Host-Specific Parasites Reveal the History and Biogeographical Contacts of Their Hosts: The Monogenea of Nearctic Cyprinoid Fishes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Parasite Collection
2.2. DNA Extraction, Amplification and Sequencing
2.3. Phylogenetic Reconstruction
3. Results
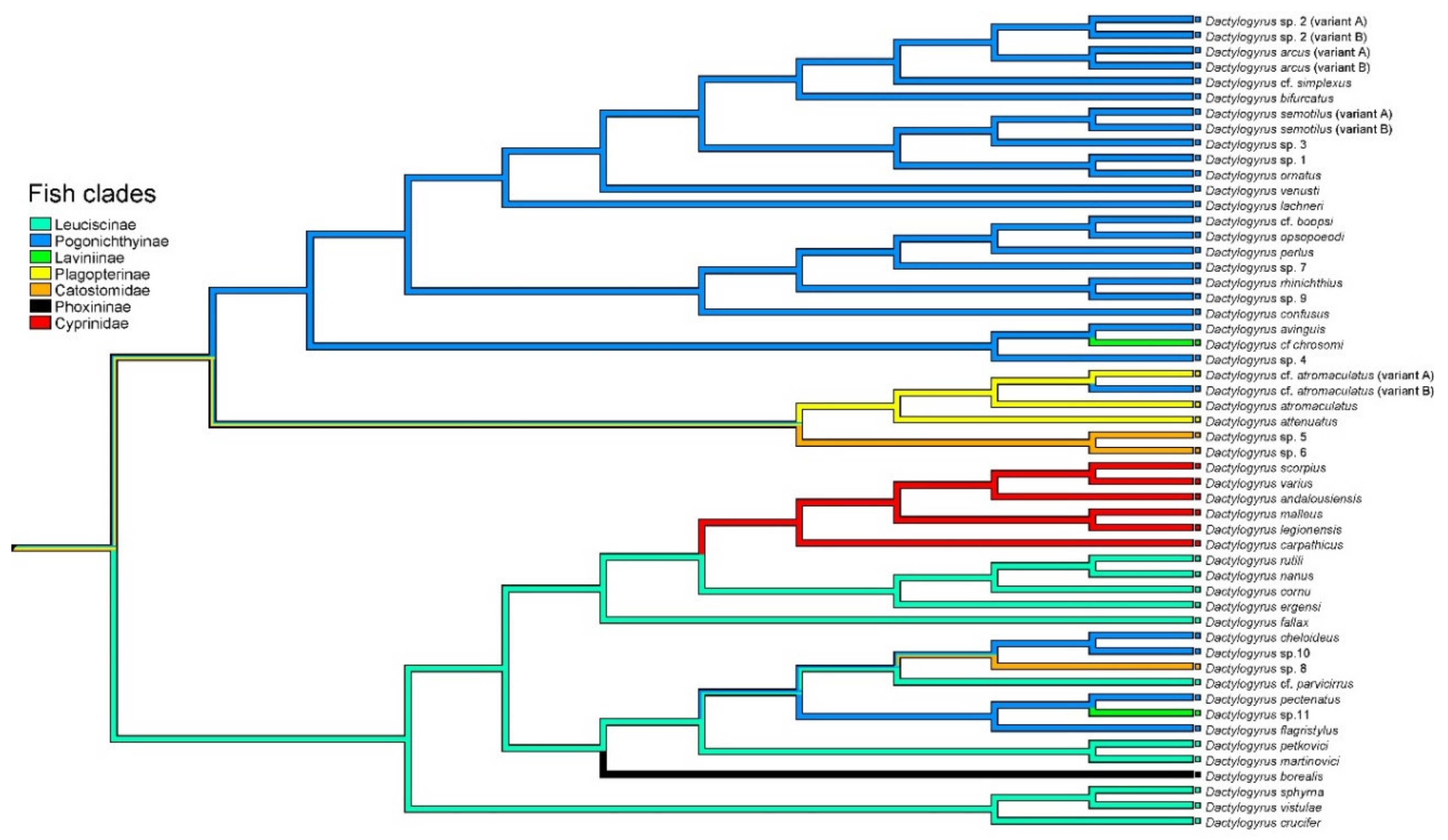
3.1. Dactylogyrus Species of Nearctic Cypriniform Fish
3.2. Phylogenetic Position of Neartic Dactylogyrus Species within the Dactylogyrus Phylogeny
3.3. Origin of Nearctic Dactylogyrus
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brooks, D.R.; Hoberg, E.P. Triage for the biosphere: The need and rationale for taxonomic inventories and phylogenetic studies and parasites. Comp. Parasitol. 2000, 67, 1–25. [Google Scholar]
- Hoberg, E.P.; Klassen, G.J. Revealing the faunal tapestry: Co-evolution and historical biogeography of hosts and parasites in marine systems. Parasitology 2002, 124, S3–S22. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.R.; McLennan, D.A. Parascript: Parasites and the Language of Evolution; Smithsonian Institution Press: Washington, DC, USA, 1993. [Google Scholar]
- Brooks, D.R.; McLennan, D.A. The Nature of Diversity: An Evolutionary Voyage of Discovery; University of Chicago Press: Chicago, IL, USA, 2002. [Google Scholar]
- Řehulková, E.; Seifertová, M.; Přikrylová, I.; Francová, K. Monogenea. In A Guide to the Parasites of African Freshwater Fishes; Scholz, T., Vanhove, M.P.M., Smit, N., Jayasundera, Z., Gelnar, M., Eds.; AbcTaxa: Brussels, Belgium, 2018; pp. 185–243. [Google Scholar]
- Rohde, K. A non-competitive mechanism responsible for restricting niches. Zool. Anz. 1977, 199, 164–172. [Google Scholar]
- Rohde, K. Simple ecological systems, simple solution to complex problems? Evol. Theor. 1989, 8, 305–350. [Google Scholar]
- Euzet, L.; Combes, C. The selection of habitats among the monogenea. Int. J. Parasitol. 1998, 28, 1645–1652. [Google Scholar] [CrossRef]
- Šimková, A.; Ondračková, M.; Gelnar, M.; Morand, S. Morphology and coexistence of congeneric ectoparasite species: Reinforcement of reproductive isolation? Biol. J. Linn. Soc. 2002, 76, 125–135. [Google Scholar] [CrossRef] [Green Version]
- Šimková, A.; Verneau, O.; Gelnar, M.; Morand, S. Specificity and specialization of congeneric monogeneans parasitizing cyprinid fish. Evolution 2006, 60, 1023–1037. [Google Scholar] [CrossRef]
- Pariselle, A.; Boeger, W.A.; Snoeks, J.; Bilong Bilong, C.F.; Morand, S.; Vanhove, M.P.M. The monogenean parasite fauna of cichlids: A potential tool for host biogeography. Int. J. Evol. Biol. 2011, 2011, 471480. [Google Scholar] [CrossRef] [Green Version]
- Benovics, M.; Desdevises, Y.; Vukić, J.; Šanda, R.; Šimková, A. The phylogenetic relationships and species richness of host-specific Dactylogyrus parasites shaped by the biogeography of Balkan cyprinids. Sci. Rep. 2018, 8, 13006. [Google Scholar] [CrossRef]
- Benovics, M.; Vukić, J.; Šanda, R.; Rahmouni, I.; Šimková, A. Disentangling the evolutionary history of peri-Mediterranean cyprinids using host-specific gill monogeneans. Int. J. Parasitol. 2020, 50, 969–984. [Google Scholar] [CrossRef]
- Benovics, M.; Desdevises, Y.; Šanda, R.; Vukić, J.; Scheifler, M.; Doadrio, I.; Sousa-Santos, C.; Šimková, A. High diversity of fish ectoparasitic monogeneans (Dactylogyrus) in the Iberian Peninsula: A case of adaptive radiation? Parasitology 2000, 147, 418–430. [Google Scholar] [CrossRef]
- Šimková, A.; Benovics, M.; Rahmouni, I.; Vukić, J. Host-specific Dactylogyrus parasites revealing new insights on the historical biogeography of Northwest African and Iberian cyprinid fish. Parasit. Vectors 2017, 10, 589. [Google Scholar] [CrossRef] [Green Version]
- Winfield, I.J.; Nelson, J.S. Cyprinid Fishes: Systematics, Biology and Exploitation; Fish and Fisheries Series No. 3; Chapman and Hall: London, UK, 1991. [Google Scholar]
- Gibson, D.I.; Timofeeva, T.A.; Gerasev, P.I. A catalogue of the nominal species of the monogenean genus Dactylogyrus Diesing, 1850 and their host genera. Syst. Parasitol. 1996, 35, 3–48. [Google Scholar] [CrossRef]
- Šimková, A.; Morand, S.; Jobet, E.; Gelnar, M.; Verneau, O. Molecular phylogeny of congeneric monogenean parasites (Dactylogyrus): A case of intrahost speciation. Evolution 2004, 58, 1001–1018. [Google Scholar] [CrossRef]
- Benovics, M.; Kičinjaová, M.L.; Šimková, A. The phylogenetic position of the enigmatic Balkan Aulopyge huegelii (Teleostei: Cyprinidae) from the perspective of host specific Dactylogyrus parasites (Monogenea), with a description of Dactylogyrus omenti n. sp. Parasit. Vectors 2017, 10, 547. [Google Scholar] [CrossRef] [Green Version]
- Kuchta, R.; Řehulková, E.; Francová, K.; Scholz, T.; Morand, S.; Šimková, A. Diversity of monogeneans and tapeworms in cypriniform fishes across two continents. Int. J. Parasitol. 2020, 50, 771–786. [Google Scholar] [CrossRef]
- Schönhuth, S.; Vukić, J.; Šanda, R.; Yang, L.; Mayden, R.L. Phylogenetic relationships and classification of the Holarctic family Leuciscidae (Cypriniformes: Cyprinoidei). Mol. Phyl. Evol. 2018, 127, 781–799. [Google Scholar] [CrossRef]
- Imoto, J.M.; Saitoh, K.; Sasaki, T.; Yonezawa, T.; Adachi, J.; Kartavtsev, Y.P.; Miya, M.; Nishida, M.; Hanzawa, N. Phylogeny and biogeography of highly diverged freshwater fish species (Leuciscinae, Cyprinidae, Teleostei) inferred from mitochondrial genome analysis. Gene 2013, 514, 112–124. [Google Scholar] [CrossRef]
- Banarescu, P. Some reconsiderations of the zoogeography of the euro-Mediterranean freshwater fish fauna. Rev. Romane Biol. Zool. 1973, 8, 257–264. [Google Scholar]
- Doadrio, I. Phylogenetic relationships and classification of western Palearctic species of the genus Barbus (Osteichthyes, Cyprinidae). Aquat. Living Resour. 1990, 3, 265–282. [Google Scholar] [CrossRef]
- Benovics, M.; Nejat, F.; Abdoli, A.; Šimková, A. Molecular and morphological phylogeny of host-specific Dactylogyrus parasites (Monogenea) sheds new light on the puzzling Middle Eastern origin of European and African lineages. Parasit. Vectors 2021, 14, 372. [Google Scholar] [CrossRef]
- Mizelle, J.D. Ectoparasites of the blunt-nosed minnow (Hyborhynchus notatus). Am. Midl. Nat. 1937, 18, 612–621. [Google Scholar] [CrossRef]
- Mizelle, J.D. New species of monogenetic flukes from Illinois fishes. Am. Midl. Nat. 1938, 19, 465–470. [Google Scholar] [CrossRef]
- Mueller, J.F. Additional species of North American Gyrodactyloidea (Trematoda). Am. Midl. Nat. 1938, 19, 220–235. [Google Scholar] [CrossRef]
- Seamster, A. Two new Dactylogyridae (Trematoda: Monogenea) from the golden shiner. J. Parasitol. 1948, 34, 111–113. [Google Scholar] [CrossRef]
- Mizelle, J.D.; Klucka, A.R. Studies on monogenetic trematodes XIV. Dactylogyridae from Wisconsin fishes. Am. Midl. Nat. 1953, 49, 720–733. [Google Scholar] [CrossRef]
- Monaco, L.H.; Mizelle, J.D. Studies on monogenetic trematodes XVII. The genus Dactylogyrus. Am. Midl. Nat. 1955, 53, 455–477. [Google Scholar] [CrossRef]
- Wood, R.A.; Mizelle, J.D. Studies on monogenetic trematodes. XXI. North American Gyrodactylinae, Dactylogyrinae and a new host record for Urocleidus dispar (Mueller, 1936). Am. Midl. Nat. 1957, 57, 183–202. [Google Scholar] [CrossRef]
- Rogers, W.A. Studies on Dactylogyrinae (Monogenea) with descriptions of 24 new species of Dactylogyrus, 5 new species of Pellucidhaptor, and the proposal of Aplodiscus gen. n. J. Parasitol. 1967, 53, 501–524. [Google Scholar] [CrossRef]
- Chien, S.-M. Dactylogyrids from North American cyprinids of the genus Nocomis. The reciprocus species group. J. Parasitol. 1971, 57, 1211–1214. [Google Scholar] [CrossRef]
- Chien, S.-M. Dactylogyrids from North American cyprinids of the genus Nocomis: The bellicus group. J. Parasitol. 1974, 60, 585–594. [Google Scholar] [CrossRef] [PubMed]
- Chien, S.-M. Dactylogyrids from North American cyprinids of the genus Nocomis: The Limulus and the Mollis groups. J. Parasitol. 1974, 60, 773–776. [Google Scholar] [CrossRef] [PubMed]
- Hanek, G.; Molnár, K.; Fernando, C.H. New and previously known Dactylogyrus spp. from southern Ontario fishes. J. Parasitol. 1975, 61, 421–426. [Google Scholar] [CrossRef]
- Mayes, M.A. New species of Gyrodactylus and Dactylogyrus (Trematoda: Monogenea) from fishes of Nebraska. J. Parasitol. 1977, 63, 805–809. [Google Scholar] [CrossRef] [PubMed]
- Cloutman, D.G. Dactylogyrus boopsi sp. n. (Monogenea: Dactylogyridae) from the bigeye shiner, Notropis boops Gilbert (Pisces: Cyprinidae). J. Helminthol. Soc. Washingt. 1994, 61, 219–220. [Google Scholar]
- Cloutman, D.G.; Rogers, W.A. Determination of the Dactylogyrus banghami complex (Monogenea: Dactylogyridae) from North American Gulf of Mexico coastal drainages with descriptions of three new species. Comp. Parasitol. 2005, 72, 10–16. [Google Scholar] [CrossRef]
- Sinnappah, N.D.; Lim, L.H.; Rohde, K.; Tinsley, R.; Combes, C.; Verneau, O. A Paedomorphic parasite associated with a neotenic amphibian host: Phylogenetic evidence suggests a revised systematic position for Sphyranuridae within anuran and turtle Polystomatoineans. Mol. Phyl. Evol. 2001, 18, 189–201. [Google Scholar] [CrossRef]
- Hassouna, N.; Michot, B.; Bachellerie, J.P. The complete nucleotide sequence of mouse 28S rRNA gene. Implications for the process of size increase of the large subunit rRNA in higher eukaryotes. Nucleic Acids Res. 1984, 12, 3563–3583. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [Green Version]
- Talavera, G.; Castresana, J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst. Biol. 2007, 56, 564–577. [Google Scholar] [CrossRef] [Green Version]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, L.T.; Schmidt, H.A.; Von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef]
- Trifinopoulos, J.; Nguyen, L.T.; von Haeseler, A.; Minh, B.Q. W-IQ-TREE: A fast online phylogenetic tool for maximum likelihood analysis. Nucleic Acids Res. 2016, 44, W232–W235. [Google Scholar] [CrossRef] [Green Version]
- Minh, B.Q.; Nguyen, M.A.T.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ronquist, F.; Teslenko, M.; Van Der Mark, P.; Ayres, D.L.; Darling, A.; Höhna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A.; Drummond, A.J.; Xie, D.; Baele, G.; Suchard, M.A. Posterior summarization in Bayesian phylogenetics using Tracer 1.7. Syst. Biol. 2018, 67, 901. [Google Scholar] [CrossRef] [Green Version]
- Rambaut, A. FigTree, Version 1.4.4; 2018. Available online: http://tree.bio.ed.ac.uk/software/figtree/ (accessed on 15 June 2021).
- Maddison, W.; Maddison, D. Mesquite 3.2. A Modular System for Evolutionary Analysis. 2019. Available online: http://mesquiteproject.org (accessed on 15 June 2021).
- Marincovich, L.; Gladenkov, A.Y. Evidence for an early opening of the Bering Strait. Nature 1999, 397, 149–151. [Google Scholar] [CrossRef]
- Marincovich, L.; Gladenkov, A.Y. New evidence for the age of Bering Strait. Quatr. Sci. Rev. 2001, 20, 20,329–335. [Google Scholar] [CrossRef]
- McKenna, M.C. Cenozoic paleontology of North Atlantic land bridges. In Structure and Development of the Greenland-Scotland Bridge: New Concepts and Methods; Bott, M.H.P., Saxov, S., Talwani, M., Thiede, J., Eds.; Plenum: New York, NY, USA, 1983. [Google Scholar]
- Tiffney, B.H. The Eocene North Atlantic land bridge: Its importance in tertiary and modern phytogeography of the northern hemisphere. J. Arnold Arbor. 1985, 66, 243–273. [Google Scholar] [CrossRef]
- Banarescu, P. The zoogeographical position of the East Asian freshwater fish fauna. Rev. Roumanie Biol. Ser. Zool. 1972, 17, 315–323. [Google Scholar]
- Briggs, J.C. Ostariophysan zoogeography: An alternative hypothesis. Copeia 1979, 1, 111–118. [Google Scholar] [CrossRef]
- Cavender, T.M. The fossil record of the Cyprinidae. In Cyprinid Fishes, Systematics, Biology and Exploitation; Winfield, I.J., Nelson, J.S., Eds.; Chapman and Hall: London, UK, 1991; pp. 34–54. [Google Scholar]
- Harris, P.M.; Hubbard, G.; Sandel, M. Catostomidae: Suckers. In Freshwater Fishes of North America: Volume 1: Petromyzontidae to Catostomidae; Warren, M.L., Burr, B.M., Eds.; Johns Hopkins University Press: Baltimore, MD, USA, 2014; pp. 451–502. [Google Scholar]
- Smith, G.R. Phylogeny and biogeography of the Catostomidae, freshwater fishes of North America and Asia. In Systematics, Historical Ecology, and North American Freshwater Fishes; Mayden, R.L., Ed.; Stanford University Press: Stanford, CA, USA, 1992; pp. 778–813. [Google Scholar]
- Bagley, J.C.; Mayden, R.L.; Harris, P.M. Phylogeny and divergence times of suckers (Cypriniformes: Catostomidae) inferred from Bayesian total evidence analyses of molecules, morphology, and fossils. Peer J. 2018, 6, e5168. [Google Scholar] [CrossRef]
- Cavender, T. Review of the fossil history of North American freshwater fishes. In The Zoogeography of North American Freshwater Fishes; Hocutt, C.H., Wiley, E.O., Eds.; John Wiley and Sons: New York, NY, USA, 1986; pp. 699–724. [Google Scholar]
- Froese, R.; Pauly, D. FishBase. World Wide Web Electronic Publication. Available online: www.fishbase.org (accessed on 15 June 2021).
- Raney, E.C.; Lachner, E.A. Age, growth, and habits of the hog sucker, Hypentelium nigricans (LeSueur). N. Y. Am. Midl. Nat. 1946, 36, 76–86. [Google Scholar] [CrossRef]




| Fish Species | Country | Locality | Body Water | Sample Size | Dactylogyrus Species |
|---|---|---|---|---|---|
| Campostoma spadiceum (Girard, 1856) | Arkansas | Polk County | Bear Creek | 11 | Dactylogyrus sp. 4 |
| Catostomus commersonii (Lacepède, 1803) | New York | Cooperstown | Oaks Creek | 12 | Dactylogyrus sp. 8 |
| Chrosomus neogaeus (Cope, 1867) | Wisconsin | Door County | Mink River | 13 | D. cf. chrosomi, Dactylogyrus sp. 11 |
| Clinostomus elongatus (Kirtland, 1840) | Wisconsin | Brown County | Baird Creek, Green Bay | 11 | D. confusus |
| Cyprinella venusta Girard, 1856 | Mississippi | Oxbow south of Cumbest Bridge landing | Pascagoula River | 13 | D. venusti |
| Mississippi | Moon Lake | Pascagoula River | 12 | Dactylogyrus sp. 7, Dactylogyrus sp. 2 variant B | |
| Cyprinella whipplei Girard, 1856 | Arkansas | Polk County | Caddo River | 2 | Dactylogyrus sp. 2 variant A |
| Hypentelium nigricans (Lesueur, 1817) | Arkansas | Montgomery County | Huddleston Creek | 3 | Dactylogyrus sp. 5, Dactylogyrus sp. 6 |
| Luxilus chrysocephalus Rafinesque, 1820 | Arkansas | Polk County | Caddo River | 3 | D. arcus variant A, D. semotilus variant A, Dactylogyrus sp. 1, Dactylogyrus sp. 3 |
| Arkansas | Polk County | Big Fork Creek | 3 | D. perlus | |
| Luxilus cornutus (Mitchill, 1817) | Wisconsin | Brown County | West Twin River | 10 | D. arcus variant B, D. semotilus variant B |
| Nocomis biguttatus (Kirtland, 1840) | Wisconsin | Brown County | West Twin River | 7 | D. avinguis, D. flagristylus, D. lachneri |
| Notemigonus crysoleucas (Mitchill, 1814) | New York | Cooperstown | Rom Hill Beaver Pond | 5 | D. cf. parvicirrus |
| Notropis petersoni Fowler, 1942 | Mississippi | Bluff Creek | 2 | D. ornatus | |
| Opsopoeodus emiliae Hay, 1881 | Mississippi | Bluff Creek | 1 | D. opsopoeodi | |
| Pimephales notatus (Rafinesque, 1820) | New York | Cooperstown | Leatherstocking Creek | 4 | D. cf. atromaculatus variant B |
| Arkansas | Polk County | Bear Creek | 2 | D. bifurcatus, D. cf. boopsi, D. cf. simplexus | |
| Pimephales promelas Rafinesque, 1820 | Wisconsin | Door County | Hickory Oak Pond | 7 | D. pectenatus |
| Rhinichthys atratulus (Hermann, 1804) | Wisconsin | Brown County | Baird Creek, Green Bay | 2 | D. cheloideus, D. rhinichthius |
| Rhinichthys cataractae (Valenciennes, 1842) | New York | Cooperstown | Leatherstocking Creek | 5 | Dactylogyrus sp. 9, Dactylogyrus sp. 10 |
| Semotilus atromaculatus (Mitchill, 1818) | Arkansas | Polk County | Big Fork Creek | 7 | D. cf. atromaculatus variant A |
| Wisconsin | Brown County | Baird Creek, Green Bay | 16 | D. atromaculatus, D. attenuatus |
| Dactylogyrus Species | Cypriniform Host Species | Cypriniform Family | Cypriniform Subfamily | Sampling Locality | GenBank Accession No. | |
|---|---|---|---|---|---|---|
| 28S rDNA | 18S rDNA and ITS1 | |||||
| Lineage I | ||||||
| D. doadrioi El Gharbi, Renaud & Lambert, 1993 | Luciobarbus guiraonis (Steindachner, 1866) | Cyprinidae | Barbinae | Spain | KY629346 | — |
| D. labei Musselius & Gusev, in Gusev, 1976 | Gibelion catla (Hamilton, 1822) | Cyprinidae | Labeoninae | India | JX566720 | — |
| D. mascomai El Gharbi, Renaud & Lambert, 1992 | Luciobarbus guiraonis (Steindachner, 1866) | Cyprinidae | Barbinae | Spain | KY629348 | — |
| D. quanfami Ha Ky, 1971 | Cirrhinus molitorella (Valenciennes, 1844) | Cyprinidae | Labeoninae | China | EF100536 | — |
| D. volutus El Gharbi, Birgi & Lambert, 1994 | Carasobarbus fritschii (Günther, 1874) | Cyprinidae | Torinae | Morocco | KY629353 | — |
| D. zatensis El Gharbi, Birgi & Lambert, 1994 | Carasobarbus fritschii | Cyprinidae | Torinae | Morocco | KY629352 | — |
| Lineage II | ||||||
| D. bicornis Malewitzkaja, 1941 | Rhodeus meridionalis Karaman, 1924 | Acheilognathidae | — | Greece | KY629345 | — |
| D. cryptomeres Bychowsky, 1934 | Gobio gobio (Linnaeus, 1758) | Gobionidae | — | Czech Republic | AJ969947 | — |
| D. hypophthalmichthys Akhmerov, 1952 | Hypophthalmichthys molitrix (Valenciennes, 1844) | Xenocyprididae | — | China | EF100532 | — |
| D. lamellatus Akhmerow, 1952 | Ctenopharyngodon idella (Valenciennes, 1844) | Xenocyprididae | — | China | AY307019 | — |
| D. parabramis Akhmerov, 1952 | Megalobrama terminalis (Richardson, 1846) | Xenocyprididae | — | China | EF100534 | — |
| D. petruschewskyi Gusev, 1955 | Megalobrama amblycephala Yih, 1955 | Xenocyprididae | — | China | AY548927 | — |
| Lineage III | ||||||
| D. anchoratus (Dujardin, 1845) Wagener, 1857 | Carassius gibelio (Bloch, 1782) | Cyprinidae | Cyprininae | Croatia | KY863555 | — |
| D. aspili Birgi & Lambert, 1987 | Enteromius macrops (Boulenger, 1911) | Cyprinidae | Smiliogastrinae | Senegal | KY629359 | — |
| D. extensus Mueller & Van Cleave, 1932 | Cyprinus carpio Linnaeus, 1758 | Cyprinidae | Cyprininae | Czech Republic | AJ969944 | — |
| D. marocanus El Gharbi, Birgi & Lambert, 1994 | Labeobarbus maroccanus (Günther, 1902) | Cyprininae | Torinae | Morocco | MW218579 | — |
| D. oligospirophallus Paperna, 1973 | Labeo coubie Rüppell, 1832 | Cyprinidae | Labeoninae | Senegal | KY629361 | — |
| D. senegalensis Paperna, 1969 | Labeo senegalensis Valenciennes, 1842 | Cyprinidae | Labeoninae | Senegal | KY629363 | — |
| D. titus Guegan, Lambert & Euzet, 1988 | Labeo senegalensis | Cyprinidae | Labeoninae | Senegal | KY629364 | — |
| D. vastator Nybelin, 1924 | Carassius gibelio (Bloch, 1782) | Cyprinidae | Cyprininae | Czech Republic | KY629366 | — |
| Lineage IV | ||||||
| Dactylogyrus andalousiensis El Gharbi, Renaud & Lambert, 1993 | Luciobarbus sclateri (Günther, 1868) | Cyprinidae | Barbinae | Portugal | KY629351 | KY629331 |
| Dactylogyrus borealis Nybelin, 1937 | Phoxinus bigerri Kottelat, 2007 | Leuciscidae | Phoxininae | Spain | MN338222 | MN365688 |
| Dactylogyrus carpathicus Zakhvatkin, 1951 | Barbus barbus (Linnaeus, 1758) | Cyprinidae | Barbinae | Czech Republic | KY201111 | KY201098 |
| Dactylogyrus cornu Linstow, 1878 | Vimba vimba (Linnaeus, 1758) | Leuciscidae | Leuciscinae | Czech Republic | KY629371 | KY629342 |
| Dactylogyrus crucifer Wagener, 1857 | Rutilus rutilus (Linnaeus, 1758) | Leuciscidae | Leuciscinae | Czech Republic | KY629374 | AJ564120 |
| Dactylogyrus ergensi Molnár, 1964 | Chondrostoma nasus (Linnaeus, 1758) | Leuciscidae | Leuciscinae | Greece | MG792989 | MG792874 |
| Dactylogyrus fallax Wagener, 1857 | Vimba vimba | Leuciscidae | Leuciscinae | Czech Republic | KY629370 | KY629341 |
| Dactylogyrus legionensis Gonzalez Lanza & Alvarez Pellitero, 1982 | Luciobarbus guiraonis (Steindachner, 1866) | Cyprinidae | Barbinae | Spain | KY629350 | KY629330 |
| Dactylogyrus malleus Linstow, 1877 | Barbus barbus | Cyprinidae | Barbinae | Czech Republic | KY201112 | KY201099 |
| Dactylogyrus martinovici Ergens, 1970 | Pachychilon pictum (Heckel & Kner, 1858) | Leuciscidae | Leuciscinae | Albania | MG793000 | MG792884 |
| Dactylogyrus nanus Dogiel & Bychowsky, 1934 | Rutilus rutilus | Leuciscidae | Leuciscinae | Czech Republic | AJ969942 | AJ564145 |
| Dactylogyrus petkovici Ergens, 1970 | Pachychilon pictum | Leuciscidae | Leuciscinae | Albania | MG793002 | MG792886 |
| Dactylogyrus rutili Gloser, 1965 | Leucos basak Heckel, 1843 | Leuciscidae | Leuciscinae | Albania | MG793020 | MG792904 |
| Dactylogyrus scorpius Rahmouni, Řehulková & Šimková, 2017 | Luciobarbus rifensis Doadrio, Casal-Lopez & Yahyaoui, 2015 | Cyprinidae | Barbinae | Morocco | KX553860 | KX578023 |
| Dactylogyrus sphyrna Linstow, 1878 | Rutilus rutilus | Leuciscidae | Leuciscinae | Czech Republic | AJ969943 | AJ564154 |
| Dactylogyrus varius Rahmouni, Řehulková & Šimková, 2017 | Luciobarbus maghrebensis Doadrio, Perea & Yahyaoui, 2015 | Cyprinidae | Barbinae | Morocco | KX553863 | KX578026 |
| Dactylogyrus vistulae Prost, 1957 | Squalius prespensis (Fowler, 1977) | Leuciscidae | Leuciscinae | Albania | KY629369 | KY629340 |
| D. arcus Rogers, 1967 (variant A) | Luxilus chrysocephalus Rafinesque, 1820 | Leuciscidae | Pogonichthyinae | Arkansas | OM108517 | OM108553 |
| D. arcus Rogers, 1967 (variant B) | Luxilus sp. | Leuciscidae | Pogonichthyinae | Wisconsin | OM108518 | OM108554 |
| D. atromaculatus Mizelle, 1938 | Semotilus atromaculatus | Leuciscidae | Plagopterinae | Wisconsin | OM108519 | OM108555 |
| D. cf. atromaculatus Mizelle, 1938 (variant A) | Semotilus atromaculatus (Mitchill, 1818) | Leuciscidae | Plagopterinae | Arkansas | OM108523 | OM108559 |
| D. cf. atromaculatus Mizelle, 1938 (variant B) | Pimephales notatus (Rafinesque, 1820) | Leuciscidae | Pogonichthyinae | New York | OM108524 | OM108560 |
| D. attenuatus Mizelle & Klucka, 1953 | Semotilus atromaculatus | Leuciscidae | Plagopterinae | Wisconsin | OM108520 | OM108556 |
| D. aviunguis Chien, 1974 | Nocomis biguttatus (Kirtland, 1840) | Leuciscidae | Pogonichthyinae | Wisconsin | OM108521 | OM108557 |
| D. bifurcatus Mizelle, 1937 | Pimephales notatus | Leuciscidae | Pogonichthyinae | Arkansas | OM108522 | OM108558 |
| D. cf. boopsi Cloutman, 1994 | Pimephales notatus | Leuciscidae | Pogonichthyinae | Arkansas | OM108525 | OM108561 |
| D. cheloideus Rogers, 1967 | Rhinichthys atratulus (Hermann, 1804) | Leuciscidae | Pogonichthyinae | Wisconsin | OM108531 | OM108567 |
| D. cf. chrosomi Hanek, Molnár & Fernando, 1975 | Chrosomus neogaeus (Cope, 1867) | Leuciscidae | Laviniinae | Wisconsin | OM108526 | OM108562 |
| D. confusus Mueller, 1938 | Clinostomus elongatus (Kirtland, 1840) | Leuciscidae | Pogonichthyinae | Wisconsin | OM108529 | OM108565 |
| D. flagristylus Chien, 1974 | Nocomis biguttatus | Leuciscidae | Pogonichthyinae | Wisconsin | OM108530 | OM108566 |
| D. lachneri Chien, 1971 | Nocomis biguttatus | Leuciscidae | Pogonichthyinae | Wisconsin | OM108532 | OM108568 |
| D. opsopoeodi Rogers, 1967 | Opsopoeodus emiliae Hay, 1881 | Leuciscidae | Pogonichthyinae | Mississippi | OM108533 | OM108569 |
| D. ornatus Rogers, 1967 | Notropis petersoni Fowler, 1942 | Leuciscidae | Pogonichthyinae | Mississippi | OM108534 | OM108570 |
| D. cf. parvicirrus Seamster, 1948 | Notemigonus crysoleucas (Mitchill, 1814) | Leuciscidae | Leuciscinae | New York | OM108527 | OM108563 |
| D. pectenatus Mayes, 1977 | Pimephales promelas Rafinesque, 1820 | Leuciscidae | Pogonichthyinae | Wisconsin | OM108535 | OM108571 |
| D. perlus Mueller, 1938 | Luxilus chrysocephalus | Leuciscidae | Pogonichthyinae | Arkansas | OM108536 | OM108572 |
| D. rhinichthius Wood & Mizelle, 1957 | Rhinichthys atratulus | Leuciscidae | Pogonichthyinae | Wisconsin | OM108537 | OM108573 |
| D. semotilus Wood & Mizelle, 1957 (variant A) | Luxilus chrysocephalus | Leuciscidae | Pogonichthyinae | Arkansas | OM108538 | OM108574 |
| D. semotilus Wood & Mizelle, 1957 (variant B) | Luxilus sp. | Leuciscidae | Pogonichthyinae | Wisconsin | OM108539 | OM108575 |
| D. cf. simplexus Monaco & Mizelle, 1955 | Pimephales notatus | Leuciscidae | Pogonichthyinae | Arkansas | OM108528 | OM108564 |
| D. venusti Rogers, 1967 | Cyprinella venusta Girard, 1856 | Leuciscidae | Pogonichthyinae | Mississippi | OM108552 | OM108588 |
| Dactylogyrus sp. 1 | Luxilus chrysocephalus | Leuciscidae | Pogonichthyinae | Arkansas | OM108540 | OM108576 |
| Dactylogyrus sp. 2 variant A | Cyprinella whipplei Girard, 1856 | Leuciscidae | Pogonichthyinae | Arkansas | OM108541 | OM108577 |
| Dactylogyrus sp. 2 variant B | Cyprinella venusta | Leuciscidae | Pogonichthyinae | Mississippi | OM108542 | OM108578 |
| Dactylogyrus sp. 3 | Luxilus chrysocephalus | Leuciscidae | Pogonichthyinae | Arkansas | OM108543 | OM108579 |
| Dactylogyrus sp. 4 | Campostoma spadiceum (Girard, 1856) | Leuciscidae | Pogonichthyinae | Arkansas | OM108544 | OM108580 |
| Dactylogyrus sp. 5 | Hypentelium nigricans (Lesueur, 1817) | Catostomidae | Catostominae | Arkansas | OM108545 | OM108581 |
| Dactylogyrus sp. 6 | Hypentelium nigricans | Catostomidae | Catostominae | Arkansas | OM108546 | OM108582 |
| Dactylogyrus sp. 7 | Cyprinella venusta | Leuciscidae | Pogonichthyinae | Mississippi | OM108547 | OM108583 |
| Dactylogyrus sp. 8 | Catostomus commersonii (Lacepède, 1803) | Catostomidae | Catostominae | New York | OM108548 | OM108584 |
| Dactylogyrus sp. 9 | Rhinichthys cataractae (Valenciennes, 1842) | Leuciscidae | Pogonichthyinae | New York | OM108549 | OM108585 |
| Dactylogyrus sp. 10 | Rhinichthys cataractae | Leuciscidae | Pogonichthyinae | New York | OM108550 | OM108586 |
| Dactylogyrus sp. 11 | Chrosomus neogaeus | Leuciscidae | Laviniinae | Wisconsin | OM108551 | OM108587 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Šimková, A.; Řehulková, E.; Choudhury, A.; Seifertová, M. Host-Specific Parasites Reveal the History and Biogeographical Contacts of Their Hosts: The Monogenea of Nearctic Cyprinoid Fishes. Biology 2022, 11, 229. https://doi.org/10.3390/biology11020229
Šimková A, Řehulková E, Choudhury A, Seifertová M. Host-Specific Parasites Reveal the History and Biogeographical Contacts of Their Hosts: The Monogenea of Nearctic Cyprinoid Fishes. Biology. 2022; 11(2):229. https://doi.org/10.3390/biology11020229
Chicago/Turabian StyleŠimková, Andrea, Eva Řehulková, Anindo Choudhury, and Mária Seifertová. 2022. "Host-Specific Parasites Reveal the History and Biogeographical Contacts of Their Hosts: The Monogenea of Nearctic Cyprinoid Fishes" Biology 11, no. 2: 229. https://doi.org/10.3390/biology11020229
APA StyleŠimková, A., Řehulková, E., Choudhury, A., & Seifertová, M. (2022). Host-Specific Parasites Reveal the History and Biogeographical Contacts of Their Hosts: The Monogenea of Nearctic Cyprinoid Fishes. Biology, 11(2), 229. https://doi.org/10.3390/biology11020229






