; Zemella, A.; Kubick, S. Cell-free protein synthesis: A promising option for future drug development. BioDrugs 2020, 34, 327–348. [Google Scholar] [CrossRef] [Green Version]
- Spirin, A.S. High-throughput cell-free systems for synthesis of functionally active proteins. Trends Biotechnol. 2004, 22, 538–545. [Google Scholar] [CrossRef]
- Mikami, S.; Kobayashi, T.; Masutani, M.; Yokoyama, S.; Imataka, H. A human cell-derived in vitro coupled transcription/translation system optimized for production of recombinant proteins. Protein Expr. Purif. 2008, 62, 190–198. [Google Scholar] [CrossRef]
- Hino, M.; Kataoka, M.; Kajimoto, K.; Yamamoto, T.; Kido, J.; Shinohara, Y.; Baba, Y. Efficiency of cell-free protein synthesis based on a crude cell extract from Escherichia coli, wheat germ, and rabbit reticulocytes. J. Biotechnol. 2008, 133, 183–189. [Google Scholar] [CrossRef] [PubMed]
- Khambhati, K.; Bhattacharjee, G.; Gohil, N.; Braddick, D.; Kulkarni, V.; Singh, V. Exploring the potential of cell-free protein synthesis for extending the abilities of biological systems. Front. Bioeng. Biotechnol. 2019, 7, 248. [Google Scholar] [CrossRef]
- Smolskaya, S.; Logashina, Y.A.; Andreev, Y.A. Escherichia coli extract-based cell-free expression system as an alternative for difficult-to-obtain protein biosynthesis. Int. J. Mol. Sci. 2020, 21, 928. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Gu, L.; Aach, J.; Church, G.M. Improved Cell-Free RNA and Protein Synthesis System. PLoS ONE 2014, 9, e106232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, A.-S.; Yao, Q.-H.; Peng, R.-H.; Duan, H.; Li, X.; Fan, H.-Q.; Cheng, Z.-M.; Li, Y. PCR-based accurate synthesis of long DNA sequences. Nat. Protoc. 2006, 1, 791–797. [Google Scholar] [CrossRef] [PubMed]
- Mohammed, Y.; EL-Baky, N.A.; Redwan, E.M. Expression, purification, and characterization of recombinant human consensus interferon-alpha in Escherichia coli under λPL promoter. Prep. Biochem. Biotechnol. 2012, 42, 426–447. [Google Scholar] [CrossRef] [PubMed]
- El-Baky, N.A.; Omar, S.H.; Redwan, E.M. The anti-cancer activity of human consensus interferon-alpha synthesized in cell-free system. Protein Expr. Purif. 2011, 80, 61–67. [Google Scholar] [CrossRef]
- Papaneophytou, C.; Kontopidis, G. A comparison of statistical approaches used for the optimization of soluble protein expression in Escherichia coli. Protein Expr. Purif. 2016, 120, 126–137. [Google Scholar] [CrossRef]
- Boyum, A. Isolation of mononuclear cells and granulocytes from human blood. Isolation of monuclear cells by one centrifugation, and of granulocytes by combining centrifugation and sedimentation at 1 g. Scand. J. Clin. Lab. Investig. 1968, 97, 77–89. [Google Scholar]
- Mosmann, T. Rapid colorimetric assay for cellular growth and survival: Application to proliferation and cytotoxicity assays. J. Immunol. Methods 1983, 65, 55–63. [Google Scholar] [CrossRef]
- Smith, S.M.; Ribble, D.; Goldstein, N.B.; Norris, D.A.; Shellman, Y.G. Chapter 20—A simple technique for quantifying apoptosis in 96-well plates. In Methods in Cell Biology; Conn, P.M., Ed.; Academic Press: Cambridge, MA, USA, 2012; Volume 112, pp. 361–368. [Google Scholar] [CrossRef] [Green Version]
- Markets and Markets. Protein Expression Market by Type (E. coli, Mammalian, Yeast, Pichia, Insect, Baculovirus, Cell-Free), Products (Competent Cells, Reagents, Instruments, Services), Application (Therapeutic, Research, Industrial) & End User—Global Forecast to 2022, Report Code BT 2435, 2017. Available online: https://www.marketsandmarkets.com/ (accessed on 1 July 2021).
- Schillberg, S.; Raven, N.; Spiegel, H.; Rasche, S.; Buntru, M. Critical Analysis of the Commercial Potential of Plants for the Production of Recombinant Proteins. Front. Plant Sci. 2019, 10, 720. [Google Scholar] [CrossRef]
- Puetz, J.; Wurm, F.M. Recombinant proteins for industrial versus pharmaceutical purposes: A review of process and pricing. Processes 2019, 7, 476. [Google Scholar] [CrossRef] [Green Version]
- Gregorio, N.E.; Levine, M.Z.; Oza, J.P. A User’s Guide to Cell-Free Protein Synthesis. Methods Protoc. 2019, 2, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quast, R.B.; Sonnabend, A.; Stech, M.; Wustenhagen, D.A.; Kubick, S. High-yield cell-free synthesis of human EGFR by IRES-mediated protein translation in a continuous exchange cell-free reaction format. Sci. Rep. 2016, 6, 30399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, K.; Choe, D.; Lee, D.H.; Cho, B.K. Engineering biology to construct microbial chassis for the production of difficult-to-express proteins. Int. J. Mol. Sci. 2020, 21, 990. [Google Scholar] [CrossRef] [Green Version]
- Nik-Pa, N.I.M.; Sobri, M.F.M.; Abd-Aziz, S.; Ibrahim, M.F.; Kamal Bahrin, E.; Mohammed Alitheen, N.B.; Ramli, N. Combined optimization of codon usage and glycine supplementation enhances the extracellular production of a β-cyclodextrin glycosyltransferase from Bacillus sp. NR5 UPM in Escherichia coli. Int. J. Mol. Sci. 2020, 21, 3919. [Google Scholar] [CrossRef]
- Binepal, G.; Ranjan, R.K.; Rajagopal, K. Expression of synthetic human tumor necrosis factor is toxic to Escherichia coli. Gene 2012, 493, 155–160. [Google Scholar] [CrossRef]
- Yin, S.; Zhang, C.; Li, Z.; Wang, Q.; Shi, H.; Yu, R.; Liu, Y.; Su, Z. Identification, characterization, and stabilization of the deamidation degradation of recombinant human tumor necrosis factor-α. Process Biochem. 2017, 53, 216–223. [Google Scholar] [CrossRef]
- Jin, H.; Uddin, M.S.; Huang, Y.L.; Teo, W.K. Purification and renaturation of recombinant human lymphotoxin (tumour necrosis factor beta) expressed in Escherichia coli as inclusion bodies. J. Chem. Technol. Biotechnol. 1994, 59, 67–72. [Google Scholar] [CrossRef]
- Wang, Y.; Ren, W.; Gao, D.; Wang, L.; Yang, Y.; Bai, Q. One-step refolding and purification of recombinant human tumor necrosis factor-α (rhTNF-α) using ion-exchange chromatography. Biomed. Chromatogr. 2015, 29, 305–311. [Google Scholar] [CrossRef]
- de Marco, A. Strategies for successful recombinant expression of disulfide bond-dependent proteins in Escherichia coli. Microb. Cell Factories 2009, 8, 26. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, Y.; Lee, C.J.; Park, J.S. Strategies for optimizing the production of proteins and peptides with multiple disulfide bonds. Antibiotics 2020, 9, 541. [Google Scholar] [CrossRef] [PubMed]
- EL-Baky, N.A.; Linjawi, M.H.; Redwan, E.M. Auto-induction expression of human consensus interferon-alpha in Escherichia coli. BMC Biotechnol. 2015, 15, 14. [Google Scholar] [CrossRef] [Green Version]
- Swartz, J. Developing cell-free biology for industrial applications. J. Ind. Microbiol. Biotechnol. 2006, 33, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Zawada, J.F.; Yin, G.; Steiner, A.R.; Yang, J.; Naresh, A.; Roy, S.M.; Gold, D.S.; Heinsohn, H.G.; Murray, C.J. Microscale to manufacturing scale-up of cell-free cytokine production-a new approach for shortening protein production development timelines. Biotechnol. Bioeng. 2011, 108, 1570–1578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Narachi, M.A.; Davis, J.M.; Hsu, Y.R.; Arakawa, T. Role of single disulfide in recombinant human tumor necrosis factor-alpha. J. Biol. Chem. 1987, 262, 13107–13110. [Google Scholar] [CrossRef]




| Rare Codon Analysis | ||
|---|---|---|
| Index | Parameter | |
| Codon adaptation index (CAI) | 0.7 | 0.8–1.0 |
| Guanine-cytosine (GC) content | 68.93% | 30–70% |
| Codon frequency distribution (CFD) | Percentage of low frequency codons is 12% | <30% |
| Primer | Vector | Sequence |
|---|---|---|
| Forward | pCR 2.1-TOPO | 5′-GGATCCATGGTGCGTAGCAGCA-3′ |
| Forward | pET101/D-TOPO | 5′-CACCGGATCCATGGTGCGTAGC-3′ |
| Reverse | pCR 2.1-TOPO and pET101/D-TOPO | 5′-CTCGAGCAGCGCAATAATGCC-3′ |
| Purification Step | Total Protein Concentration (µg/mL) | Recovery (%) |
|---|---|---|
| Soluble fraction of CFPS reaction | 320.7 | 100 |
| Q-Sepharose | 160.3 | 49.98 |
| Heparin–Sepharose | 70.5 | 21.98 |
| Independent Variables | Levels | ||||
|---|---|---|---|---|---|
| −− | − | 0 | + | ++ | |
| A: Incubation temperature ( °C) | 20 | 25 | 30 | 35 | 40 |
| B: Incubation time (h) | 2 | 4 | 6 | 8 | 10 |
| Run Order | Independent Variables | Response Human TNF-α Concentration (µg/mL) | Standardized Residual | ||
|---|---|---|---|---|---|
| A | B | Actual | Predicted | ||
| 1 | 0 | 0 | 191.7 | 218.8 | −0.66 |
| 2 | ++ | 0 | 57.0 | 95.5 | −1.86 |
| 3 | 0 | 0 | 242.2 | 218.8 | 0.57 |
| 4 | − | + | 217.0 | 253.1 | −1.12 |
| 5 | 0 | ++ | 260.9 | 244.0 | 0.81 |
| 6 | 0 | 0 | 191.0 | 218.8 | −0.67 |
| 7 | 0 | 0 | 240.0 | 218.8 | 0.52 |
| 8 | − | − | 55.6 | 39.0 | 0.52 |
| 9 | −− | 0 | 7.2 | −12.1 | 0.93 |
| 10 | 0 | −− | 145.3 | 181.3 | −1.74 |
| 11 | 0 | 0 | 190.7 | 218.8 | −0.68 |
| 12 | + | + | 145.9 | 124.2 | 0.67 |
| 13 | + | − | 350.0 | 275.6 | 2.31 |
| Term | Coefficient | Standard Error of Coefficient (SE Coef) | T | P |
|---|---|---|---|---|
| Constant | −3242.42 | 551.14 | −5.88 | 0.001 |
| A | 166.48 | 26.56 | 6.27 | 0 |
| B | 286.61 | 73.84 | 3.88 | 0.006 |
| A × A | −1.77 | 0.38 | −4.68 | 0.002 |
| B × B | −0.38 | 2.36 | −0.16 | 0.876 |
| A × B | −9.14 | 2.26 | −4.04 | 0.005 |
| Source | Degree of Freedom (DF) | Sequential Sum of Squares (Seq SS) | Adjusted Sum of Squares (Adj SS) | Adjusted Mean Squares (Adj MS) | F | P |
|---|---|---|---|---|---|---|
| Regression | 5 | 93,002 | 93,002 | 18,600.4 | 9.09 | 0.006 |
| Linear | 2 | 11,638 | 80,774 | 40,387.2 | 19.73 | 0.001 |
| Square | 2 | 47,956 | 47,956 | 23,978.0 | 11.71 | 0.006 |
| Interaction | 1 | 33,409 | 33,409 | 33,408.5 | 16.32 | 0.005 |
| Residual error | 7 | 14,329 | 14,329 | 2047.0 | ||
| Lack of fit | 3 | 11,326 | 11,326 | 3775.4 | 5.03 | 0.076 |
| Pure error | 4 | 3003 | 3003 | 750.7 | ||
| Total | 12 | 107,331 |
| Independent Variables | Levels | Human TNF-α Concentration (µg/mL) Estimated by Quantitative ELISA | ||
|---|---|---|---|---|
| Basal | Optimum | Basal | Optimum | |
| Incubation temperature (°C) | 30 | 40 | 200 | 390 |
| Incubation time (h) | 6 | 2 | ||
| Human TNF-α Concentration (ng/mL) | Percentage Cell Viability |
|---|---|
| 250 | 99.99 ± 0.01 |
| 500 | 99.96 ± 0.003 |
| 750 | 99.37 ± 0.56 |
| 1000 | 89.53 ± 0.48 |
| 1500 | 77.45 ± 1.09 |
| Human TNF-α Concentration (ng/mL) | |
|---|---|
| IC50 | EC100 |
| 1728.24 ± 10.74 | 250.08 ± 0.62 |
| Human TNF-α Concentration (ng/mL) | Percentage Cytotoxicity | ||
|---|---|---|---|
| Caco-2 | HepG-2 | MCF-7 | |
| 40 | 31.50 ± 0.10 | 43.88 ± 0.14 | 26.82 ± 0.91 |
| 90 | 32.01 ± 0.60 | 44.17 ± 0.39 | 28.53 ± 0.94 |
| 180 | 34.28 ± 0.59 | 45.58 ± 0.29 | 34.20 ± 1.27 |
| 350 | 58.81 ± 1.56 | 60.86 ± 0.30 | 46.80 ± 0.80 |
| 700 | 62.33 ± 0.15 | 63.11 ± 0.97 | 61.82 ± 0.68 |
| Human TNF-α Concentration (ng/mL) | Human Cancer Cell Lines | ||
|---|---|---|---|
| Caco-2 | HepG-2 | MCF-7 | |
| IC50 | 297.6 ± 10 | 197.4 ± 0.05 | 373.9 ± 3.5 |
| EC100 | 17.71 ± 0.07 | 10 ± 0.02 | 10 ± 1.63 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-Baky, N.A.; EL-Fakharany, E.M.; Sabry, S.A.; El-Helow, E.R.; Redwan, E.M.; Sabry, A. A De Novo Optimized Cell-Free System for the Expression of Soluble and Active Human Tumor Necrosis Factor-Alpha. Biology 2022, 11, 157. https://doi.org/10.3390/biology11020157
El-Baky NA, EL-Fakharany EM, Sabry SA, El-Helow ER, Redwan EM, Sabry A. A De Novo Optimized Cell-Free System for the Expression of Soluble and Active Human Tumor Necrosis Factor-Alpha. Biology. 2022; 11(2):157. https://doi.org/10.3390/biology11020157
Chicago/Turabian StyleEl-Baky, Nawal Abd, Esmail M. EL-Fakharany, Soraya A. Sabry, Ehab R. El-Helow, Elrashdy Mustafa Redwan, and Amira Sabry. 2022. "A De Novo Optimized Cell-Free System for the Expression of Soluble and Active Human Tumor Necrosis Factor-Alpha" Biology 11, no. 2: 157. https://doi.org/10.3390/biology11020157
APA StyleEl-Baky, N. A., EL-Fakharany, E. M., Sabry, S. A., El-Helow, E. R., Redwan, E. M., & Sabry, A. (2022). A De Novo Optimized Cell-Free System for the Expression of Soluble and Active Human Tumor Necrosis Factor-Alpha. Biology, 11(2), 157. https://doi.org/10.3390/biology11020157








