Application of Loop-Mediated Isothermal Amplification (LAMP) in Sex Identification of Parrots Bred in Egypt
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Methods
2.1. Feather Samples
2.2. DNA Isolation
2.3. Sex Identification with PCR
2.4. Optimization of LAMP for Sex Identification
2.5. PCR and LAMP Amplification Detection
2.6. Figures and Statistical Analysis
3. Results
3.1. Sample Collection and DNA Isolation
3.2. Parrot Sex Identification with PCR
3.3. Parrot Sex Identification with LAMP
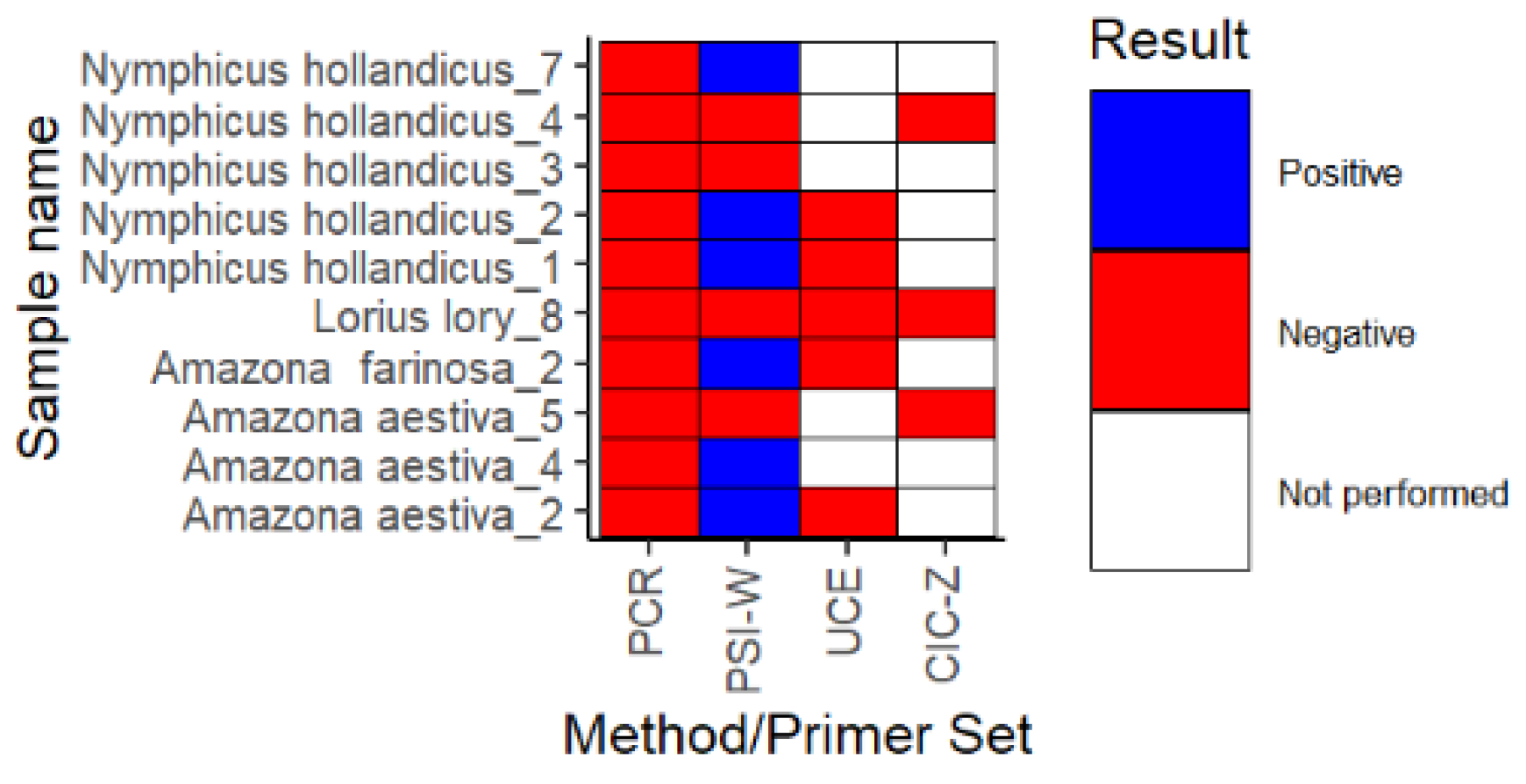
3.4. Comparison of PCR and LAMP Techniques for Sex Identification in Parrots
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Díaz Casana, C.F.; Vivas Ruíz, D.E.; Sandoval Peña, G.A.; Chimoy Effio, P.J. Molecular Sexing of the White-Winged Guan (Penelope albipennis) and Other Wild Birds of the North of Peru. Sex. Dev. Genet. Mol. Biol. Evol. Endocrinol. Embryol. Pathol. Sex Determ. Differ. 2019, 13, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Olah, G.; Butchart SH, M.; Symes, A.; Guzmán, I.M.; Cunningham, R.; Brightsmith, D.J.; Heinsohn, R. Ecological and socio-economic factors affecting extinction risk in parrots. Biodivers. Conserv. 2016, 25, 205–223. [Google Scholar] [CrossRef]
- Snyder, N.; McGowan PJ, K.; Gilardi, J.; Grajal, A. Parrots: Status Survey and Conservation Action Plan 2000–2004; IUCN: Gland, Switzerland, 2000; Available online: https://portals.iucn.org/library/node/7687 (accessed on 10 January 2021).
- Griffiths, R.; Double, M.C.; Orr, K.; Dawson, R.J. A DNA test to sex most birds. Mol. Ecol. 1998, 7, 1071–1075. [Google Scholar] [CrossRef] [PubMed]
- Cerit, H.; Avanus, K. Sex determination by CHDW and CHDZ genes of avian sex chromosomes in Nymphicus hollandicus. Turk. J. Vet. Anim. Sci. (Turk.) 2008, 31, 371–374. Available online: https://agris.fao.org/agris-search/search.do?recordID=TR2010000804 (accessed on 10 January 2021).
- Cerit, H.; Avanus, K. Sex identification in avian species using DNA typing methods. World’s Poult. Sci. J. 2007, 63, 91–100. [Google Scholar] [CrossRef]
- Olah, G.; Heinsohn, R.G.; Brightsmith, D.J.; Espinoza, J.R.; Peakall, R. Validation of non-invasive genetic tagging in two large macaw species (Ara macao and A. chloropterus) of the Peruvian Amazon. Conserv. Genet. Resour. 2016, 8, 499–509. [Google Scholar] [CrossRef]
- Jensen, T.; Pernasetti, F.M.; Durrant, B. Conditions for rapid sex determination in 47 avian species by PCR of genomic DNA from blood, shell-membrane blood vessels, and feathers. Zoo Biol. 2003, 22, 561–571. [Google Scholar] [CrossRef]
- Segelbacher, G.; Steinbrück, G. Bird faeces for sex identification and microsatellite analysis. Vogelwarte 2001, 41, 139–142. [Google Scholar]
- Bosnjak, J.; Stevanov-Pavlovic, M.; Vucicevic, M.; Stevanovic, J.; Simeunovic, P.; Resanovic, R.; Stanimirovic, Z. Feasibility of Non-Invasive Molecular Method for Sexing of Parrots. Pak. J. Zool. 2013, 45. Available online: https://go.gale.com/ps/i.do?p=AONE&sw=w&issn=00309923&v=2.1&it=r&id=GALE%7CA336556829&sid=googleScholar&linkaccess=abs (accessed on 10 January 2021).
- Griffiths, R.; Tlwarl, B. Sex of the last wild Spix’s macaw. Nature 1995, 375, 454. [Google Scholar] [CrossRef]
- Vucicevic, M.; Stevanov-Pavlovic, M.; Stevanovic, J.; Bosnjak, J.; Gajic, B.; Aleksic, N.; Stanimirovic, Z. Sex determination in 58 bird species and evaluation of CHD gene as a universal molecular marker in bird sexing. Zoo Biol. 2013, 32, 269–276. [Google Scholar] [CrossRef] [PubMed]
- Fridolfsson, A.-K.; Ellegren, H. A Simple and Universal Method for Molecular Sexing of Non-Ratite Birds. J. Avian Biol. 1999, 30, 116–121. [Google Scholar] [CrossRef]
- Kroczak, A.; Wołoszyńska, M.; Wierzbicki, H.; Kurkowski, M.; Grabowski, K.A.; Piasecki, T.; Galosi, L.; Urantówka, A.D. New Bird Sexing Strategy Developed in the Order Psittaciformes Involves Multiple Markers to Avoid Sex Misidentification: Debunked Myth of the Universal DNA Marker. Genes 2021, 12, 878. [Google Scholar] [CrossRef] [PubMed]
- Mataragka, A.; Balaskas, C.; Sotirakoglou, K.; Ikonomopoulos, J. Comparative evaluation of the performance of the PCR assays commonly used for the determination of sex in avian species. J. King Saud Univ.-Sci. 2020, 32, 228–234. [Google Scholar] [CrossRef]
- Wang, L.-C.; Chen, C.-T.; Lee, H.-Y.; Li, S.-H.; Lir, J.-T.; Chin, S.-C.; Pu, C.-E.; Wang, C.-H. Sexing a wider range of avian species based on two CHD1 introns with a unified reaction condition. Zoo Biol. 2007, 26, 425–431. [Google Scholar] [CrossRef]
- Centeno-Cuadros, A.; Abbasi, I.; Nathan, R. Sex determination in the wild: A field application of loop-mediated isothermal amplification successfully determines sex across three raptor species. Mol. Ecol. Resour. 2017, 17, 153–160. [Google Scholar] [CrossRef] [Green Version]
- Dhama, K.; Karthik, K.; Chakraborty, S.; Tiwari, R.; Kapoor, S.; Kumar, A.; Thomas, P. Loop-mediated isothermal amplification of DNA (LAMP): A new diagnostic tool lights the world of diagnosis of animal and human pathogens: A review. Pak. J. Biol. Sci. PJBS 2014, 17, 151–166. [Google Scholar] [CrossRef] [Green Version]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, E63. [Google Scholar] [CrossRef] [Green Version]
- Parida, M.; Sannarangaiah, S.; Dash, P.K.; Rao PV, L.; Morita, K. Loop mediated isothermal amplification (LAMP): A new generation of innovative gene amplification technique; perspectives in clinical diagnosis of infectious diseases. Rev. Med. Virol. 2008, 18, 407–421. [Google Scholar] [CrossRef]
- Mori, Y.; Nagamine, K.; Tomita, N.; Notomi, T. Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 2001, 289, 150–154. [Google Scholar] [CrossRef]
- Tomita, N.; Mori, Y.; Kanda, H.; Notomi, T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008, 3, 877–882. [Google Scholar] [CrossRef] [PubMed]
- Tanner, N.A.; Zhang, Y.; Evans, T.C. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. BioTechniques 2015, 58, 59–68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Centeno-Cuadros, A.; Tella, J.L.; Delibes, M.; Edelaar, P.; Carrete, M. Validation of loop-mediated isothermal amplification for fast and portable sex determination across the phylogeny of birds. Mol. Ecol. Resour. 2018, 18, 251–263. [Google Scholar] [CrossRef] [PubMed]
- Fair, J.M.; Paul, E.; Jones, J.; Clark, A.B.; Davie, C.; Kaiser, G. Guidelines to the Use of Wild Birds in Research, 3rd ed.; BIRDNET: Ithaca, NY, USA, 2010; Available online: https://birdnet.org/info-for-ornithologists/guidelines-english-3rd-edition-2010/ (accessed on 15 January 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis, 2nd ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- McNemar, Q. Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika 1947, 12, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Settele, J.; Brondízio, E.S.; Ngo, H.T.; Guèze, M.; Agard, J.; Arneth, A.; Balvanera, P.; Brauman, K.A.; Butchart, S.H.M.; et al. (Eds.) IPBES (2019): Summary for Policymakers of the Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; IPBES Secretariat: Bonn, Germany, 2019. [Google Scholar] [CrossRef]
- Teletchea, F. Wildlife Conservation: Is Domestication a Solution? In Global Exposition of Wildlife Management; InTech: London, UK, 2017; pp. 1–22. [Google Scholar] [CrossRef] [Green Version]
- Sulandart, S.; Zein, M.S.A. Application of Two Molecular Sexing Methods for Indonesian Bird Species: Implication for Captive Breeding Programs in Indonesia. HAYATI J. Biosci. 2012, 19, 183–190. [Google Scholar] [CrossRef] [Green Version]
- Purwaningrum, M.; Nugroho, H.A.; Asvan, M.; Karyanti, K.; Alviyanto, B.; Kusuma, R.; Haryanto, A. Molecular techniques for sex identification of captive birds. Vet. World 2019, 12, 1506–1513. [Google Scholar] [CrossRef]
- Çakmak, E.; Akın Pekşen, Ç.; Bilgin, C.C. Comparison of three different primer sets for sexing birds. J. Vet. Diagn. Investig. Off. Publ. Am. Assoc. Vet. Lab. Diagn. Inc. 2017, 29, 59–63. [Google Scholar] [CrossRef]
- Olah, G.; Heinsohn, R.G.; Brightsmith, D.J.; Peakall, R. The application of non-invasive genetic tagging reveals new insights into the clay lick use by macaws in the Peruvian Amazon. Conserv. Genet. 2017, 18, 1037–1046. [Google Scholar] [CrossRef]
- Campos, P.F.; Gilbert, T.M.P. DNA Extraction from Keratin and Chitin. In Ancient DNA: Methods and Protocols; Shapiro, B., Hofreiter, M., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 43–49. [Google Scholar] [CrossRef]
- Fernández, D.; Rodríguez, E.M.; Arroyo, G.H.; Padola, N.L.; Parma, A.E. Seasonal variation of Shiga toxin-encoding genes (stx) and detection of E. coli O157 in dairy cattle from Argentina. J. Appl. Microbiol. 2009, 106, 1260–1267. [Google Scholar] [CrossRef]
- Fernández-Soto, P.; Mvoulouga, P.O.; Akue, J.P.; Abán, J.L.; Santiago, B.V.; Sánchez, M.C.; Muro, A. Development of a highly sensitive loop-mediated isothermal amplification (LAMP) method for the detection of Loa loa. PLoS ONE 2014, 9, e94664. [Google Scholar] [CrossRef]
- Kaneko, H.; Kawana, T.; Fukushima, E.; Suzutani, T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. J. Biochem. Biophys. Methods 2007, 70, 499–501. [Google Scholar] [CrossRef] [PubMed]
- Nagamine, K.; Hase, T.; Notomi, T. Accelerated Reaction by Loop-Mediated Isothermal Amplification Using Loop Primers. Mol. Cell Probes 2002, 16, 223–229. [Google Scholar] [CrossRef] [PubMed]


| Family | Genus | Species | Common Name | Status by IUCN 2019 | Number of Birds |
|---|---|---|---|---|---|
| Psittacidae | Ara | Ara ararauna | Blue-and-yellow Macaw | Near Threatened | 5 |
| Ara chloropterus | Red-and-green Macaw | Least Concern | 8 | ||
| Ara militaris | Military Macaw | Vulnerable | 2 | ||
| Amazona | Amazona aestiva | Blue-fronted Amazon | Least Concern | 5 | |
| Amazona amazonica | Orange-winged Amazon | --------- | 5 | ||
| Amazona farinosa | Southern mealy Amazon | Near Threatened | 3 | ||
| Aratinga | Aratinga solstitialis | Sun Parakeet (Sun conure) | Endangered | 5 | |
| Diopsittaca | Diopsittaca nobilis | Northern Red-shouldered Macaw | Least Concern | 8 | |
| Lorius | Lorius lory | Lory | --------- | 10 | |
| Myiopsitta | Myiopsitta monachus | Monk Parakeet | Least Concern | 12 | |
| Poicephalus meyeri | Meyer’s Parrot (red eyes) | Least Concern | 4 | ||
| Poicephalus senegalus | Senegal Parrot | Least Concern | 10 | ||
| Psittacus | Psittacus erithacus timneh | Timneh Parrot | Endangered | 4 | |
| Psittacus erithacus | African Grey Parrot | Endangered | 4 | ||
| Pyrrhura | Pyrrhura molinae | Green-cheeked Parakeet | Least Concern | 11 | |
| Cacatuidae | Nymphicus | Nymphicus hollandicus | Cockatiel | Least Concern | 12 |
| Cacatua | Cacatua sulphurea | Yellow-crested Cockatoo | Critically Endangered | 5 | |
| Psittaculidae | Melopsittacus | Melopsittacus undulatus | Budgerigar | Least Concern | 10 |
| Agaprins | Agapornis fischeri | Fischer’s lovebird | Least Concern | 15 | |
| Psittacula | Psittacula eupatria | Alexandrine Parakeet | Near Threatened | 4 | |
| Psittacula krameri | Rose-ringed Parakeet | Least Concern | 8 |
| Primer Set | Primer Name | Oligonucleotide Sequence (5′→3′) | Source |
|---|---|---|---|
| PCR | 2550F | GTTACTGATTCGTCTACGAGA | [13] |
| 2718R | ATTGAAATGATCCAGTGCTTG | ||
| UCE | UCE-F3 | GGGAAACAAGGATAAAATTACTCC | [24] |
| UCE-B3 | TGCCCAGAAAATTCCATTC | ||
| UCE-FIP | CGAGTGTGTTAAGCACAGTTTTATTTTTTATGGTTAATGA CCTATAGTATCTCC | ||
| UCE-BIP | GAGGACTGTTCTGCAGGGTATTTTTTTGCTATCTGATTCGAAAAGTC | ||
| PSI-W | PSI-W-F3 | CAGTTTCCCTTTCAGGTAAG | [24] |
| PSI-W-B3 | TCAGTTGCCAAAACAATGG | ||
| PSI-W-FIP | TTCTTCACAAAGGACACTTTTCTTTTTGTAGTAGCCAAGAAGCCTT | ||
| PSI-W-BIP | AGGAAAAGACTGGCAATTACTATATGCTAATTTTGGGGAGATAAGATTAATGTAACA | ||
| CIC-Z | CIC-Z-F3 | TCACAGAAGATGGAGATTCC | [24] |
| CIC-Z-B3 | CAACAGAGTTCTGATTTTCTCA | ||
| CIC-Z-FIP | GCCAAGAAGCTTTGGTCTTGAACTTTTCTCTGGACAACTTGTTCAGT | ||
| CIC-Z-BIP | ACTACCACCAAGATTCATACCTGATTTTCAGATGGTGAGGATGCTG |
| Family | Species | Common Name | Positive Control UCE | Sexing | PCR Optimal Annealing Temperature | |
|---|---|---|---|---|---|---|
| PSI-W | CIC-Z | |||||
| Psittacidae | Ara ararauna | Blue-and-yellow Macaw | --- | 57 °C/1 h | --- | 56 °C |
| Ara chloropterus | Red-and-green Macaw | --- | 57 °C/1 h | 63 °C/1 h | 56 °C | |
| Ara militaris | Military Macaw | --- | 57 °C/1 h | --- | 56 °C | |
| Amazona aestiva | Blue-fronted Amazon | --- | 63 °C/1 h | --- | 50 °C | |
| Amazona amazonica | Orange-winged Amazon | 57 °C/1 h | 63 °C/1 h | 63 °C/1 h | 56 °C | |
| Amazona farinosa | Southern mealy Amazon | --- | 63 °C/1 h | --- | 56 °C | |
| Aratinga solstitialis | Sun Parakeet (sun conure) | 57 °C/1 h | 60 °C/1 h | --- | 52 °C | |
| Diopsittaca nobilis | Northern Red-shouldered Macaw | --- | 63 °C/1 h | --- | 52 °C | |
| Lorius lory | Lory | 57 °C/1 h | 63 °C/1 h | 63 °C/1 h | 52 °C | |
| Myiopsitta monachus | Monk Parakeet | --- | 57 °C/1 h | 63 °C/1 h | 52 °C | |
| Poicephalus meyeri | Meyer’s Parrot (red eyes) | --- | 57 °C/1 h | 63 °C/1 h | 56 °C | |
| Poicephalus senegalus | Senegal Parrot | --- | 63 °C/1 h | 63 °C/1 h | 56 °C | |
| Psittacus erithacus | African Grey Parrot | --- | 60 °C/1 h | 63 °C/1 h | 50 °C | |
| Psittacus erithacus timneh | Timneh Parrot | --- | 57 °C/1 h | --- | 50 °C | |
| Pyrrhura molinae | Green-cheeked Parakeet | --- | 57 °C/1 h | 63 °C/1 h | 52 °C | |
| Cacatuidae | Nymphicus hollandicus | Cockatiel | --- | 57 °C/1 h | --- | 50 °C |
| Cacatua sulphurea | Yellow-crested Cockatoo | --- | 57 °C/1 h | 63 °C/1 h | 56 °C | |
| Psittaculidae | Melopsittacus undulatus | Budgerigar | --- | 63 °C/1 h | 63 °C/1 h | 52 °C |
| Agapornis fischeri | Fischer’s lovebird | 57 °C/1 h | 57 °C/1 h | 63 °C/1 h | 52 °C | |
| Psittacula eupatria | Alexandrine Parakeet | --- | 60 °C/1 h | --- | 56 °C | |
| Psittacula krameri | Rose-ringed Parakeet | 57 °C/1 h | 57 °C/1 h | --- | 56 °C | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elnomrosy, S.M.; Hagag, N.M.; AbdAllah, M.I.; Kolenda, R.; Zacharski, M. Application of Loop-Mediated Isothermal Amplification (LAMP) in Sex Identification of Parrots Bred in Egypt. Biology 2022, 11, 565. https://doi.org/10.3390/biology11040565
Elnomrosy SM, Hagag NM, AbdAllah MI, Kolenda R, Zacharski M. Application of Loop-Mediated Isothermal Amplification (LAMP) in Sex Identification of Parrots Bred in Egypt. Biology. 2022; 11(4):565. https://doi.org/10.3390/biology11040565
Chicago/Turabian StyleElnomrosy, Sara M., Naglaa M. Hagag, Mohamed I. AbdAllah, Rafał Kolenda, and Maciej Zacharski. 2022. "Application of Loop-Mediated Isothermal Amplification (LAMP) in Sex Identification of Parrots Bred in Egypt" Biology 11, no. 4: 565. https://doi.org/10.3390/biology11040565






