Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Structure Retrieval and Modeling
2.2. SANCDB Screening for Drug-Like Molecules
2.3. Computational Molecular Screening of the SANCDB
2.4. Protein–Ligand Complexes MD Simulation
2.5. Post-Simulation Validation of the Top Hits
2.5.1. The Binding Free Energy Calculations
2.5.2. Prediction of Bioactivity of the Top Hits
2.5.3. Determination of KD (Dissociation Constant)
3. Results and Discussion
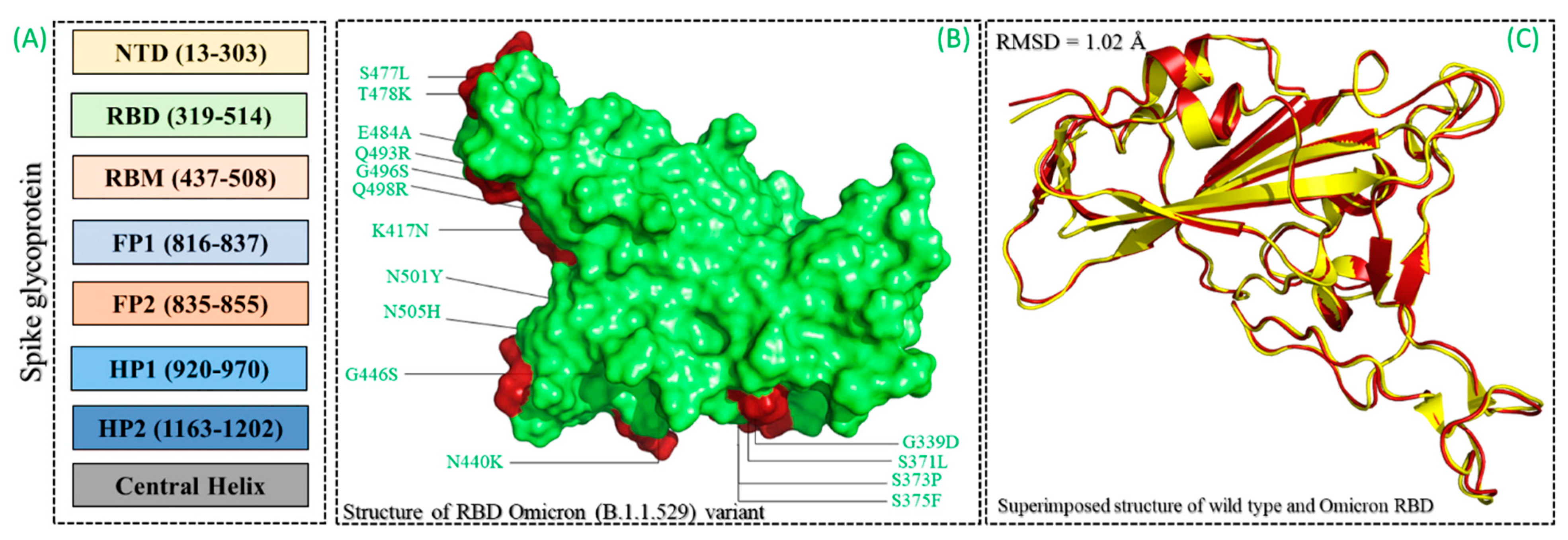
3.1. Structural Modeling and Screening
3.2. Binding Mode of 1,2,3,6-Tetragalloylglucose (SANC00944)
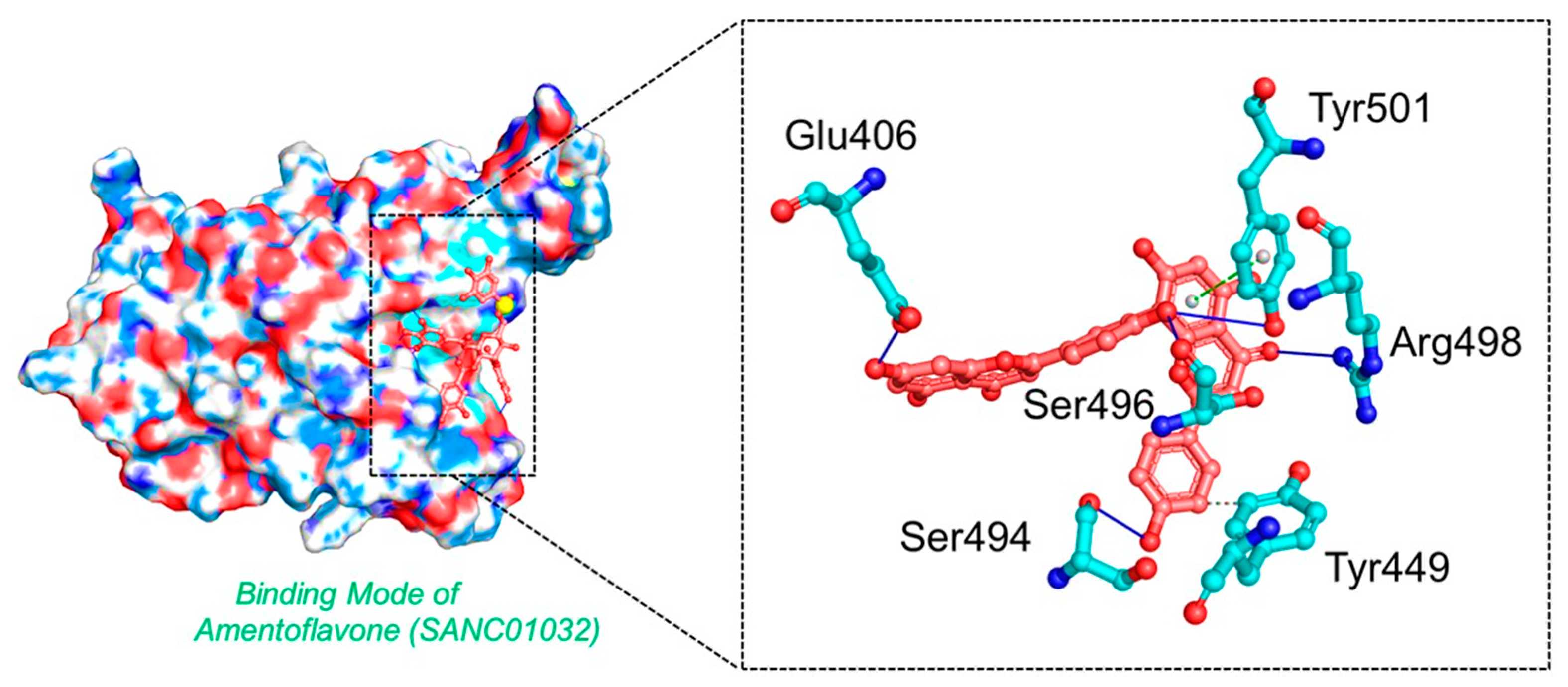
3.3. Binding Mode of Amentoflavone (SANC01032)
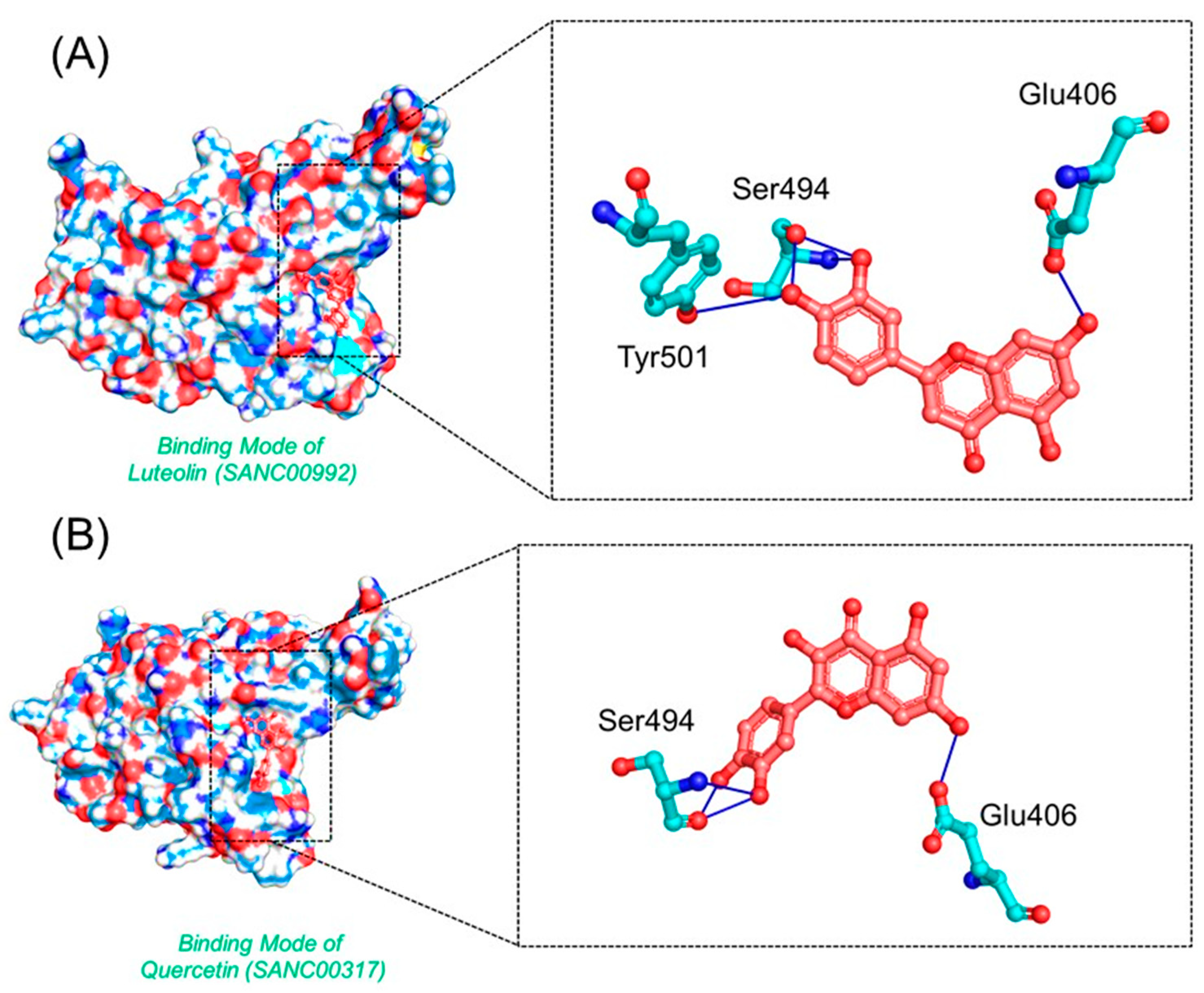
3.4. Binding Modes of Luteolin (SANC00992) and Quercetin (SANC00317)
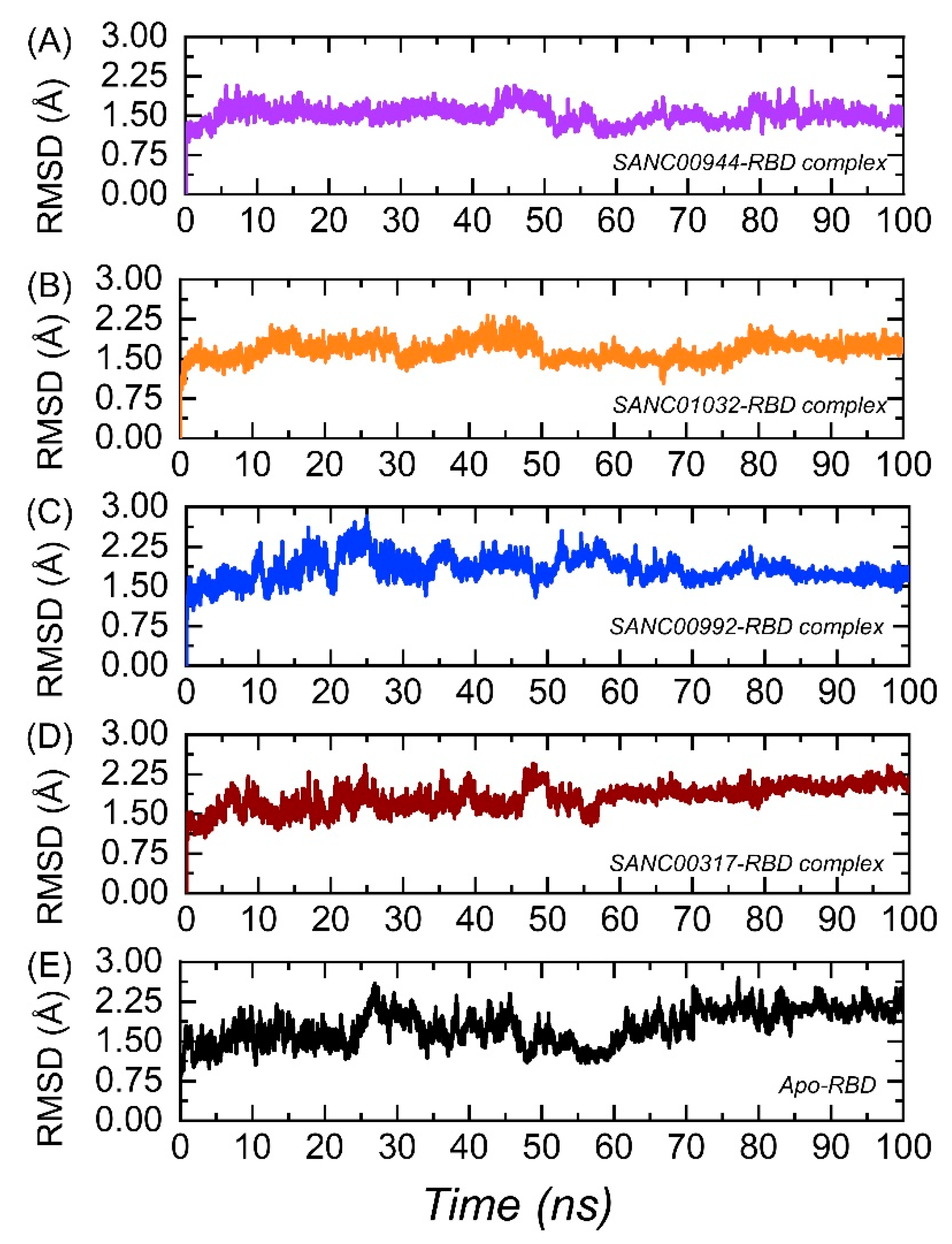
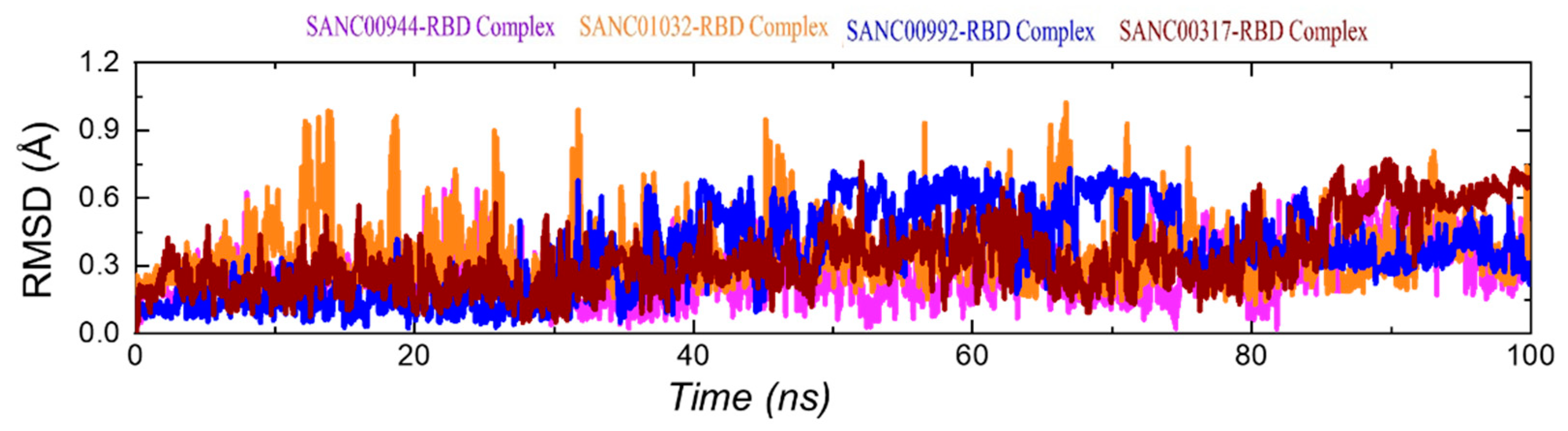
3.5. Dynamic Stability of the Top Hits
3.6. Structural Compactness Analysis of the Top Hits
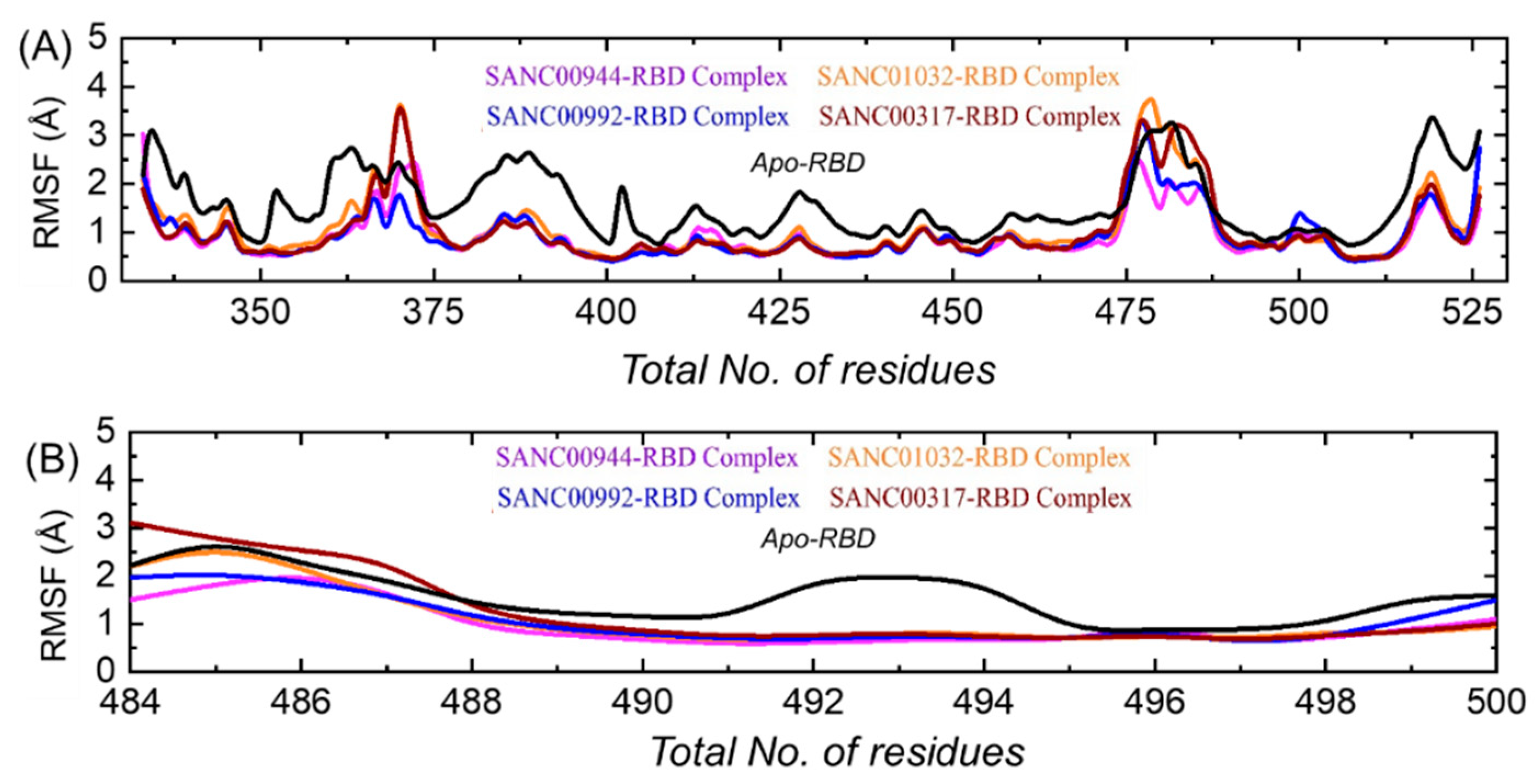
3.7. Residual Flexibility Analysis
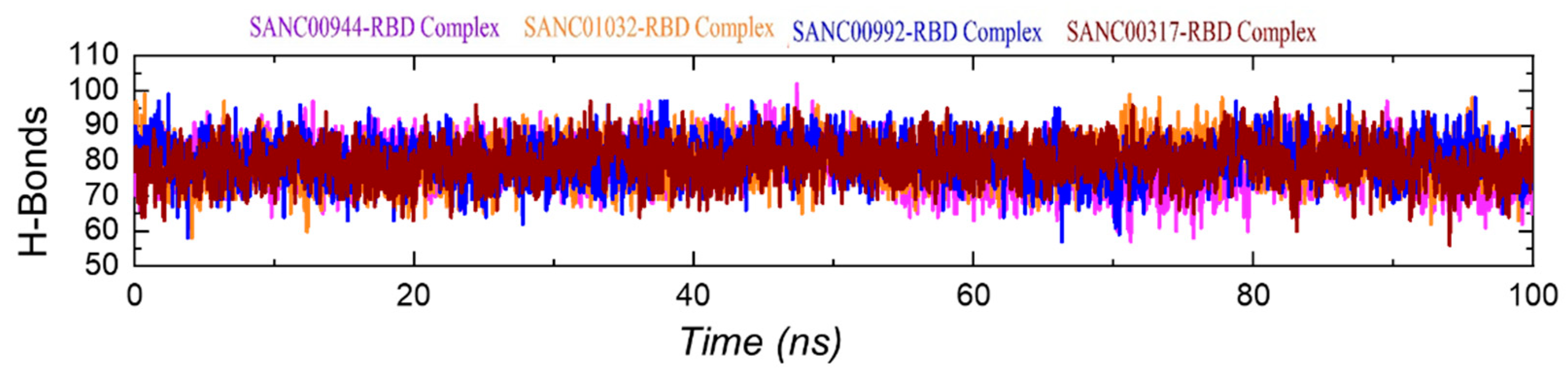
3.8. Hydrogen Bonding Analysis
3.9. Binding Free Energy Estimation
3.10. Bioactivity Prediction and Dissociation Constant (KD)
4. Conclusions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ul Qamar, M.T.; Alqahtani, S.M.; Alamri, M.A.; Chen, L.-L. Structural basis of SARS-CoV-2 3CLpro and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020, 10, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Hasöksüz, M.; Kiliç, S.; Saraç, F. Coronaviruses and SARS-COV-2. Turk. J. Med. Sci. 2020, 50, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Tahir ul Qamar, M.; Rehman, A.; Tusleem, K.; Ashfaq, U.A.; Qasim, M.; Zhu, X.; Fatima, I.; Shahid, F.; Chen, L.-L. Designing of a next generation multiepitope based vaccine (MEV) against SARS-COV-2: Immunoinformatics and in silico approaches. PLoS ONE 2020, 15, e0244176. [Google Scholar] [CrossRef] [PubMed]
- Harvey, W.T.; Carabelli, A.M.; Jackson, B.; Gupta, R.K.; Thomson, E.C.; Harrison, E.M.; Ludden, C.; Reeve, R.; Rambaut, A.; Peacock, S.J. SARS-CoV-2 variants, spike mutations and immune escape. Nat. Rev. Microbiol. 2021, 19, 409–424. [Google Scholar] [CrossRef]
- Abdool Karim, S.S.; de Oliveira, T. New SARS-CoV-2 variants—clinical, public health, and vaccine implications. N. Engl. J. Med. 2021, 384, 1866–1868. [Google Scholar] [CrossRef]
- Choi, J.Y.; Smith, D.M. SARS-CoV-2 Variants of Concern. Yonsei Med. J. 2021, 62, 961. [Google Scholar] [CrossRef]
- Muhseen, Z.T.; Hameed, A.R.; Al-Hasani, H.M.; ul Qamar, M.T.; Li, G. Promising terpenes as SARS-CoV-2 spike receptor-binding domain (RBD) attachment inhibitors to the human ACE2 receptor: Integrated computational approach. J. Mol. Liq. 2020, 320, 114493. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Shahid, F.; Aslam, S.; Ashfaq, U.A.; Aslam, S.; Fatima, I.; Fareed, M.M.; Zohaib, A.; Chen, L.-L. Reverse vaccinology assisted designing of multiepitope-based subunit vaccine against SARS-CoV-2. Infect. Dis. Poverty 2020, 9, 1–14. [Google Scholar] [CrossRef]
- Tortorici, M.A.; Veesler, D. Structural insights into coronavirus entry. Adv. Virus Res. 2019, 105, 93–116. [Google Scholar] [CrossRef]
- Tu, H.; Avenarius, M.R.; Kubatko, L.; Hunt, M.; Pan, X.; Ru, P.; Garee, J.; Thomas, K.; Mohler, P.; Pancholi, P.; et al. Distinct Patterns of Emergence of SARS-CoV-2 Spike Variants including N501Y in Clinical Samples in Columbus Ohio. bioRxiv 2021. [Google Scholar] [CrossRef]
- Walls, A.C.; Park, Y.-J.; Tortorici, M.A.; Wall, A.; McGuire, A.T.; Veesler, D. Structure, function, and antigenicity of the SARS-CoV-2 spike glycoprotein. Cell 2020, 181, 281–292.e286. [Google Scholar] [CrossRef]
- Khan, A.; Wei, D.-Q.; Kousar, K.; Abubaker, J.; Ahmad, S.; Ali, J.; Al-Mulla, F.; Ali, S.S.; Nizam-Uddin, N.; Sayaf, A.M. Preliminary Structural Data Revealed that the SARS-CoV-2 B. 1.617 Variant’s RBD binds to ACE2 receptor stronger than the Wild Type to Enhance the Infectivity. ChemBioChem 2021, 22, 2641–2649. [Google Scholar] [CrossRef]
- Khan, A.; Gui, J.; Ahmad, W.; Haq, I.; Shahid, M.; Khan, A.A.; Shah, A.; Khan, A.; Ali, L.; Anwar, Z.; et al. The SARS-CoV-2 B.1.618 variant slightly alters the spike RBD–ACE2 binding affinity and is an antibody escaping variant: A computational structural perspective. RSC Adv. 2021, 11, 30132–30147. [Google Scholar] [CrossRef]
- Khan, A.; Khan, T.; Ali, S.; Aftab, S.; Wang, Y.; Qiankun, W.; Khan, M.; Suleman, M.; Ali, S.; Heng, W. SARS-CoV-2 new variants: Characteristic features and impact on the efficacy of different vaccines. Biomed. Pharmacother. 2021, 143, 112176. [Google Scholar] [CrossRef] [PubMed]
- Khan, A.; Zia, T.; Suleman, M.; Khan, T.; Ali, S.S.; Abbasi, A.A.; Mohammad, A.; Wei, D.Q. Higher infectivity of the SARS-CoV-2 new variants is associated with K417N/T, E484K, and N501Y mutants: An insight from structural data. J. Cell Physiol. 2021, 236, 7045–7057. [Google Scholar] [CrossRef]
- Pulliam, J.R.; van Schalkwyk, C.; Govender, N.; von Gottberg, A.; Cohen, C.; Groome, M.J.; Dushoff, J.; Mlisana, K.; Moultrie, H. Increased risk of SARS-CoV-2 reinfection associated with emergence of the Omicron variant in South Africa. medRxiv 2021. [Google Scholar] [CrossRef]
- Wilhelm, A.; Widera, M.; Grikscheit, K.; Toptan, T.; Schenk, B.; Pallas, C.; Metzler, M.; Kohmer, N.; Hoehl, S.; Helfritz, F.A. Reduced neutralization of SARS-CoV-2 Omicron variant by vaccine sera and monoclonal antibodies. medRxiv 2021. [Google Scholar] [CrossRef]
- Roessler, A.; Riepler, L.; Bante, D.; von Laer, D.; Kimpel, J. SARS-CoV-2 B. 1.1. 529 variant (Omicron) evades neutralization by sera from vaccinated and convalescent individuals. medRxiv 2021. [Google Scholar] [CrossRef]
- Alamri, M.A.; Tahir Ul Qamar, M.; Mirza, M.U.; Bhadane, R.; Alqahtani, S.M.; Muneer, I.; Froeyen, M.; Salo-Ahen, O.M. Pharmacoinformatics and molecular dynamics simulation studies reveal potential covalent and FDA-approved inhibitors of SARS-CoV-2 main protease 3CLpro. J. Biomol. Struct. Dyn. 2021, 39, 4936–4948. [Google Scholar] [CrossRef]
- Alamri, M.A.; ul Qamar, M.T.; Mirza, M.U.; Alqahtani, S.M.; Froeyen, M.; Chen, L.-L. Discovery of human coronaviruses pan-papain-like protease inhibitors using computational approaches. J. Pharm. Anal. 2020, 10, 546–559. [Google Scholar] [CrossRef]
- Li, F. Structure, function, and evolution of coronavirus spike proteins. Annu. Rev. Virol. 2016, 3, 237–261. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, F.; Li, W.; Farzan, M.; Harrison, S.C. Structure of SARS coronavirus spike receptor-binding domain complexed with receptor. Science 2005, 309, 1864–1868. [Google Scholar] [CrossRef] [PubMed]
- Goddard, T.D.; Huang, C.C.; Ferrin, T.E. Software extensions to UCSF chimera for interactive visualization of large molecular assemblies. Structure 2005, 13, 473–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [Green Version]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Meng, E.C.; Couch, G.S.; Croll, T.I.; Morris, J.H.; Ferrin, T.E. UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 2021, 30, 70–82. [Google Scholar] [CrossRef]
- Song, W.; Gui, M.; Wang, X.; Xiang, Y. Cryo-EM structure of the SARS coronavirus spike glycoprotein in complex with its host cell receptor ACE2. PLoS Pathog. 2018, 14, e1007236. [Google Scholar] [CrossRef]
- Diallo, B.N.t.; Glenister, M.; Musyoka, T.M.; Lobb, K.; Tastan Bishop, Ö. SANCDB: An update on South African natural compounds and their readily available analogs. J. Cheminform. 2021, 13, 37. [Google Scholar] [CrossRef]
- Lagorce, D.; Bouslama, L.; Becot, J.; Miteva, M.A.; Villoutreix, B.O. FAF-Drugs4: Free ADME-tox filtering computations for chemical biology and early stages drug discovery. Bioinformatics 2017, 33, 3658–3660. [Google Scholar] [CrossRef] [Green Version]
- Dallakyan, S.; Olson, A.J. Small-Molecule Library Screening by Docking with PyRx. In Chemical Biology; Springer: Berlin/Heidelberg, Germany, 2015; pp. 243–250. [Google Scholar]
- Ravindranath, P.A.; Forli, S.; Goodsell, D.S.; Olson, A.J.; Sanner, M.F. AutoDockFR: Advances in Protein-Ligand Docking with Explicitly Specified Binding Site Flexibility. PLOS Comput. Biol. 2015, 11, e1004586. [Google Scholar] [CrossRef] [Green Version]
- Case, D.A.; Cheatham III, T.E.; Darden, T.; Gohlke, H.; Luo, R.; Merz Jr, K.M.; Onufriev, A.; Simmerling, C.; Wang, B.; Woods, R.J. The Amber biomolecular simulation programs. J. Comput. Chem. 2005, 26, 1668–1688. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, W.; Kollman, P.A.; Case, D.A. Antechamber: An accessory software package for molecular mechanical calculations. J. Am. Chem. Soc. 2001, 222, U403. [Google Scholar]
- Roe, D.R.; Cheatham, T.E., III. PTRAJ and CPPTRAJ: Software for processing and analysis of molecular dynamics trajectory data. J. Chem. Theory Comput. 2013, 9, 3084–3095. [Google Scholar] [CrossRef]
- Ahmad, F.; Albutti, A.; Tariq, M.H.; Din, G.; Tahir ul Qamar, M.; Ahmad, S. Discovery of Potential Antiviral Compounds against Hendra Virus by Targeting Its Receptor-Binding Protein (G) Using Computational Approaches. Molecules 2022, 27, 554. [Google Scholar] [CrossRef]
- Tahir ul Qamar, M.; Mirza, M.U.; Song, J.-M.; Rao, M.J.; Zhu, X.; Chen, L.-L. Probing the structural basis of Citrus phytochrome B using computational modelling and molecular dynamics simulation approaches. J. Mol. Liq. 2021, 340, 116895. [Google Scholar] [CrossRef]
- Muneer, I.; Ahmad, S.; Naz, A.; Abbasi, S.W.; Alblihy, A.; Aloliqi, A.A.; Alkhayl, F.F.; Alrumaihi, F.; Ahmad, S.; El Bakri, Y. Discovery of Novel Inhibitors from Medicinal Plants for V-Domain Ig Suppressor of T-Cell Activation (VISTA). Front. Mol. Biosci. 2021, 951. [Google Scholar] [CrossRef]
- ul Qamar, M.T.; Ahmad, S.; Khan, A.; Mirza, M.U.; Ahmad, S.; Abro, A.; Chen, L.-L.; Almatroudi, A.; Wei, D.-Q. Structural probing of HapR to identify potent phytochemicals to control Vibrio cholera through integrated computational approaches. Comput. Biol. Med. 2021, 138, 104929. [Google Scholar] [CrossRef]
- Altharawi, A.; Ahmad, S.; Alamri, M.A.; ul Qamar, M.T. Structural insight into the binding pattern and interaction mechanism of chemotherapeutic agents with Sorcin by docking and molecular dynamic simulation. Colloids Surf. B Biointerfaces 2021, 208, 112098. [Google Scholar] [CrossRef]
- Khan, A.; Ali, S.S.; Khan, M.T.; Saleem, S.; Ali, A.; Suleman, M.; Babar, Z.; Shafiq, A.; Khan, M.; Wei, D.-Q. Combined drug repurposing and virtual screening strategies with molecular dynamics simulation identified potent inhibitors for SARS-CoV-2 main protease (3CLpro). J. Biomol. Struct. Dyn. 2020, 1–12. [Google Scholar] [CrossRef]
- Khan, A.; Heng, W.; Wang, Y.; Qiu, J.; Wei, X.; Peng, S.; Saleem, S.; Khan, M.; Ali, S.S.; Wei, D.-Q. In silico and in vitro evaluation of kaempferol as a potential inhibitor of the SARS-CoV-2 main protease (3CLpro). Phytother. Res. PTR 2021, 35, 2841–2845. [Google Scholar] [CrossRef]
- Khan, A.; Kaushik, A.C.; Ali, S.S.; Ahmad, N.; Wei, D.-Q. Deep-learning-based target screening and similarity search for the predicted inhibitors of the pathways in Parkinson’s disease. RSC Adv. 2019, 9, 10326–10339. [Google Scholar] [CrossRef] [Green Version]
- Khan, A.; Khan, M.; Saleem, S.; Babar, Z.; Ali, A.; Khan, A.A.; Sardar, Z.; Hamayun, F.; Ali, S.S.; Wei, D.-Q. Phylogenetic analysis and structural perspectives of RNA-dependent RNA-polymerase inhibition from SARs-CoV-2 with natural products. Interdiscip. Sci. Comput. Life Sci. 2020, 12, 335–348. [Google Scholar] [CrossRef]
- Wang, Y.; Khan, A.; Chandra Kaushik, A.; Junaid, M.; Zhang, X.; Wei, D.-Q. The systematic modeling studies and free energy calculations of the phenazine compounds as anti-tuberculosis agents. J. Biomol. Struct. Dyn. 2019, 37, 4051–4069. [Google Scholar] [CrossRef] [PubMed]
- Xue, L.C.; Rodrigues, J.P.; Kastritis, P.L.; Bonvin, A.M.; Vangone, A. PRODIGY: A web server for predicting the binding affinity of protein–protein complexes. Bioinformatics 2016, 32, 3676–3678. [Google Scholar] [CrossRef] [PubMed]
- Vangone, A.; Schaarschmidt, J.; Koukos, P.; Geng, C.; Citro, N.; Trellet, M.E.; Xue, L.C.; Bonvin, A. Large-scale prediction of binding affinity in protein-small ligand complexes: The PRODIGY-LIG web server. Bioinformatics 2019, 35, 1585–1587. [Google Scholar] [CrossRef] [Green Version]
- Mehmood, I.; Ijaz, M.; Ahmad, S.; Ahmed, T.; Bari, A.; Abro, A.; Allemailem, K.S.; Almatroudi, A.; Tahir ul Qamar, M. SARS-CoV-2: An update on genomics, risk assessment, potential therapeutics and vaccine development. Int. J. Environ. Res. Public Health 2021, 18, 1626. [Google Scholar] [CrossRef] [PubMed]
- Lall, N.; Kishore, N.; Momtaz, S.; Hussein, A.; Naidoo, S.; Nqephe, M.; Crampton, B. Extract from Ceratonia siliqua Exhibits Depigmentation Properties. Phytother. Res. 2015, 29, 1729–1736. [Google Scholar] [CrossRef]
- Ayers, S.; Zink, D.L.; Mohn, K.; Powell, J.S.; Brown, C.M.; Murphy, T.; Brand, R.; Pretorius, S.; Stevenson, D.; Thompson, D.; et al. Flavones from Struthiola argentea with anthelmintic activity in vitro. Phytochemistry 2008, 69, 541–545. [Google Scholar] [CrossRef]
- Tshikalange, T.E.; Meyer, J.J.M.; Hussein, A.A. Antimicrobial activity, toxicity and the isolation of a bioactive compound from plants used to treat sexually transmitted diseases. J. Ethnopharmacol. 2005, 96, 515–519. [Google Scholar] [CrossRef]
- van Heerden, F.R.; Viljoen, A.M.; van Wyk, B.E. The major flavonoid of Dodonaea angustifolia. Fitoterapia 2000, 71, 602–604. [Google Scholar] [CrossRef]
- Adelekan, A.M.; Prozesky, E.A.; Hussein, A.A.; Ureña, L.D.; van Rooyen, P.H.; Liles, D.C.; Meyer, J.J.M.; Rodríguez, B. Bioactive Diterpenes and Other Constituents of Croton steenkampianus. J. Nat. Prod. 2008, 71, 1919–1922. [Google Scholar] [CrossRef]
- Sun, H.; Li, Y.; Tian, S.; Xu, L.; Hou, T. Assessing the performance of MM/PBSA and MM/GBSA methods. 4. Accuracies of MM/PBSA and MM/GBSA methodologies evaluated by various simulation protocols using PDBbind data set. Phys. Chem. Chem. Phys. 2014, 16, 16719–16729. [Google Scholar] [CrossRef] [PubMed]




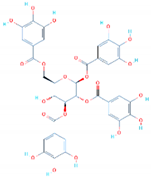
| Compound 2D Structure | Compound ID | Interactions | Docking Score (kcal/mol) |
|---|---|---|---|
 | SANC00944 | Tyr453, Leu455, Phe456, Arg403, Glu406, Glu406, Asn417, Tyr449, Tyr453, Arg493, Ser496, Ser496, | −9.35 |
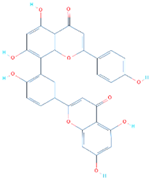 | SANC01032 | Tyr449, Glu406, Ser494, Ser496, Arg498, Tyr501 | −8.41 |
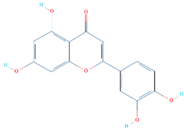 | SANC00992 | Glu406, Ser496, Ser496, Ser496, Tyr501 | −6.99 |
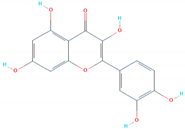 | SANC00317 | Glu406, Ser496, Ser496, Ser496 | −6.93 |
| Complexes Name | Glu406 | Asn417 | Ser446 | Ser494 | Arg498 | Asn501 |
|---|---|---|---|---|---|---|
| SANC00944–RBD | 1.8 Å | 2.3 Å | 3.1 Å | 2.96 Å | 3.45 Å | 2.56 Å |
| SANC01032–RBD | 2.16 Å | - | - | 2.46 Å | 3.52 Å | - |
| SANC00992–RBD | 2.03 Å | 2.14 Å | - | 2.88 Å | - | - |
| SANC00317–RBD | 2.93 Å | - | 3.58 Å | 2.64 Å | - | - |
| Complexes Name | vdW | Electrostatic | SA | GB | Total |
|---|---|---|---|---|---|
| SANC00944–RBD | −48.39 | −18.35 | −11.32 | 31.52 | −46.54 |
| SANC01032–RBD | −41.27 | −17.66 | −8.24 | 25.29 | −41.88 |
| SANC00992–RBD | −36.41 | −10.39 | −10.02 | 27.77 | −29.05 |
| SANC00317–RBD | −32.28 | −14.23 | −9.66 | 25.14 | −31.03 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hakami, A.R. Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach. Biology 2022, 11, 258. https://doi.org/10.3390/biology11020258
Hakami AR. Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach. Biology. 2022; 11(2):258. https://doi.org/10.3390/biology11020258
Chicago/Turabian StyleHakami, Abdulrahim R. 2022. "Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach" Biology 11, no. 2: 258. https://doi.org/10.3390/biology11020258
APA StyleHakami, A. R. (2022). Targeting the RBD of Omicron Variant (B.1.1.529) with Medicinal Phytocompounds to Abrogate the Binding of Spike Glycoprotein with the hACE2 Using Computational Molecular Search and Simulation Approach. Biology, 11(2), 258. https://doi.org/10.3390/biology11020258






