Specificities and Dynamics of Transposable Elements in Land Plants
Abstract
:Simple Summary
Abstract
1. Introduction
2. Plant TE Landscape
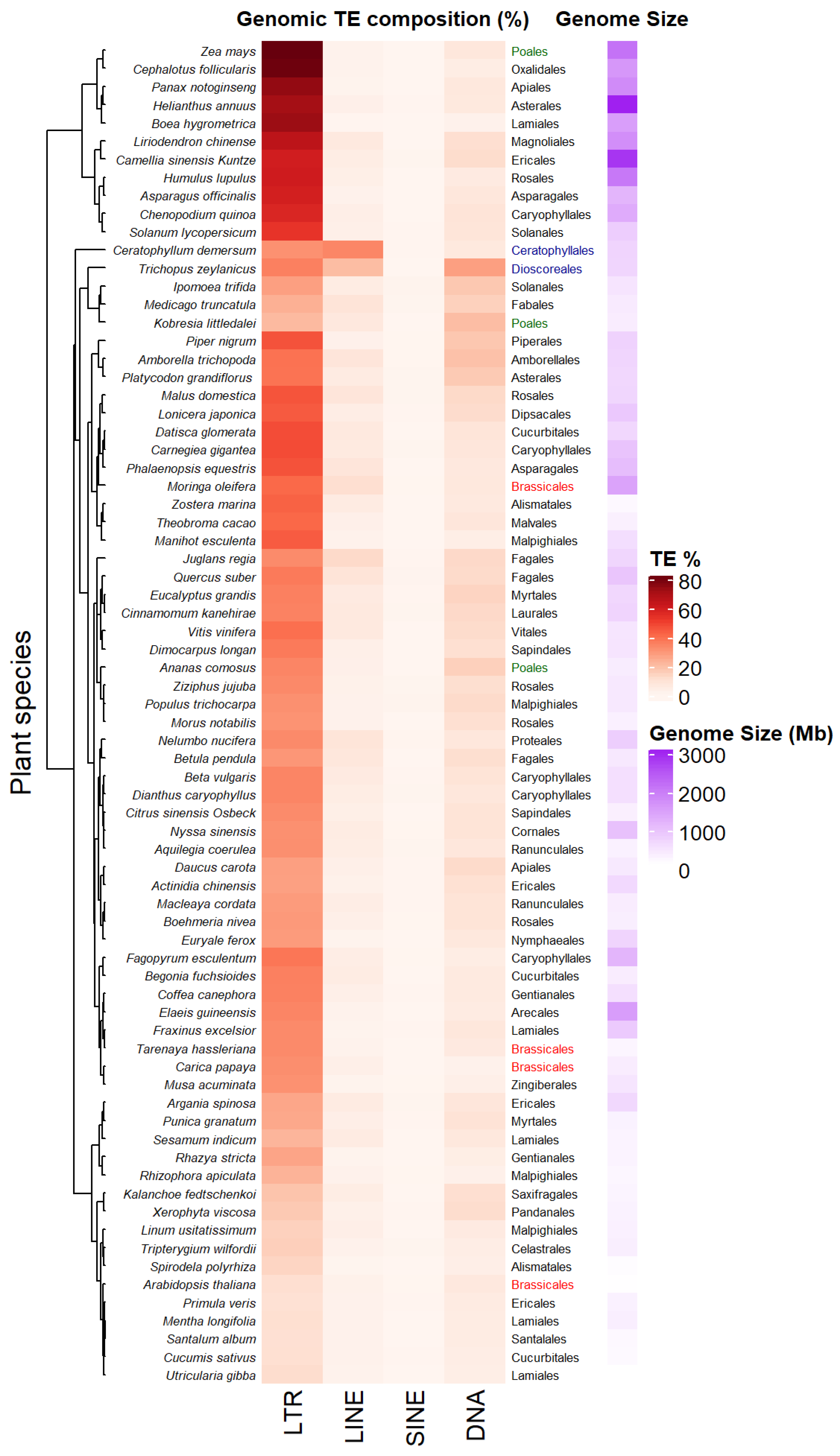
2.1. A Highly Variable TE Abundance
2.2. Challenging Evaluation of Plant TE Diversity and Classification
3. Regulating Factors of TE Transposition Control at the Molecular Level
3.1. TE Regulatory Motifs Involved in TE Transcription/Activation
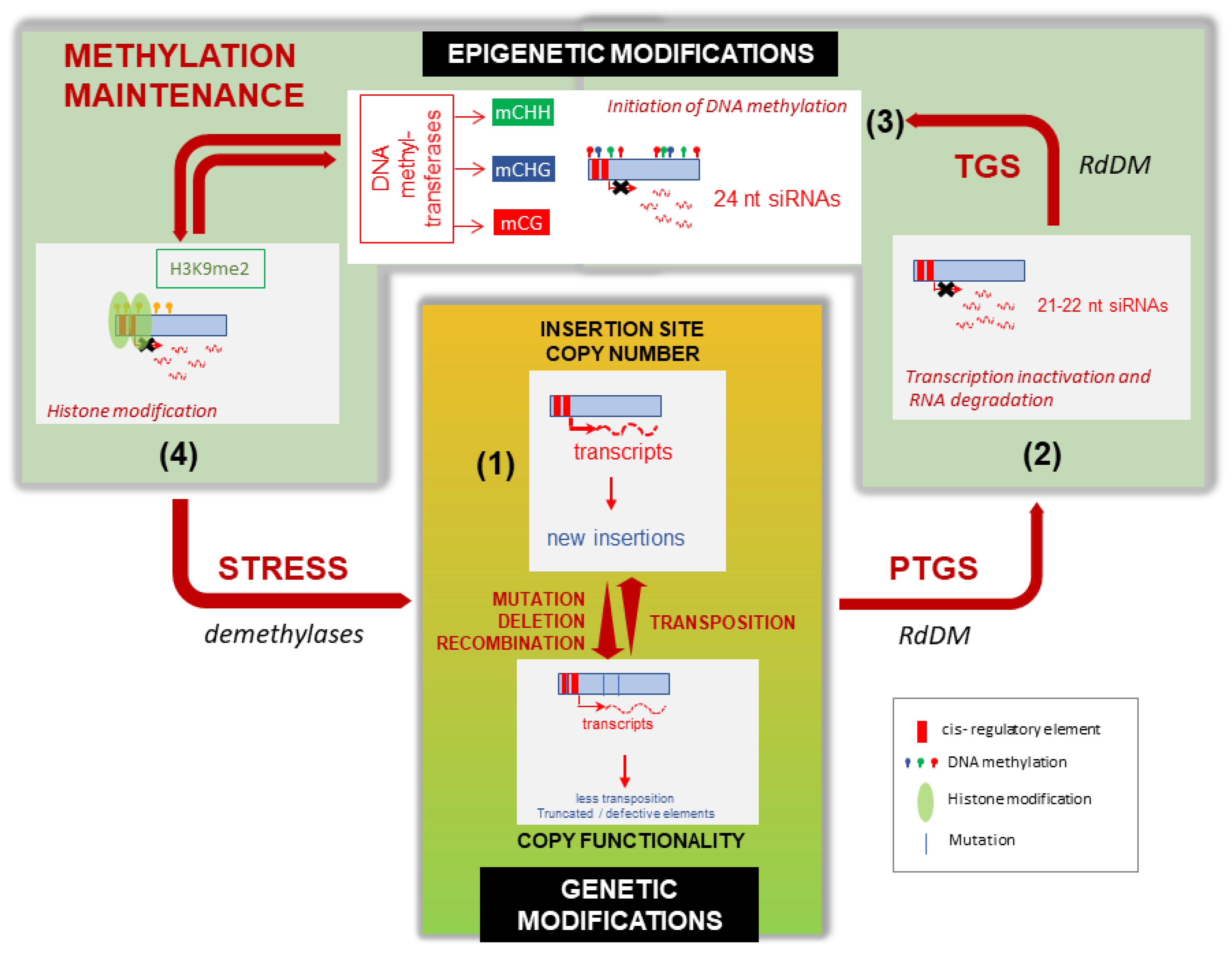
3.2. Epigenetic Control of TEs
3.3. Transposable Element Biology
3.4. Other Key-Players of Plant Genome Architecture
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- McClintock, B. Mutable Loci in Maize. Annu. Rep. Dir. Dep. Genet. Carnegie Inst. Wash. 1951, 50, 174–181. [Google Scholar]
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 Maize Genome: Complexity, Diversity, and Dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doolittle, W.F.; Sapienza, C. Selfish Genes, the Phenotype Paradigm and Genome Evolution. Nature 1980, 284, 601–603. [Google Scholar] [CrossRef] [PubMed]
- Orgel, L.E.; Crick, F.H.C. Selfish DNA: The Ultimate Parasite. Nature 1980, 284, 604–607. [Google Scholar] [CrossRef]
- Oliver, K.R.; Greene, W.K. Transposable Elements: Powerful Facilitators of Evolution. BioEssays 2009, 31, 703–714. [Google Scholar] [CrossRef]
- Kazazian, H.H. Mobile Elements: Drivers of Genome Evolution. Science 2004, 303, 1626–1632. [Google Scholar] [CrossRef] [Green Version]
- Biémont, C.; Vieira, C. Junk DNA as an Evolutionary Force. Nature 2006, 443, 521–524. [Google Scholar] [CrossRef]
- Hua-Van, A.; Le Rouzic, A.; Boutin, T.S.; Filée, J.; Capy, P. The Struggle for Life of the Genome’s Selfish Architects. Biol. Direct 2011, 6, 19. [Google Scholar] [CrossRef] [Green Version]
- Lisch, D. How Important Are Transposons for Plant Evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef]
- Schulman, A.H.; Wicker, T. A Field Guide to Transposable Elements. In Plant Transposons and Genome Dynamics in Evolution; Fedoroff, N.V., Ed.; Wiley-Blackwell: Oxford, UK, 2013; pp. 15–40. ISBN 978-1-118-50015-6. [Google Scholar]
- Bennetzen, J.L.; Wang, H. The Contributions of Transposable Elements to the Structure, Function, and Evolution of Plant Genomes. Annu. Rev. Plant Biol. 2014, 65, 505–530. [Google Scholar] [CrossRef]
- Ibarra-Laclette, E.; Lyons, E.; Hernández-Guzmán, G.; Pérez-Torres, C.A.; Carretero-Paulet, L.; Chang, T.-H.; Lan, T.; Welch, A.J.; Juárez, M.J.A.; Simpson, J.; et al. Architecture and Evolution of a Minute Plant Genome. Nature 2013, 498, 94–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wicker, T.; Gundlach, H.; Spannagl, M.; Uauy, C.; Borrill, P.; Ramírez-González, R.H.; De Oliveira, R.; International Wheat Genome Sequencing Consortium; Mayer, K.F.X.; Paux, E.; et al. Impact of Transposable Elements on Genome Structure and Evolution in Bread Wheat. Genome Biol. 2018, 19, 103. [Google Scholar] [CrossRef] [PubMed]
- Döring, H.P.; Tillmann, E.; Starlinger, P. DNA Sequence of the Maize Transposable Element Dissociation. Nature 1984, 307, 127–130. [Google Scholar] [CrossRef] [PubMed]
- Pohlman, R.F.; Fedoroff, N.V.; Messing, J. The Nucleotide Sequence of the Maize Controlling Element Activator. Cell 1984, 37, 635–643. [Google Scholar] [CrossRef]
- Wicker, T.; Sabot, F.; Hua-Van, A.; Bennetzen, J.L.; Capy, P.; Chalhoub, B.; Flavell, A.; Leroy, P.; Morgante, M.; Panaud, O.; et al. A Unified Classification System for Eukaryotic Transposable Elements. Nat. Rev. Genet. 2007, 8, 973–982. [Google Scholar] [CrossRef] [PubMed]
- Finnegan, D.J. Eukaryotic Transposable Elements and Genome Evolution. Trends Genet. 1989, 5, 103–107. [Google Scholar] [CrossRef]
- Bourque, G.; Burns, K.H.; Gehring, M.; Gorbunova, V.; Seluanov, A.; Hammell, M.; Imbeault, M.; Izsvák, Z.; Levin, H.L.; Macfarlan, T.S.; et al. Ten Things You Should Know about Transposable Elements. Genome Biol. 2018, 19, 199. [Google Scholar] [CrossRef]
- Wells, J.N.; Feschotte, C. A Field Guide to Eukaryotic Transposable Elements. Annu. Rev. Genet. 2020, 54, 539–561. [Google Scholar] [CrossRef]
- Neumann, P.; Novák, P.; Hoštáková, N.; Macas, J. Systematic Survey of Plant LTR-Retrotransposons Elucidates Phylogenetic Relationships of Their Polyprotein Domains and Provides a Reference for Element Classification. Mob. DNA 2019, 10, 1. [Google Scholar] [CrossRef]
- Jiang, N.; Jordan, I.K.; Wessler, S.R. Dasheng and RIRE2. A Nonautonomous Long Terminal Repeat Element and Its Putative Autonomous Partner in the Rice Genome. Plant Physiol. 2002, 130, 1697–1705. [Google Scholar] [CrossRef] [Green Version]
- Wicker, T.; Guyot, R.; Yahiaoui, N.; Keller, B. CACTA Transposons in Triticeae. A Diverse Family of High-Copy Repetitive Elements. Plant Physiol. 2003, 132, 52–63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stritt, C.; Thieme, M.; Roulin, A.C. Rare Transposable Elements Challenge the Prevailing View of Transposition Dynamics in Plants. Am. J. Bot. 2021, 108, 1310–1314. [Google Scholar] [CrossRef] [PubMed]
- Arkhipova, I.R. Distribution and Phylogeny of Penelope-Like Elements in Eukaryotes. Syst. Biol. 2006, 55, 875–885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craig, R.J.; Yushenova, I.A.; Rodriguez, F.; Arkhipova, I.R. An Ancient Clade of Penelope-Like Retroelements with Permuted Domains Is Present in the Green Lineage and Protists, and Dominates Many Invertebrate Genomes. Mol. Biol. Evol. 2021, 38, 5005–5020. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Faridi, N.; Casola, C. An Ancient Transkingdom Horizontal Transfer of Penelope -like Retroelements from Arthropods to Conifers. Genome Biol. Evol. 2016, 8, 1252–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heitkam, T.; Holtgräwe, D.; Dohm, J.C.; Minoche, A.E.; Himmelbauer, H.; Weisshaar, B.; Schmidt, T. Profiling of Extensively Diversified Plant LINEs Reveals Distinct Plant-Specific Subclades. Plant J. 2014, 79, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Wenke, T.; Döbel, T.; Sörensen, T.R.; Junghans, H.; Weisshaar, B.; Schmidt, T. Targeted Identification of Short Interspersed Nuclear Element Families Shows Their Widespread Existence and Extreme Heterogeneity in Plant Genomes. Plant Cell 2011, 23, 3117–3128. [Google Scholar] [CrossRef] [Green Version]
- Seibt, K.M.; Schmidt, T.; Heitkam, T. The Conserved 3′ Angio-domain Defines a Superfamily of Short Interspersed Nuclear Elements (SINEs) in Higher Plants. Plant J. 2020, 101, 681–699. [Google Scholar] [CrossRef]
- Bao, W.; Jurka, M.G.; Kapitonov, V.V.; Jurka, J. New Superfamilies of Eukaryotic DNA Transposons and Their Internal Divisions. Mol. Biol. Evol. 2009, 26, 983–993. [Google Scholar] [CrossRef] [Green Version]
- De Assis, R.; Baba, V.Y.; Cintra, L.A.; Gonçalves, L.S.A.; Rodrigues, R.; Vanzela, A.L.L. Genome Relationships and LTR-Retrotransposon Diversity in Three Cultivated Capsicum L. (Solanaceae) Species. BMC Genom. 2020, 21, 237. [Google Scholar] [CrossRef] [Green Version]
- Feschotte, C.; Pritham, E.J. DNA Transposons and the Evolution of Eukaryotic Genomes. Annu. Rev. Genet. 2007, 41, 331–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapitonov, V.V.; Jurka, J. Helitrons on a Roll: Eukaryotic Rolling-Circle Transposons. Trends Genet. 2007, 23, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Piednoël, M.; Gonçalves, I.R.; Higuet, D.; Bonnivard, E. Eukaryote DIRS1-like Retrotransposons: An Overview. BMC Genom. 2011, 12, 621. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, D.; Zheng, Z.; Li, Y.; Hu, H.; Wang, Z.; Du, X.; Zhang, S.; Zhu, M.; Dong, L.; Ren, G.; et al. Which Factors Contribute Most to Genome Size Variation within Angiosperms? Ecol. Evol. 2021, 11, 2660–2668. [Google Scholar] [CrossRef]
- Piegu, B.; Guyot, R.; Picault, N.; Roulin, A.; Saniyal, A.; Kim, H.; Collura, K.; Brar, D.S.; Jackson, S.; Wing, R.A.; et al. Doubling Genome Size without Polyploidization: Dynamics of Retrotransposition-Driven Genomic Expansions in Oryza Australiensis, a Wild Relative of Rice. Genome Res. 2006, 16, 1262–1269. [Google Scholar] [CrossRef] [Green Version]
- Macas, J.; Novák, P.; Pellicer, J.; Čížková, J.; Koblížková, A.; Neumann, P.; Fuková, I.; Doležel, J.; Kelly, L.J.; Leitch, I.J. In Depth Characterization of Repetitive DNA in 23 Plant Genomes Reveals Sources of Genome Size Variation in the Legume Tribe Fabeae. PLoS ONE 2015, 10, e0143424. [Google Scholar] [CrossRef]
- Hawkins, J.S.; Kim, H.; Nason, J.D.; Wing, R.A.; Wendel, J.F. Differential Lineage-Specific Amplification of Transposable Elements Is Responsible for Genome Size Variation in Gossypium. Genome Res. 2006, 16, 1252–1261. [Google Scholar] [CrossRef] [Green Version]
- Bertioli, D.J.; Cannon, S.B.; Froenicke, L.; Huang, G.; Farmer, A.D.; Cannon, E.K.S.; Liu, X.; Gao, D.; Clevenger, J.; Dash, S.; et al. The Genome Sequences of Arachis Duranensis and Arachis Ipaensis, the Diploid Ancestors of Cultivated Peanut. Nat. Genet. 2016, 48, 438–446. [Google Scholar] [CrossRef]
- Chuong, E.B.; Elde, N.C.; Feschotte, C. Regulatory Activities of Transposable Elements: From Conflicts to Benefits. Nat. Rev. Genet. 2017, 18, 71–86. [Google Scholar] [CrossRef] [Green Version]
- Vicient, C.M. Transcriptional Activity of Transposable Elements in Maize. BMC Genom. 2010, 11, 601. [Google Scholar] [CrossRef] [Green Version]
- Johns, M.A.; Mottinger, J.; Freeling, M. A Low Copy Number, Copia-like Transposon in Maize. EMBO J. 1985, 4, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Grandbastien, M.-A. LTR Retrotransposons, Handy Hitchhikers of Plant Regulation and Stress Response. Biochim. Biophys. Acta BBA Gene Regul. Mech. 2015, 1849, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Naito, K.; Zhang, F.; Tsukiyama, T.; Saito, H.; Hancock, C.N.; Richardson, A.O.; Okumoto, Y.; Tanisaka, T.; Wessler, S.R. Unexpected Consequences of a Sudden and Massive Transposon Amplification on Rice Gene Expression. Nature 2009, 461, 1130–1134. [Google Scholar] [CrossRef] [PubMed]
- Orozco-Arias, S.; Isaza, G.; Guyot, R. Retrotransposons in Plant Genomes: Structure, Identification, and Classification through Bioinformatics and Machine Learning. Int. J. Mol. Sci. 2019, 20, 3837. [Google Scholar] [CrossRef] [Green Version]
- Casacuberta, J.M.; Grandbastien, M.-A. Characterisation of LTR Sequences Involved in the Protoplast Specific Expression of the Tobacco Tntl Retrotransposon. Nucleic Acids Res. 1993, 21, 2087–2093. [Google Scholar] [CrossRef] [Green Version]
- Takeda, S.; Sugimoto, K.; Otsuki, H.; Hirochika, H. A 13-Bp Cis-Regulatory Element in the LTR Promoter of the Tobacco Retrotransposon Tto1 Is Involved in Responsiveness to Tissue Culture, Wounding, Methyl Jasmonate and Fungal Elicitors. Plant J. 1999, 18, 383–393. [Google Scholar] [CrossRef]
- Grandbastien, M.-A.; Audeon, C.; Bonnivard, E.; Casacuberta, J.M.; Chalhoub, B.; Costa, A.-P.P.; Le, Q.H.; Melayah, D.; Petit, M.; Poncet, C.; et al. Stress Activation and Genomic Impact of Tnt1 Retrotransposons in Solanaceae. Cytogenet. Genome Res. 2005, 110, 229–241. [Google Scholar] [CrossRef]
- Galindo-González, L.; Mhiri, C.; Deyholos, M.K.; Grandbastien, M.-A. LTR-Retrotransposons in Plants: Engines of Evolution. Gene 2017, 626, 14–25. [Google Scholar] [CrossRef]
- Beguiristain, T.; Grandbastien, M.-A.; Puigdomènech, P.; Casacuberta, J.M. Three Tnt1 Subfamilies Show Different Stress-Associated Patterns of Expression in Tobacco. Consequences for Retrotransposon Control and Evolution in Plants. Plant Physiol. 2001, 127, 212–221. [Google Scholar] [CrossRef] [Green Version]
- Lisch, D. Epigenetic Regulation of Transposable Elements in Plants. Annu. Rev. Plant Biol. 2009, 60, 43–66. [Google Scholar] [CrossRef] [Green Version]
- Lanciano, S.; Mirouze, M. Transposable Elements: All Mobile, All Different, Some Stress Responsive, Some Adaptive? Curr. Opin. Genet. Dev. 2018, 49, 106–114. [Google Scholar] [CrossRef] [PubMed]
- Sigman, M.J.; Slotkin, R.K. The First Rule of Plant Transposable Element Silencing: Location, Location, Location. Plant Cell 2016, 28, 304–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Lang, Z.; Zhu, J.-K. Dynamics and Function of DNA Methylation in Plants. Nat. Rev. Mol. Cell Biol. 2018, 19, 489–506. [Google Scholar] [CrossRef] [PubMed]
- Borges, F.; Martienssen, R.A. The Expanding World of Small RNAs in Plants. Nat. Rev. Mol. Cell Biol. 2015, 16, 727–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fultz, D.; Choudury, S.G.; Slotkin, R.K. Silencing of Active Transposable Elements in Plants. Curr. Opin. Plant Biol. 2015, 27, 67–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matzke, M.A.; Mosher, R.A. RNA-Directed DNA Methylation: An Epigenetic Pathway of Increasing Complexity. Nat. Rev. Genet. 2014, 15, 394–408. [Google Scholar] [CrossRef]
- Hernández-Pinzón, I.; Cifuentes, M.; Hénaff, E.; Santiago, N.; Espinás, M.L.; Casacuberta, J.M. The Tnt1 Retrotransposon Escapes Silencing in Tobacco, Its Natural Host. PLoS ONE 2012, 7, e33816. [Google Scholar] [CrossRef] [Green Version]
- Ito, H.; Gaubert, H.; Bucher, E.; Mirouze, M.; Vaillant, I.; Paszkowski, J. An SiRNA Pathway Prevents Transgenerational Retrotransposition in Plants Subjected to Stress. Nature 2011, 472, 115–119. [Google Scholar] [CrossRef]
- Mirouze, M.; Reinders, J.; Bucher, E.; Nishimura, T.; Schneeberger, K.; Ossowski, S.; Cao, J.; Weigel, D.; Paszkowski, J.; Mathieu, O. Selective Epigenetic Control of Retrotransposition in Arabidopsis. Nature 2009, 461, 427–430. [Google Scholar] [CrossRef]
- Quadrana, L.; Etcheverry, M.; Gilly, A.; Caillieux, E.; Madoui, M.-A.; Guy, J.; Bortolini Silveira, A.; Engelen, S.; Baillet, V.; Wincker, P.; et al. Transposition Favors the Generation of Large Effect Mutations That May Facilitate Rapid Adaption. Nat. Commun. 2019, 10, 3421. [Google Scholar] [CrossRef]
- Marí-Ordóñez, A.; Marchais, A.; Etcheverry, M.; Martin, A.; Colot, V.; Voinnet, O. Reconstructing de Novo Silencing of an Active Plant Retrotransposon. Nat. Genet. 2013, 45, 1029–1039. [Google Scholar] [CrossRef] [PubMed]
- Rigal, M.; Becker, C.; Pélissier, T.; Pogorelcnik, R.; Devos, J.; Ikeda, Y.; Weigel, D.; Mathieu, O. Epigenome Confrontation Triggers Immediate Reprogramming of DNA Methylation and Transposon Silencing in Arabidopsis thaliana F1 Epihybrids. Proc. Natl. Acad. Sci. USA 2016, 113, E2083–E2092. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nozawa, K.; Chen, J.; Jiang, J.; Leichter, S.M.; Yamada, M.; Suzuki, T.; Liu, F.; Ito, H.; Zhong, X. DNA Methyltransferase CHROMOMETHYLASE3 Prevents ONSEN Transposon Silencing under Heat Stress. PLoS Genet. 2021, 17, e1009710. [Google Scholar] [CrossRef]
- Zemach, A.; Kim, M.Y.; Hsieh, P.-H.; Coleman-Derr, D.; Eshed-Williams, L.; Thao, K.; Harmer, S.L.; Zilberman, D. The Arabidopsis Nucleosome Remodeler DDM1 Allows DNA Methyltransferases to Access H1-Containing Heterochromatin. Cell 2013, 153, 193–205. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirochika, H.; Okamoto, H.; Kakutani, T. Silencing of Retrotransposons in Arabidopsis and Reactivation by the Ddm1 Mutation. Plant Cell 2000, 12, 357–368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pérez-Hormaeche, J.; Potet, F.; Beauclair, L.; Le Masson, I.; Courtial, B.; Bouché, N.; Lucas, H. Invasion of the Arabidopsis Genome by the Tobacco Retrotransposon Tnt1 Is Controlled by Reversible Transcriptional Gene Silencing. Plant Physiol. 2008, 147, 1264–1278. [Google Scholar] [CrossRef] [Green Version]
- Reinders, J.; Mirouze, M.; Nicolet, J.; Paszkowski, J. Parent-of-origin Control of Transgenerational Retrotransposon Proliferation in Arabidopsis. EMBO Rep. 2013, 14, 823–828. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.C.; Ernst, E.; Berube, B.; Borges, F.; Parent, J.-S.; Ledon, P.; Schorn, A.; Martienssen, R.A. Arabidopsis Retrotransposon Virus-like Particles and Their Regulation by Epigenetically Activated Small RNA. Genome Res. 2020, 30, 576–588. [Google Scholar] [CrossRef]
- Saha, A.; Mitchell, J.A.; Nishida, Y.; Hildreth, J.E.; Ariberre, J.A.; Gilbert, W.V.; Garfinkel, D.J. A Trans -Dominant Form of Gag Restricts Ty1 Retrotransposition and Mediates Copy Number Control. J. Virol. 2015, 89, 3922–3938. [Google Scholar] [CrossRef] [Green Version]
- Misra, S.; Rio, D.C. Cytotype Control of Drosophila P Element Transposition: The 66 Kd Protein Is a Repressor of Transposase Activity. Cell 1990, 62, 269–284. [Google Scholar] [CrossRef]
- Lampe, D.J.; Akerley, B.J.; Rubin, E.J.; Mekalanos, J.J.; Robertson, H.M. Hyperactive Transposase Mutants of the Himar1 Mariner Transposon. Proc. Natl. Acad. Sci. USA 1999, 96, 11428–11433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sultana, T.; Zamborlini, A.; Cristofari, G.; Lesage, P. Integration Site Selection by Retroviruses and Transposable Elements in Eukaryotes. Nat. Rev. Genet. 2017, 18, 292–308. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-F.; Su, T.; Cheng, G.-Q.; Wang, B.-X.; Li, X.; Deng, C.-L.; Gao, W.-J. Chromosome Evolution in Connection with Repetitive Sequences and Epigenetics in Plants. Genes 2017, 8, 290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jedlicka, P.; Lexa, M.; Vanat, I.; Hobza, R.; Kejnovsky, E. Nested Plant LTR Retrotransposons Target Specific Regions of Other Elements, While All LTR Retrotransposons Often Target Palindromes and Nucleosome-Occupied Regions: In Silico Study. Mob. DNA 2019, 10, 50. [Google Scholar] [CrossRef] [Green Version]
- Roquis, D.; Robertson, M.; Yu, L.; Thieme, M.; Julkowska, M.; Bucher, E. Genomic Impact of Stress-Induced Transposable Element Mobility in Arabidopsis. Nucleic Acids Res. 2021, 49, 10431–10447. [Google Scholar] [CrossRef]
- Wendel, J.F.; Lisch, D.; Hu, G.; Mason, A.S. The Long and Short of Doubling down: Polyploidy, Epigenetics, and the Temporal Dynamics of Genome Fractionation. Curr. Opin. Genet. Dev. 2018, 49, 1–7. [Google Scholar] [CrossRef]
- Jiao, Y.; Wickett, N.J.; Ayyampalayam, S.; Chanderbali, A.S.; Landherr, L.; Ralph, P.E.; Tomsho, L.P.; Hu, Y.; Liang, H.; Soltis, P.S.; et al. Ancestral Polyploidy in Seed Plants and Angiosperms. Nature 2011, 473, 97–100. [Google Scholar] [CrossRef]
- Alix, K.; Gérard, P.R.; Schwarzacher, T.; Heslop-Harrison, J.S. (Pat) Polyploidy and Interspecific Hybridization: Partners for Adaptation, Speciation and Evolution in Plants. Ann. Bot. 2017, 120, 183–194. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The Evolutionary Significance of Polyploidy. Nat. Rev. Genet. 2017, 18, 411–424. [Google Scholar] [CrossRef]
- McClintock, B. The Significance of Responses of the Genome to Challenge. Science 1984, 226, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and Genome Evolution in Plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vicient, C.M.; Casacuberta, J.M. Impact of Transposable Elements on Polyploid Plant Genomes. Ann. Bot. 2017, 120, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Kashkush, K.; Feldman, M.; Levy, A.A. Transcriptional Activation of Retrotransposons Alters the Expression of Adjacent Genes in Wheat. Nat. Genet. 2003, 33, 102–106. [Google Scholar] [CrossRef]
- Madlung, A.; Tyagi, A.P.; Watson, B.; Jiang, H.; Kagochi, T.; Doerge, R.W.; Martienssen, R.; Comai, L. Genomic Changes in Synthetic Arabidopsis Polyploids: Genomic Changes in Arabidopsis Polyploids. Plant J. 2004, 41, 221–230. [Google Scholar] [CrossRef]
- Sarilar, V.; Palacios, P.M.; Rousselet, A.; Ridel, C.; Falque, M.; Eber, F.; Chèvre, A.; Joets, J.; Brabant, P.; Alix, K. Allopolyploidy Has a Moderate Impact on Restructuring at Three Contrasting Transposable Element Insertion Sites in Resynthesized Brassica napus Allotetraploids. New Phytol. 2013, 198, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Senerchia, N.; Felber, F.; Parisod, C. Genome Reorganization in F1 Hybrids Uncovers the Role of Retrotransposons in Reproductive Isolation. Proc. R. Soc. B Biol. Sci. 2015, 282, 20142874. [Google Scholar] [CrossRef] [Green Version]
- Mhiri, C.; Parisod, C.; Daniel, J.; Petit, M.; Lim, K.Y.; Dorlhac de Borne, F.; Kovarik, A.; Leitch, A.R.; Grandbastien, M. Parental Transposable Element Loads Influence Their Dynamics in Young Nicotiana Hybrids and Allotetraploids. New Phytol. 2019, 221, 1619–1633. [Google Scholar] [CrossRef] [Green Version]
- Lukens, L.N.; Pires, J.C.; Leon, E.; Vogelzang, R.; Oslach, L.; Osborn, T. Patterns of Sequence Loss and Cytosine Methylation within a Population of Newly Resynthesized Brassica Napus Allopolyploids. Plant Physiol. 2006, 140, 336–348. [Google Scholar] [CrossRef] [Green Version]
- Koh, J.; Soltis, P.S.; Soltis, D.E. Homeolog Loss and Expression Changes in Natural Populations of the Recently and Repeatedly Formed Allotetraploid Tragopogon mirus (Asteraceae). BMC Genom. 2010, 11, 97. [Google Scholar] [CrossRef] [Green Version]
- Parisod, C.; Alix, K.; Just, J.; Petit, M.; Sarilar, V.; Mhiri, C.; Ainouche, M.; Chalhoub, B.; Grandbastien, M.-A. Impact of Transposable Elements on the Organization and Function of Allopolyploid Genomes: Research Review. New Phytol. 2010, 186, 37–45. [Google Scholar] [CrossRef]
- Martienssen, R.A. Heterochromatin, Small RNA and Post-Fertilization Dysgenesis in Allopolyploid and Interploid Hybrids of Arabidopsis: Research Review. New Phytol. 2010, 186, 46–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borges, F.; Parent, J.-S.; Van Ex, F.; Wolff, P.; Martínez, G.; Köhler, C.; Martienssen, R.A. Transposon-Derived Small RNAs Triggered by MiR845 Mediate Genome Dosage Response in Arabidopsis. Nat. Genet. 2018, 50, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Martinez, G.; Wolff, P.; Wang, Z.; Moreno-Romero, J.; Santos-González, J.; Conze, L.L.; DeFraia, C.; Slotkin, R.K.; Köhler, C. Paternal EasiRNAs Regulate Parental Genome Dosage in Arabidopsis. Nat. Genet. 2018, 50, 193–198. [Google Scholar] [CrossRef]
- Vitte, C.; Fustier, M.-A.; Alix, K.; Tenaillon, M.I. The Bright Side of Transposons in Crop Evolution. Brief. Funct. Genom. 2014, 13, 276–295. [Google Scholar] [CrossRef] [Green Version]
- Kejnovsky, E.; Leitch, I.J.; Leitch, A.R. Contrasting Evolutionary Dynamics between Angiosperm and Mammalian Genomes. Trends Ecol. Evol. 2009, 24, 572–582. [Google Scholar] [CrossRef]
- Murat, F.; de Peer, Y.V.; Salse, J. Decoding Plant and Animal Genome Plasticity from Differential Paleo-Evolutionary Patterns and Processes. Genome Biol. Evol. 2012, 4, 917–928. [Google Scholar] [CrossRef] [Green Version]
- Dubin, M.J.; Mittelsten Scheid, O.; Becker, C. Transposons: A Blessing Curse. Curr. Opin. Plant Biol. 2018, 42, 23–29. [Google Scholar] [CrossRef]
- Springer, N.M.; Lisch, D.; Li, Q. Creating Order from Chaos: Epigenome Dynamics in Plants with Complex Genomes. Plant Cell 2016, 28, 314–325. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Zhang, B.; Lisch, D.; Ma, J. Patterns and Consequences of Subgenome Differentiation Provide Insights into the Nature of Paleopolyploidy in Plants. Plant Cell 2017, 29, 2974–2994. [Google Scholar] [CrossRef] [Green Version]

| Class | Order (Non-Autonomous TE Name) | Superfamily | Family/Lineage | Plant Family Examples |
|---|---|---|---|---|
| Class I | LTR-Retrotransposons | Copia | Osser | Volvox canteri Osser |
| (retrotransposons) | (LARD) | Bryco | representatives in moss species | |
| (TRIM/SMART) | Lyco | representatives in clubmosses species (Lycopodiaceae) | ||
| Gymco-I | representatives in gymnosperms species | |||
| Gymco-II | representatives in gymnosperms species | |||
| Gymco-III | representatives in gymnosperms species | |||
| Gymco-IV | representatives in gymnosperms species | |||
| Ale/Retrofit | Oryza longistaminata Retrofit, Oryza sativa Hopscotch | |||
| Ivana | Oryza sativa Oryko1-1 and Ilona, Hordeum vulgare HORPIA, Nicotiana tabacum Queenti | |||
| Ikeros | Zea mays Sto-4 | |||
| Tork | Nicotiana tabacum Tnt1, Tto1 and Tnt2, Solanum lycopersicum Tork4, Ipomea batatas Batata | |||
| Alesia | low copy number representatives in many Angiosperms, close to the Ale lineage | |||
| Angela | Triticum aestivum Angela, Oryza sativa RIRE1, Hordeum vulgare BARE1 | |||
| Bianca | Triticeae Bianca, Arabidopsis thaliana RomaniAT5 | |||
| SIRE/Maximus | Solanum lycopersicum ToRTL1, Zea mays Opie-2, Glycine max SIRE1 | |||
| TAR | Oryza spp. Houba and Osr-1, Arabidopsis thaliana ATcopia95 | |||
| Gypsy (Chromovirus) | Galadriel | Solanum esculentum Galadriel, Musa Monkey, Tntom1 | ||
| Tekay | Hordeum vulgare Bagy-1, Arabidopsis thaliana Legolas Peabody, Oryza sativa RIRE3, Lilium henryi Del | |||
| Reina | Zea mays Reina, Arabidopsis thaliana Gloin or Gimli | |||
| CRM | Zea mays CRM (centromeric retrotransposon of maize), Beta vulgaris Beetle1, Oryza sativa RIRE7 | |||
| (Non-chromovirus) | Phygy | Phycomitrella patens Chr21 (4035670,4045566) | ||
| Selgy | Selaginella moellendorffii LTR-RT | |||
| Athila | Arabidopsis thaliana Athila4-1, Diaspora, Hordeum vulgare Bagy-2 | |||
| TatI | Selaginella moellendorffii LTR-RT | |||
| TatII | Picea abies, Picea glauca LTR-RTs | |||
| TatIII | Picea abies, Picea glauca LTR-RTs | |||
| Ogre/TatIV + TatV | Pisum sativum Ogre | |||
| Retand/TatVI | Zea mays Cinful-1, Arabidopsis thaliana Tat4-1, Oryza sativa RIRE2, Sorghum bicolor RetroSor1, Silene latifolia Retand | |||
| Non-LTR retrotransposons PLE | Penelope/Poseidon | Pinus taeda (loblolly pine) and Picea abies (Norway spruce) Dryad PLEs by horizontal transfer | ||
| EN(-)PLE | Selaginella moellendorffii spike moss, Pinus taeda and Picea abies EN(-)PLEs | |||
| LINE | L1 | Llb | sweet potato Llb, Beta vulgaris BvL1 | |
| LINE-CS | Cannabis sativa LINE-CS, Beta vulgaris Belline2, Belline5 | |||
| BNR | Beta vulgaris Belline1/BNR | |||
| PUR | Carica papaya L1-26_Cpa, Solanum tuberosum L1-3_Stu, Vitis vinifera | |||
| Cin4 | Zea mays Cin4 | |||
| Karma | Oryza sativa Karma | |||
| nubo | Oryza sativa LINE-1 or OSLINE1-4, Zea mays L1-2_ZM | |||
| RTE | plant RTE | Malus x domestica RTE-1_Mad, Solanum tuberosum RTE-1_Stu | ||
| SINE | tRNA | Nicotiana tabacum TS, Au, Solanales SolS-II, Brassicale BraS-I, SB families, mainly found in Angiosperm | ||
| Class II Subclass 1 | TIR (MITE) | Tc1-Mariner | Stowaway (MITE): Sorghum bicolor Stowaway, Brassica BraSto | |
| hAT | Zea mays Ac/Ds, Antirrhinum majus Tam3, Nicotiana tabacum Slide | |||
| Sola | Physcomitrella Sola1, found also in Capsicum annuum and C. baccatum | |||
| (MULE) | MuDR-Foldback | Zea mays Mu, MULEs | ||
| PIF-Harbinger | Zea mays PIFa, Oryza sativa Pong; Tourist (MITE): mPing/Ping; mPIF/PIFa | |||
| CACTA | Zea mays En/Spm, Arabidopsis thaliana CAC1, Antirrhinum majus Tam1, Petunia hybrida PsI | |||
| Subclass 2 | Helitron | Helitron | Oryza sativa, Arabidopsis thaliana AthE1 Atrep, Ipomoea tricolor Hel-It1 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mhiri, C.; Borges, F.; Grandbastien, M.-A. Specificities and Dynamics of Transposable Elements in Land Plants. Biology 2022, 11, 488. https://doi.org/10.3390/biology11040488
Mhiri C, Borges F, Grandbastien M-A. Specificities and Dynamics of Transposable Elements in Land Plants. Biology. 2022; 11(4):488. https://doi.org/10.3390/biology11040488
Chicago/Turabian StyleMhiri, Corinne, Filipe Borges, and Marie-Angèle Grandbastien. 2022. "Specificities and Dynamics of Transposable Elements in Land Plants" Biology 11, no. 4: 488. https://doi.org/10.3390/biology11040488
APA StyleMhiri, C., Borges, F., & Grandbastien, M.-A. (2022). Specificities and Dynamics of Transposable Elements in Land Plants. Biology, 11(4), 488. https://doi.org/10.3390/biology11040488






