High Proportions of Radiation-Resistant Strains in Culturable Bacteria from the Taklimakan Desert
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
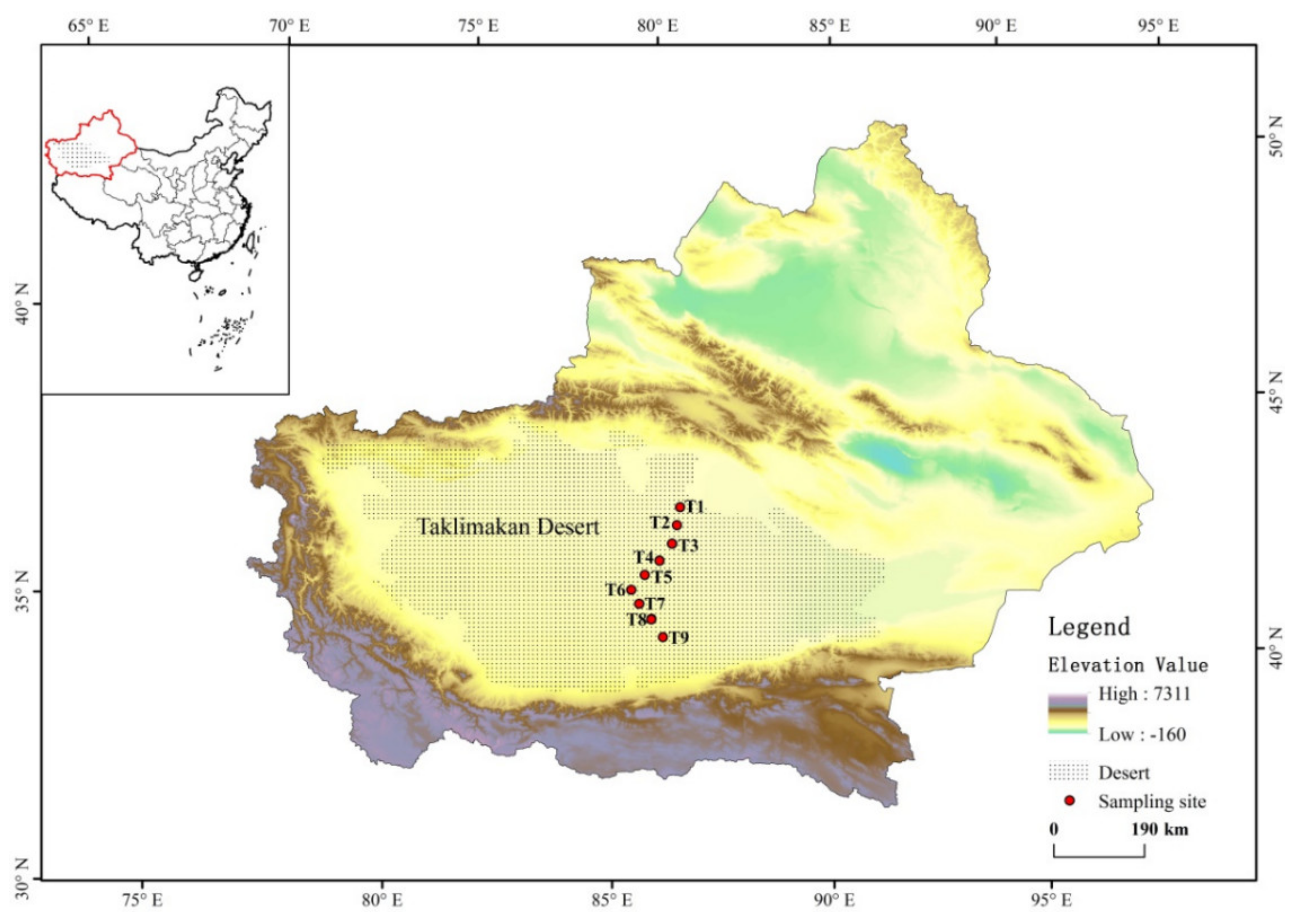
2.1. Site Description and Sample Collection
2.2. Measurement of Soil Physico-Chemical Properties
2.3. Microbial Enumeration
2.4. 16S rRNA Gene Sequencing and Phylogenetic Analyses
2.5. Screening Radiation-Resistant Strains
2.6. Data Analysis
3. Results
3.1. The Physico-Chemical Properties of Sand Soil from the Taklimakan Desert
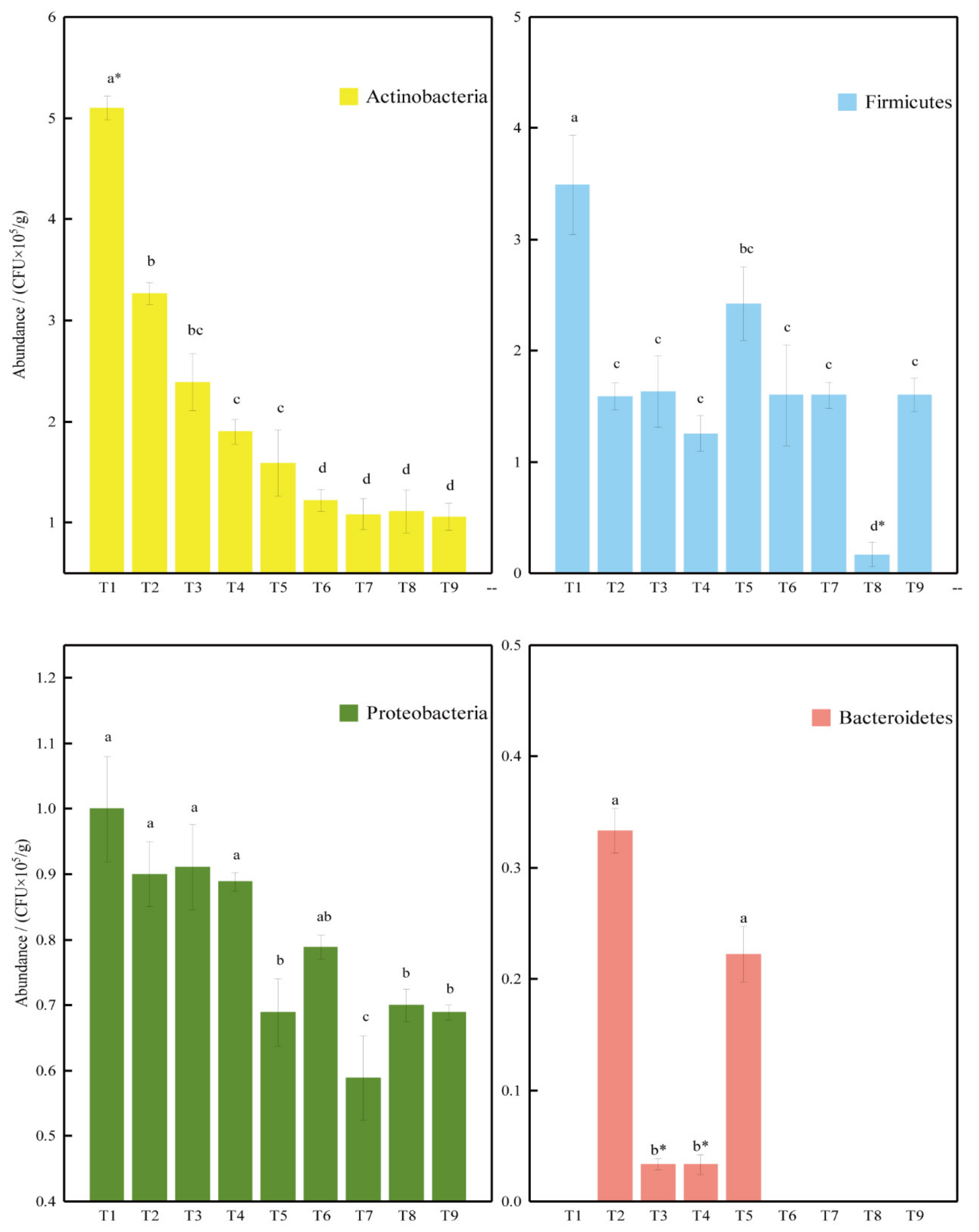
3.2. Abundance and Diversity of Culturable Bacteria
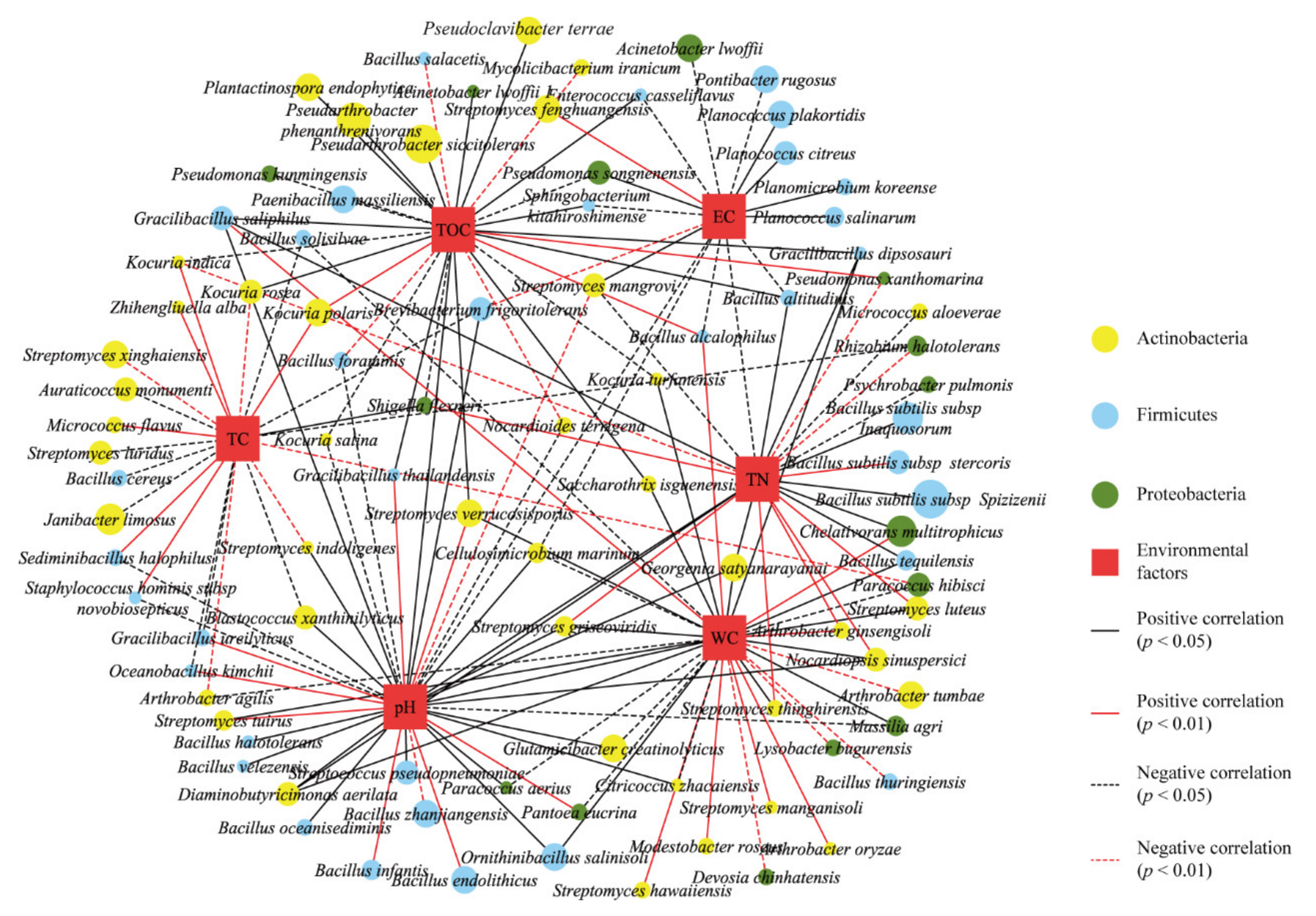
3.3. Distribution of Culturable Bacteria and Correlations with Environmental Factors
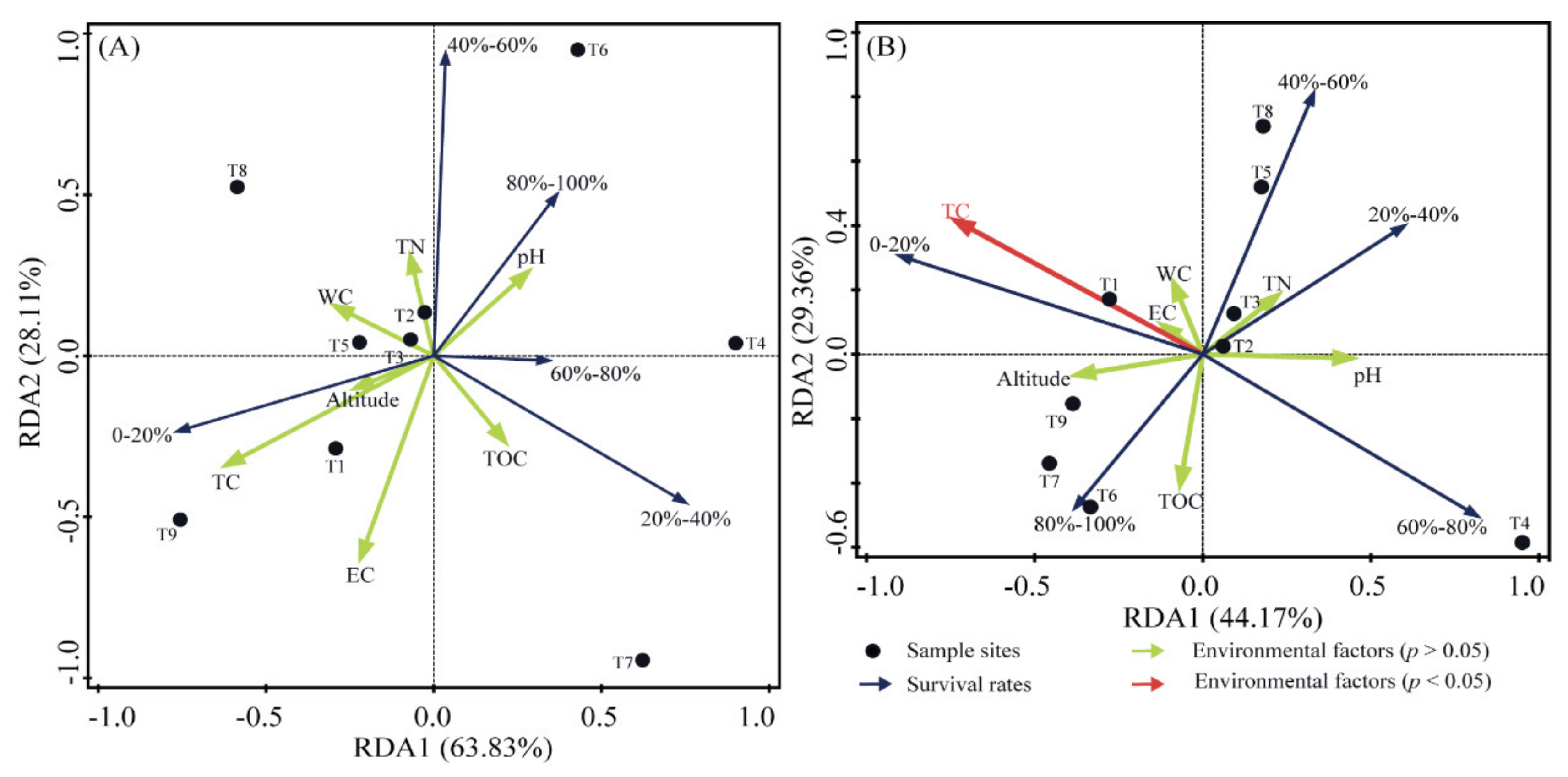
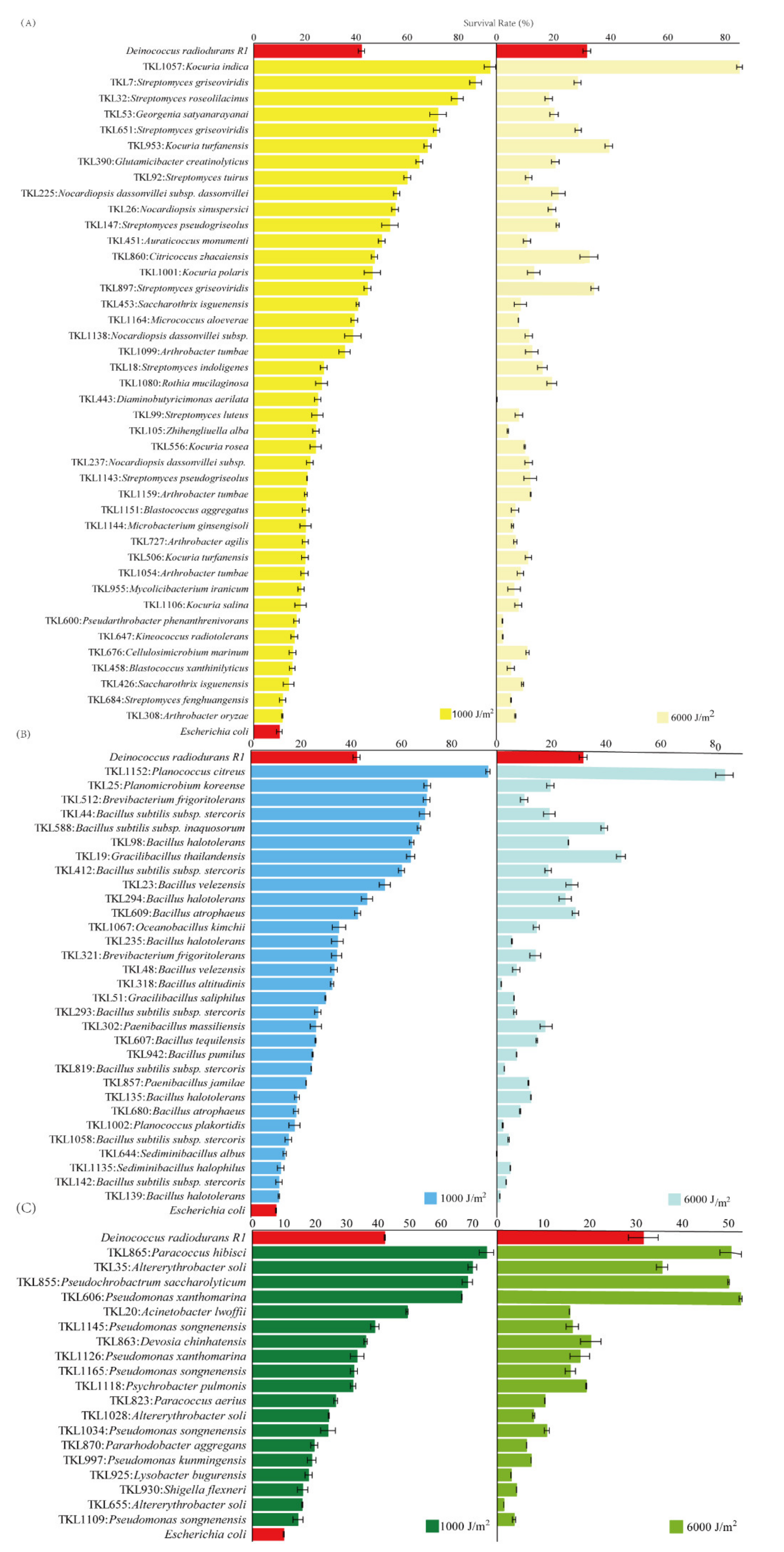
3.4. Survival Rates after Exposure to UV-C and γ-rays Radiation
4. Discussion
4.1. Diversity of Distribution of Culturable Bacteria in the Taklimakan Desert
4.2. The Proportions of Radiation-Resistant Strains in the Culturable Bacteria
4.3. The Traits of the Strongly Radiation-Resistant Bacteria in the Taklimakan Desert
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saleh, Y.G.; Mayo, M.S.; Ahearn, D.G. Resistance of some common fungi to gamma irradiation. Appl. Environ. Microbiol. 1988, 54, 2134–2135. [Google Scholar] [CrossRef] [Green Version]
- Gérard, J.C.; Gustin, J.; Grodent, D.; Delamere, P.; Clarke, J.T. Excitation of the FUV Io tail on Jupiter: Characterization of the electron precipitation. J. Geophys. Res. Space Phys. 2002, 107, SMP-30. [Google Scholar] [CrossRef]
- Jolivet, E.; Corre, E.; L’Haridon, S.; Forterre, P.; Prieur, D. Thermococcus marinus sp. nov. and Thermococcus radiotolerans sp. nov., two hyperthermophilic archaea from deep-sea hydrothermal vents that resist ionizing radiation. Extrem. Life Under Extrem. Cond. 2004, 8, 219–227. [Google Scholar] [CrossRef]
- Yu, L.Z.; Luo, X.S.; Liu, M.; Huang, Q. Diversity of ionizing radiation-resistant bacteria obtained from the Taklimakan Desert. J. Basic Microbiol. 2015, 55, 135–140. [Google Scholar] [CrossRef]
- Mattimore, V.; Battista, J.R. Radioresistance of Deinococcus radiodurans: Functions necessary to survive ionizing radiation are also necessary to survive prolonged desiccation. J. Bacteriol. 1996, 178, 633–637. [Google Scholar] [CrossRef] [Green Version]
- Billi, D.; Wright, D.J.; Helm, R.F.; Prickett, T.; Potts, M.; Crowe, J.H. Engineering desiccation tolerance in Escherichia coli. Appl. Environ. Microbiol. 2000, 66, 1680–1684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shukla, M.; Chaturvedi, R.; Tamhane, D.; Vyas, P.; Archana, G.; Apte, S.; Bandekar, J.; Desai, A. Multiple-stress tolerance of ionizing radiation-resistant bacterial isolates obtained from various habitats: Correlation between stresses. Curr. Microbiol. 2007, 54, 142–148. [Google Scholar] [CrossRef] [PubMed]
- Bauermeister, A.; Moeller, R.; Reitz, G.; Sommer, S.; Rettberg, P. Effect of relative humidity on Deinococcus radiodurans’ resistance to prolonged desiccation, heat, ionizing, germicidal, and environmentally relevant UV radiation. Microb. Ecol. 2011, 61, 715–722. [Google Scholar] [CrossRef]
- Rainey, F.A.; Ray, K.; Ferreira, M.; Gatz, B.Z.; Nobre, M.F.; Bagaley, D.; Rash, B.A.; Park, M.J.; Earl, A.M.; Shank, N.C.; et al. Extensive diversity of ionizing-radiation-resistant bacteria recovered from Sonoran Desert soil and description of nine new species of the genus Deinococcus obtained from a single soil sample. Appl. Environ. Microbiol. 2005, 71, 5225–5235. [Google Scholar] [CrossRef] [Green Version]
- Paulino-Lima, I.G.; Azua-Bustos, A.; Vicuna, R.; Gonzalez-Silva, C.; Salas, L.; Teixeira, L.; Rosado, A.; Leitao, A.A.; Lage, C. Isolation of UVC-tolerant bacteria from the hyperarid Atacama Desert, Chile. Microb. Ecol. 2013, 65, 325–335. [Google Scholar] [CrossRef]
- Paulino-Lima, I.G.; Fujishima, K.; Navarrete, J.U.; Galante, D.; Rodrigues, F.; Azua-Bustos, A.; Rothschild, L.J. Extremely high UV-C radiation resistant microorganisms from desert environments with different manganese concentrations. J. Photochem. Photobiol. B Biol. 2016, 163, 327–336. [Google Scholar] [CrossRef]
- de Groot, A.; Chapon, V.; Servant, P.; Christen, R.; Saux, M.F.; Sommer, S.; Heulin, T. Deinococcus deserti sp. nov., a gamma-radiation-tolerant bacterium isolated from the Sahara Desert. Int. J. Syst. Evol. Microbiol. 2005, 55, 2441–2446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero-Calasanz Mdel, C.; Göker, M.; Broughton, W.J.; Cattaneo, A.; Favet, J.; Pötter, G.; Rohde, M.; Spröer, C.; Schumann, P.; Klenk, H.P.; et al. Geodermatophilus tzadiensis sp. nov., a UV radiation-resistant bacterium isolated from sand of the Saharan desert. Syst. Appl. Microbiol. 2013, 36, 177–182. [Google Scholar] [CrossRef] [PubMed]
- Hezbri, K.; Ghodhbane-Gtari, F.; Montero-Calasanz, M.D.C.; Nouioui, I.; Rohde, M.; Spröer, C.; Schumann, P.; Klenk, H.P.; Gtari, M. Geodermatophilus pulveris sp. nov., a gamma-radiation-resistant actinobacterium isolated from the Sahara desert. Int. J. Syst. Evol. Microbiol. 2016, 66, 3828–3834. [Google Scholar] [CrossRef]
- Baqué, M.; Viaggiu, E.; Scalzi, G.; Billi, D. Endurance of the endolithic desert cyanobacterium Chroococcidiopsis under UVC radiation. Extrem. Life Under Extrem. Cond. 2013, 17, 161–169. [Google Scholar] [CrossRef]
- Mohseni, M.; Abbaszadeh, J.; Nasrollahi Omran, A. Radiation resistant of native Deinococcus spp. isolated from the Lout desert of Iran “the hottest place on Earth”. Int. J. Environ. Sci. Technol. 2014, 11, 1939–1946. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Liu, C.; Tang, Y.; Zhou, G.; Shen, P.; Fang, C.; Yokota, A. Hymenobacter xinjiangensis sp. nov., a radiation-resistant bacterium isolated from the desert of Xinjiang, China. Int. J. Syst. Evol. Microbiol. 2007, 57, 1752–1756. [Google Scholar] [CrossRef] [Green Version]
- Liu, M.; Dai, J.; Liu, Y.; Cai, F.; Wang, Y.; Rahman, E.; Fang, C. Desertibacter roseus gen. nov., sp. nov., a gamma radiation-resistant bacterium in the family Rhodospirillaceae, isolated from desert sand. Int. J. Syst. Evol. Microbiol. 2011, 61, 1109–1113. [Google Scholar] [CrossRef] [Green Version]
- Peng, F.; Zhang, L.; Luo, X.; Dai, J.; An, H.; Tang, Y.; Fang, C. Deinococcus xinjiangensis sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 709–713. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Kim, M.C.; Wang, L.; Zhu, G.; Zhang, Y.; Huang, Y.; Wei, Z.; Danzeng, W.; Peng, F. Deinococcus taklimakanensis sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 4311–4316. [Google Scholar] [CrossRef]
- Yuan, L.L.; Zhang, L.L.; Luo, X.X.; Xia, Z.F.; Sun, B.B.; Zeng, H. Streptomyces taklimakanensis sp. nov., an actinomycete isolated from the Taklimakan desert. Antonie Van Leeuwenhoek 2020, 113, 1023–1031. [Google Scholar] [CrossRef] [PubMed]
- Zhao, L.; Zhou, Y.; Li, J.; Xia, Y.; Wang, W.; Luo, X.; Yin, J.; Zhong, J. Transcriptional response of Bacillus megaterium FDU301 to PEG200-mediated arid stress. BMC Microbiol. 2020, 20, 351. [Google Scholar] [CrossRef]
- Jiang, H.; Liu, N.N.; Liu, G.L.; Chi, Z.; Wang, J.M.; Zhang, L.L.; Chi, Z.M. Melanin production by a yeast strain XJ5-1 of Aureobasidium melanogenum isolated from the Taklimakan desert and its role in the yeast survival in stress environments. Extremophiles 2016, 20, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Chen, T.J.; Chi, Z.; Hu, Z.; Liu, G.L.; Sun, Y.; Zhang, S.H.; Chi, Z.M. Macromolecular pullulan produced by Aureobasidium melanogenum 13-2 isolated from the Taklimakan desert and its crucial roles in resistance to the stress treatments. Int. J. Biol. Macromol. 2019, 135, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Beblo-Vranesevic, K.; Bohmeier, M.; Perras, A.K.; Schwendner, P.; Rabbow, E.; Moissl-Eichinger, C.; Cockell, C.S.; Vannier, P.; Marteinsson, V.T.; Monaghan, E.P.; et al. Lack of correlation of desiccation and radiation tolerance in microorganisms from diverse extreme environments tested under anoxic conditions. FEMS Microbiol. Lett. 2018, 365–371. [Google Scholar] [CrossRef]
- Imlay, J.A. The molecular mechanisms and physiological consequences of oxidative stress: Lessons from a model bacterium. Nat. Rev. Microbiol. 2013, 11, 443–454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lushchak, V.I. Adaptive response to oxidative stress: Bacteria, fungi, plants and animals. Comp. Biochem. Physiology. Toxicol. Pharmacol. CBP 2011, 153, 175–190. [Google Scholar] [CrossRef]
- Narumi, I. Unlocking radiation resistance mechanisms: Still a long way to go. Trends Microbiol. 2003, 11, 422–425. [Google Scholar] [CrossRef]
- Georgiou, C.D.; Sun, H.J.; McKay, C.P.; Grintzalis, K.; Papapostolou, I.; Zisimopoulos, D.; Panagiotidis, K.; Zhang, G.; Koutsopoulou, E.; Christidis, G.E.; et al. Evidence for photochemical production of reactive oxygen species in desert soils. Nat. Commun. 2015, 6, 7100. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Yu, Y.; Xia, D.; Yin, D.; He, J.; Liu, N.; Li, F. Urban particle size distributions during two contrasting dust events originating from Taklimakan and Gobi Deserts. Environ. Pollut. 2015, 207, 107–122. [Google Scholar] [CrossRef]
- Yang, S.; Yang, T. Exploration of the dynamic water resource carrying capacity of the Keriya River Basin on the southern margin of the Taklimakan Desert, China. Reg. Sustain. 2021, 2, 73–82. [Google Scholar] [CrossRef]
- Keren, R.; Lavy, A.; Ilan, M. Erratum to: Increasing the Richness of Culturable Arsenic-Tolerant Bacteria from Theonella swinhoei by Addition of Sponge Skeleton to the Growth Medium. Microb. Ecol. 2016, 72, 496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mclean, E.O. Soil pH and Lime Requirement. In Methods of Soil Analysis; American Society of Agronomy, Inc. Soil Science Society of America, Inc.: Madison, WI, USA, 1983; pp. 199–224. [Google Scholar]
- Bremner, J.M.; Mulvaney, C.S. Nitrogen—Total. In Methods of Soil Analysis; Soil Science Society of America, American Society of Agronomy: Madison, WI, USA, 1983; pp. 595–624. [Google Scholar]
- Nelson, D.W.; Sommers, L.E. Total Carbon, Organic Carbon, and Organic Matter. In Methods of Soil Analysis; Soil Science Society of America, American Society of Agronomy: Madison, WI, USA, 1983; pp. 539–579. [Google Scholar]
- Liebner, S.; Rublack, K.; Stuehrmann, T.; Wagner, D. Diversity of aerobic methanotrophic bacteria in a permafrost active layer soil of the Lena Delta, Siberia. Microb. Ecol. 2009, 57, 25–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lane, D.J. 16S/23S rRNA Sequencing. In Nucleic Acid Techniques in Bacterial Systematics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 1991. [Google Scholar]
- Yoon, S.H.; Ha, S.M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; McGettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [Green Version]
- Nishimaki, T.; Sato, K. An Extension of the Kimura Two-Parameter Model to the Natural Evolutionary Process. J. Mol. Evol. 2019, 87, 60–67. [Google Scholar] [CrossRef] [Green Version]
- Felsenstein, J. Confidence Limits on Phylogenies: An Approach Using the Bootstrap. Evol. Int. J. Org. Evol. 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Yan, H.; Feng, L.; Zhao, Y.; Feng, L.; Wu, D.; Zhu, C. Prediction of the spatial distribution of Alternanthera philoxeroides in China based on ArcGIS and MaxEnt. Glob. Ecol. Conserv. 2020, 21, e00856. [Google Scholar] [CrossRef]
- White, J.R.; Nagarajan, N.; Pop, M. Statistical methods for detecting differentially abundant features in clinical metagenomic samples. PLoS Comput. Biol. 2009, 5, e1000352. [Google Scholar] [CrossRef]
- Symeonidou, V.; Fotopoulou, F.; Anderson, R.; Finch, A.; Ottersbach, K. 2017–Exploiting the foetal origin of mll-af4-driven acute lymphoblastic leukaemia. Exp. Hematol. 2020, 88, S33–S34. [Google Scholar] [CrossRef]
- Ter Braak, C.; Šmilauer, P. Canoco Reference Manual and User’s Guide: Software of Ordination (Version 5.0); Microcomputer Power: Ithaca, NY, USA, 2012. [Google Scholar]
- Shuryak, I.; Matrosova, V.Y.; Gaidamakova, E.K.; Tkavc, R.; Grichenko, O.; Klimenkova, P.; Volpe, R.P.; Daly, M.J. Microbial cells can cooperate to resist high-level chronic ionizing radiation. PLoS ONE 2017, 12, e0189261. [Google Scholar] [CrossRef]
- An, S.; Couteau, C.; Luo, F.; Neveu, J.; DuBow, M.S. Bacterial diversity of surface sand samples from the Gobi and Taklamaken deserts. Microb. Ecol 2013, 4, 850–860. [Google Scholar] [CrossRef]
- An, S.; Sin, H.H.; DuBow, M.S. Modification of atmospheric sand-associated bacterial communities during Asian sandstorms in China and South Korea. Heredity 2015, 114, 460–467. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Kong, W.; Wu, N.; Zhang, Y. Bacterial diversity and community along the succession of biological soil crusts in the Gurbantunggut Desert, Northern China. J. Basic Microbiol. 2016, 56, 670–679. [Google Scholar] [CrossRef]
- Nan, W.; Yuanming, Z.; Huixia, P.; Dong, Q. Culture-dependent bacteria diversity of moss crusts in the Gurbantunggut Desert. Arid Land Geogr. 2014, 37, 250–258. [Google Scholar] [CrossRef]
- Lu, X.; Xiaolu, C.; Shiyan, W.; Lei, S.; Lubing, L. The Diversity of Cultivable Bacteria of Soil in Aqik Valley, the North Boundary of Kumtag Desert. J. Nucl. Agric. Sci. 2017, 31, 342–349. [Google Scholar] [CrossRef]
- Lin, Y.-T.; Whitman, W.B.; Coleman, D.C.; Chen, T.-H.; Chiu, C.-Y. Composition of bacterial communities in sand dunes of subtropical coastal forests. Biol. Fertil. Soils 2014, 50, 809–814. [Google Scholar] [CrossRef]
- Shet, S.A.; Garg, S. Prokaryotic diversity of tropical coastal sand dunes ecosystem using metagenomics. 3 Biotech 2021, 11, 252. [Google Scholar] [CrossRef]
- Bahadur, A.; Zhang, W.; Sajjad, W.; Nasir, F.; Zhang, G.; Liu, G.; Chen, T. Bacterial diversity patterns of desert dunes in the northeastern Qinghai-Tibet Plateau, China. Arch. Microbiol. 2021, 203, 2809–2823. [Google Scholar] [CrossRef]
- Mandakovic, D.; Maldonado, J.; Pulgar, R.; Cabrera, P.; Gaete, A.; Urtuvia, V.; Seeger, M.; Cambiazo, V.; González, M. Microbiome analysis and bacterial isolation from Lejía Lake soil in Atacama Desert. Extremophiles 2018, 22, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Contador, C.A.; Veas-Castillo, L.; Tapia, E.; Antipán, M.; Miranda, N.; Ruiz-Tagle, B.; García-Araya, J.; Andrews, B.A.; Marin, M.; Dorador, C.; et al. Atacama Database: A platform of the microbiome of the Atacama Desert. Antonie Van Leeuwenhoek 2020, 113, 185–195. [Google Scholar] [CrossRef] [PubMed]
- Okoro, C.K.; Brown, R.; Jones, A.L.; Andrews, B.A.; Asenjo, J.A.; Goodfellow, M.; Bull, A.T. Diversity of culturable actinomycetes in hyper-arid soils of the Atacama Desert, Chile. Antonie Van Leeuwenhoek 2009, 95, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Arocha-Garza, H.F.; Canales-Del Castillo, R.; Eguiarte, L.E.; Souza, V.; De la Torre-Zavala, S. High diversity and suggested endemicity of culturable Actinobacteria in an extremely oligotrophic desert oasis. PeerJ 2017, 5, e3247. [Google Scholar] [CrossRef] [Green Version]
- Molina-Menor, E.; Gimeno-Valero, H.; Pascual, J.; Peretó, J.; Porcar, M. High Culturable Bacterial Diversity from a European Desert: The Tabernas Desert. Front. Microbiol. 2020, 11, 583120. [Google Scholar] [CrossRef]
- Chowdhury, S.P.; Schmid, M.; Hartmann, A.; Tripathi, A.K. Identification of diazotrophs in the culturable bacterial community associated with roots of Lasiurus sindicus, a perennial grass of Thar Desert, India. Microb. Ecol. 2007, 54, 82–90. [Google Scholar] [CrossRef]
- Bachate, S.P.; Cavalca, L.; Andreoni, V. Arsenic-resistant bacteria isolated from agricultural soils of Bangladesh and characterization of arsenate-reducing strains. J. Appl. Microbiol. 2009, 107, 145–156. [Google Scholar] [CrossRef]
- Aljohani, R.; Samarasinghe, H.; Ashu, T.; Xu, J. Diversity and relationships among strains of culturable yeasts in agricultural soils in Cameroon. Sci. Rep. 2018, 8, 15687. [Google Scholar] [CrossRef]
- Belov, A.A.; Cheptsov, V.S.; Vorobyova, E.A. Soil bacterial communities of Sahara and Gibson deserts: Physiological and taxonomical characteristics. AIMS Microbiol. 2018, 4, 685–710. [Google Scholar] [CrossRef]
- Yu, J.; Steinberger, Y. Soil microbial metabolic profiles in two geomorphological units in a semistable sand-dune ecosystem. Soil Biol. Biochem. 2012, 45, 71–78. [Google Scholar] [CrossRef]
- Abdul Majid, S.; Graw, M.F.; Chatziefthimiou, A.D.; Nguyen, H.; Richer, R.; Louge, M.; Sultan, A.A.; Schloss, P.; Hay, A.G. Microbial Characterization of Qatari Barchan Sand Dunes. PLoS ONE 2016, 11, e0161836. [Google Scholar] [CrossRef] [Green Version]
- Matsukawa, E.; Nakagawa, Y.; Iimura, Y.; Hayakawa, M. Stimulatory effect of indole-3-acetic acid on aerial mycelium formation and antibiotic production in Streptomyces spp. Actinomycetologica 2007, 21, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Wu, X.; Tai, X.; Sun, L.; Wu, M.; Zhang, W.; Chen, X.; Zhang, G.; Chen, T.; Liu, G.; et al. Variation in Actinobacterial Community Composition and Potential Function in Different Soil Ecosystems Belonging to the Arid Heihe River Basin of Northwest China. Front. Microbiol. 2019, 10, 2209. [Google Scholar] [CrossRef] [PubMed]
- Fierer, N.; Lauber, C.L.; Ramirez, K.S.; Zaneveld, J.; Bradford, M.A.; Knight, R. Comparative metagenomic, phylogenetic and physiological analyses of soil microbial communities across nitrogen gradients. ISME J. 2012, 6, 1007–1017. [Google Scholar] [CrossRef] [Green Version]
- Lauber, C.L.; Hamady, M.; Knight, R.; Fierer, N. Pyrosequencing-based assessment of soil pH as a predictor of soil bacterial community structure at the continental scale. Appl. Environ. Microbiol. 2009, 75, 5111–5120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Riquelme, C.; Marshall Hathaway, J.J.; Enes Dapkevicius Mde, L.; Miller, A.Z.; Kooser, A.; Northup, D.E.; Jurado, V.; Fernandez, O.; Saiz-Jimenez, C.; Cheeptham, N. Actinobacterial Diversity in Volcanic Caves and Associated Geomicrobiological Interactions. Front. Microbiol. 2015, 6, 1342. [Google Scholar] [CrossRef] [Green Version]
- Hauschild, P.; Röttig, A.; Madkour, M.H.; Al-Ansari, A.M.; Almakishah, N.H.; Steinbüchel, A. Lipid accumulation in prokaryotic microorganisms from arid habitats. Appl. Microbiol. Biotechnol. 2017, 101, 2203–2216. [Google Scholar] [CrossRef]
- Abadi, V.; Sepehri, M.; Rahmani, H.A.; Dolatabad, H.K.; Shamshiripour, M.; Khatabi, B. Diversity and abundance of culturable nitrogen-fixing bacteria in the phyllosphere of maize. J. Appl. Microbiol. 2021, 131, 898–912. [Google Scholar] [CrossRef]
- Amin, A.; Ahmed, I.; Khalid, N.; Khan, I.U.; Ali, A.; Dahlawi, S.M.; Li, W.J. Insights on comparative bacterial diversity between different arid zones of Cholistan Desert, Pakistan. 3 Biotech 2020, 10, 224. [Google Scholar] [CrossRef]
- Genderjahn, S.; Alawi, M.; Mangelsdorf, K.; Horn, F.; Wagner, D. Desiccation- and Saline-Tolerant Bacteria and Archaea in Kalahari Pan Sediments. Front. Microbiol. 2018, 9, 2082. [Google Scholar] [CrossRef]
- Gao, J.; Luo, Y.; Wei, Y.; Huang, Y.; Zhang, H.; He, W.; Sheng, H.; An, L. Effect of aridity and dune type on rhizosphere soil bacterial communities of Caragana microphylla in desert regions of northern China. PLoS ONE 2019, 14, e0224195. [Google Scholar] [CrossRef] [Green Version]
- Rosenstein, B.S. Solar-UV Actions on Living Cells. Praeger Special Studies; Jagger, J., Ed.; Praeger Publishing: New York, NY, USA, 1985. [Google Scholar]
- Kim, S.W.; Achana, F.; Petrou, S. A bootstrapping approach for generating an inverse distance weight matrix when multiple observations have an identical location in large health surveys. Int. J. Health Geogr. 2019, 18, 27. [Google Scholar] [CrossRef] [PubMed]
- Lee, C.; Choi, N.; Bae, M.K.; Choo, K.; Lee, S.J. Transposition of Insertion Sequences was Triggered by Oxidative Stress in Radiation-Resistant Bacterium Deinococcus geothermalis. Microorganisms 2019, 7, 446. [Google Scholar] [CrossRef] [Green Version]
- Thirkell, D.; Summerfield, M. Variation in pigment production by Planococcus citreus Migula with cultural age and with sea salt concentration in the medium. Antonie Van Leeuwenhoek 1980, 46, 51–57. [Google Scholar] [CrossRef]
- Wang, K.; Zhang, L.; Li, J.; Pan, Y.; Meng, L.; Xu, T.; Zhang, C.; Liu, H.; Hong, S.; Huang, H.; et al. Planococcus dechangensis sp. nov., a moderately halophilic bacterium isolated from saline and alkaline soils in Dechang Township, Zhaodong City, China. Antonie Van Leeuwenhoek 2015, 107, 1075–1083. [Google Scholar] [CrossRef] [PubMed]
- Mehrabadi, J.F.; Mirzaie, A.; Ahangar, N.; Rahimi, A.; Rokni-Zadeh, H. Draft Genome Sequence of Kocuria rhizophila RF, a Radiation-Resistant Soil Isolate. Genome Announc. 2016, 4, e00095-16. [Google Scholar] [CrossRef] [Green Version]
- Guesmi, S.; Pujic, P.; Nouioui, I.; Dubost, A.; Najjari, A.; Ghedira, K.; Igual, J.M.; Miotello, G.; Cherif, A.; Armengaud, J.; et al. Ionizing-radiation-resistant Kocuria rhizophila PT10 isolated from the Tunisian Sahara xerophyte Panicum turgidum: Polyphasic characterization and proteogenomic arsenal. Genomics 2021, 113, 317–330. [Google Scholar] [CrossRef]
- Melin, A.M.; Perromat, A.; Lorin, C.; Déléris, G. Gamma irradiation and cellular damage in Kocuria rosea: Investigation by one- and two-dimensional infrared spectroscopy. Arch. Biochem. Biophys. 2002, 408, 211–219. [Google Scholar] [CrossRef]
- Liu, L.; Choi, S. A Paper-Based Biological Solar Cell. SLAS Technol. 2019, 25, 75–81. [Google Scholar] [CrossRef]
- Pulschen, A.A.; Rodrigues, F.; Duarte, R.T.; Araujo, G.G.; Santiago, I.F.; Paulino-Lima, I.G.; Rosa, C.A.; Kato, M.J.; Pellizari, V.H.; Galante, D. UV-resistant yeasts isolated from a high-altitude volcanic area on the Atacama Desert as eukaryotic models for astrobiology. MicrobiologyOpen 2015, 4, 574–588. [Google Scholar] [CrossRef]
- Jackson, E.; Heidl, M.; Imfeld, D.; Meeus, L.; Schuetz, R.; Campiche, R. Discovery of a Highly Selective MC1R Agonists Pentapeptide to Be Used as a Skin Pigmentation Enhancer and with Potential Anti-Aging Properties. Int. J. Mol. Sci. 2019, 20, 6143. [Google Scholar] [CrossRef] [Green Version]
- Dib, J.R.; Weiss, A.; Neumann, A.; Ordonez, O.; Estevez, M.C.; Farias, M.E. Isolation of bacteria from remote high altitude Andean lakes able to grow in the presence of antibiotics. Recent Pat. Anti-Infect. Drug Discov. 2009, 4, 66–76. [Google Scholar] [CrossRef]
- Farci, D.; Slavov, C.; Tramontano, E.; Piano, D. The S-layer Protein DR_2577 Binds Deinoxanthin and under Desiccation Conditions Protects against UV-Radiation in Deinococcus radiodurans. Front. Microbiol. 2016, 7, 155. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Samples | Altitude (m) | WC (%) | pH | EC (S/m) | TN (%) | TC (%) | TOC (%) |
|---|---|---|---|---|---|---|---|
| T1 | 868 ± 3.2 | 0.218 ± 0.010 | 8.58 ± 0.11 | 310 ± 0.12 | 1.352 ± 0.11 | 2.321 ± 0.12 | 0.732 ± 0.016 |
| T2 | 876 ± 3.2 | 0.276 ± 0.020 | 8.92 ± 0.18 | 386 ± 2.23 | 1.352 ± 0.11 | 2.122 ± 0.25 | 0.712 ± 0.008 |
| T3 | 901 ± 3.2 | 0.118 ± 0.008 | 8.62 ± 0.24 | 297 ± 0.58 | 1.221 ± 0.05 | 1.655 ± 0.31 | 0.733 ± 0.012 |
| T4 | 938 ± 3.2 | 0.145 ± 0.020 | 8.67 ± 0.21 | 458 ± 0.45 | 1.022 ± 0.15 | 1.135 ± 0.15 | 0.659 ± 0.037 |
| T5 | 982 ± 3.2 | 0.153 ± 0.020 | 8.82 ± 0.13 | 633 ± 0.34 | 0.998 ± 0.08 | 1.685 ± 0.08 | 0.652 ± 0.034 |
| T6 | 1045 ± 3.2 | 0.162 ± 0.012 | 8.43 ± 0.18 | 248 ± 1.22 | 0.998 ± 0.09 | 1.674 ± 0.09 | 0.632 ± 0.034 |
| T7 | 1068 ± 3.2 | 0.134 ± 0.025 | 8.01 ± 0.22 | 671 ± 1.34 | 0.668 ± 0.02 | 2.122 ± 0.08 | 0.598 ± 0.019 |
| T8 | 1080 ± 3.2 | 0.185 ± 0.008 | 7.92 ± 0.31 | 496 ± 1.38 | 0.889 ± 0.02 | 2.135 ± 0.05 | 0.351 ± 0.034 |
| T9 | 1155 ± 3.2 | 0.165 ± 0.010 | 7.92 ± 0.18 | 733 ± 0.38 | 0.768 ± 0.08 | 2.134 ± 0.01 | 0.653 ± 0.016 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, Y.; Chen, T.; Li, J.; Wu, M.; Liu, G.; Zhang, W.; Zhang, B.; Zhang, S.; Zhang, G. High Proportions of Radiation-Resistant Strains in Culturable Bacteria from the Taklimakan Desert. Biology 2022, 11, 501. https://doi.org/10.3390/biology11040501
Liu Y, Chen T, Li J, Wu M, Liu G, Zhang W, Zhang B, Zhang S, Zhang G. High Proportions of Radiation-Resistant Strains in Culturable Bacteria from the Taklimakan Desert. Biology. 2022; 11(4):501. https://doi.org/10.3390/biology11040501
Chicago/Turabian StyleLiu, Yang, Tuo Chen, Juan Li, Minghui Wu, Guangxiu Liu, Wei Zhang, Binglin Zhang, Songlin Zhang, and Gaosen Zhang. 2022. "High Proportions of Radiation-Resistant Strains in Culturable Bacteria from the Taklimakan Desert" Biology 11, no. 4: 501. https://doi.org/10.3390/biology11040501
APA StyleLiu, Y., Chen, T., Li, J., Wu, M., Liu, G., Zhang, W., Zhang, B., Zhang, S., & Zhang, G. (2022). High Proportions of Radiation-Resistant Strains in Culturable Bacteria from the Taklimakan Desert. Biology, 11(4), 501. https://doi.org/10.3390/biology11040501






