Transcriptome Profiling to Dissect the Role of Genome Duplication on Graft Compatibility Mechanisms in Watermelon
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Grafting Methods, Growth Conditions, and Sampling
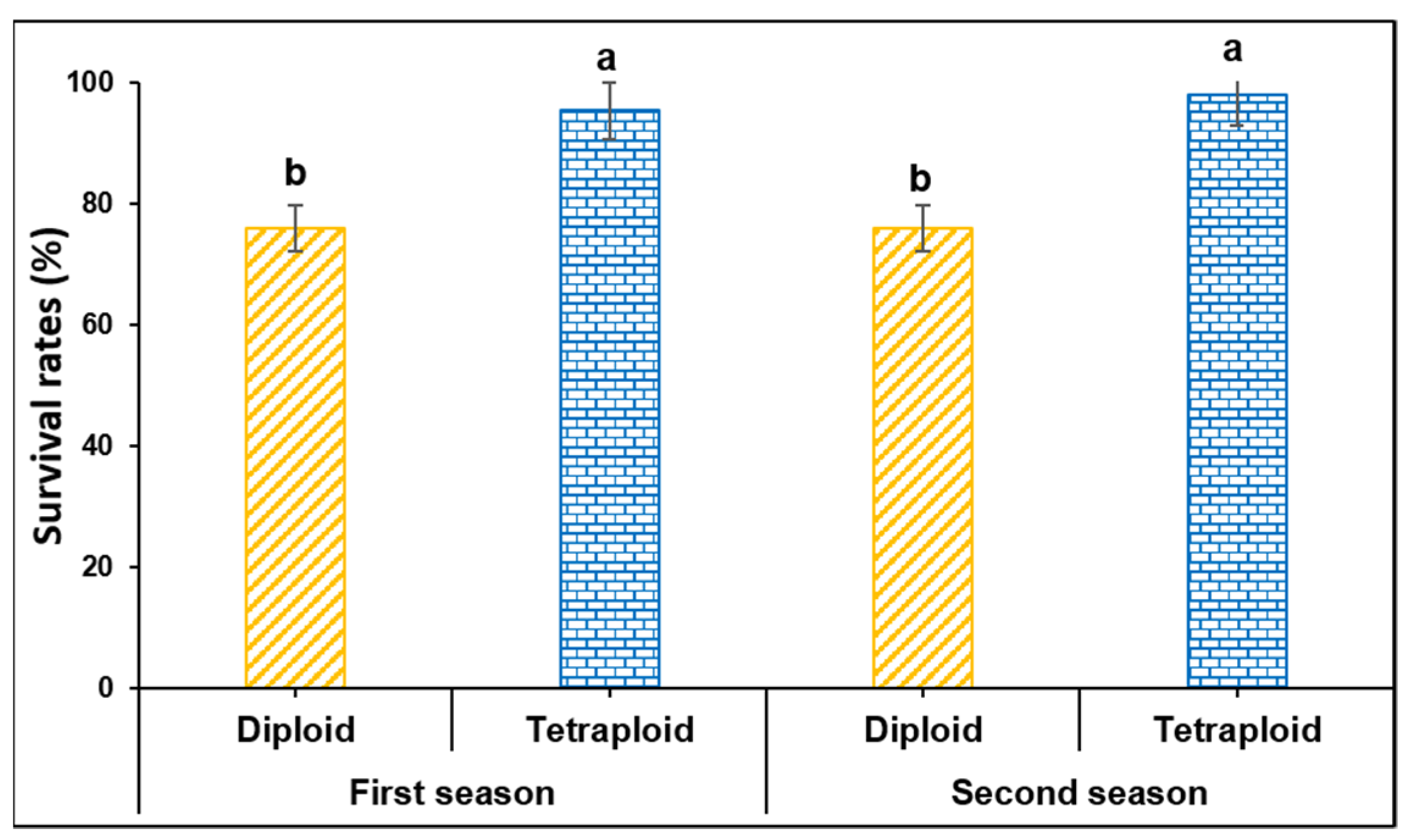
2.3. Determination of Survival Rate
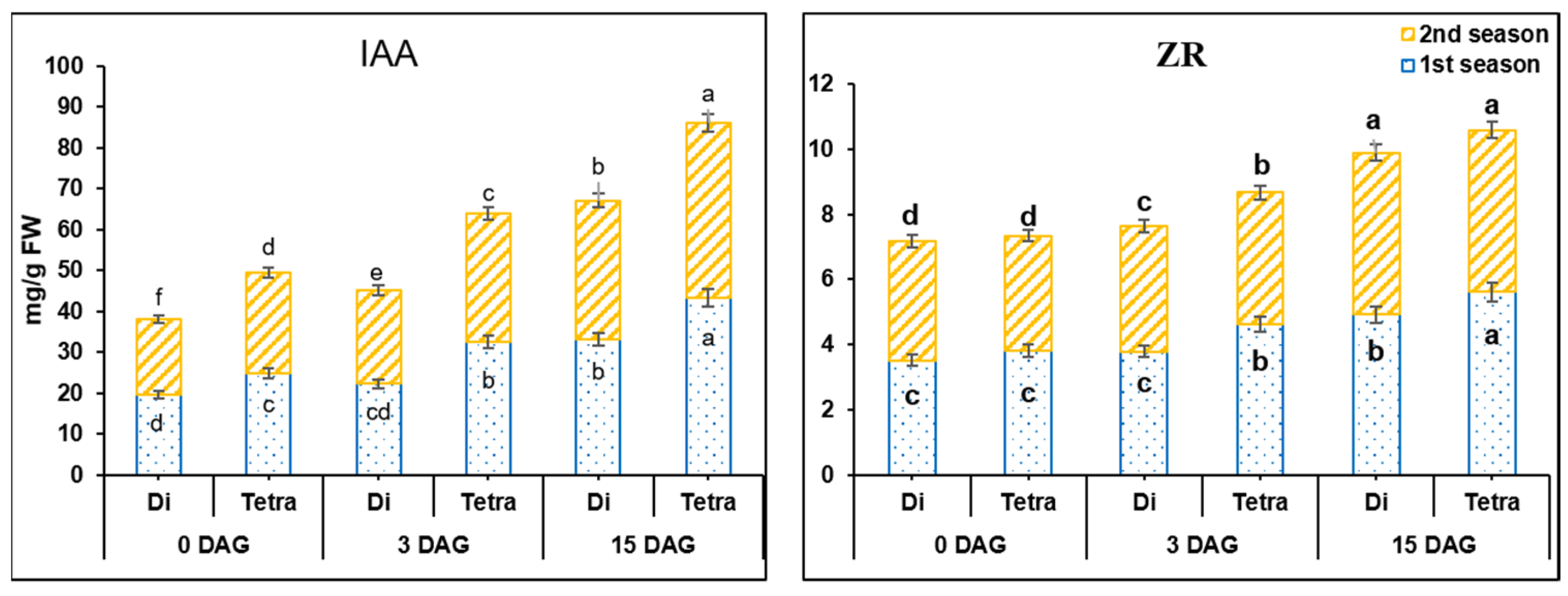
2.4. Phytohormone Determinations
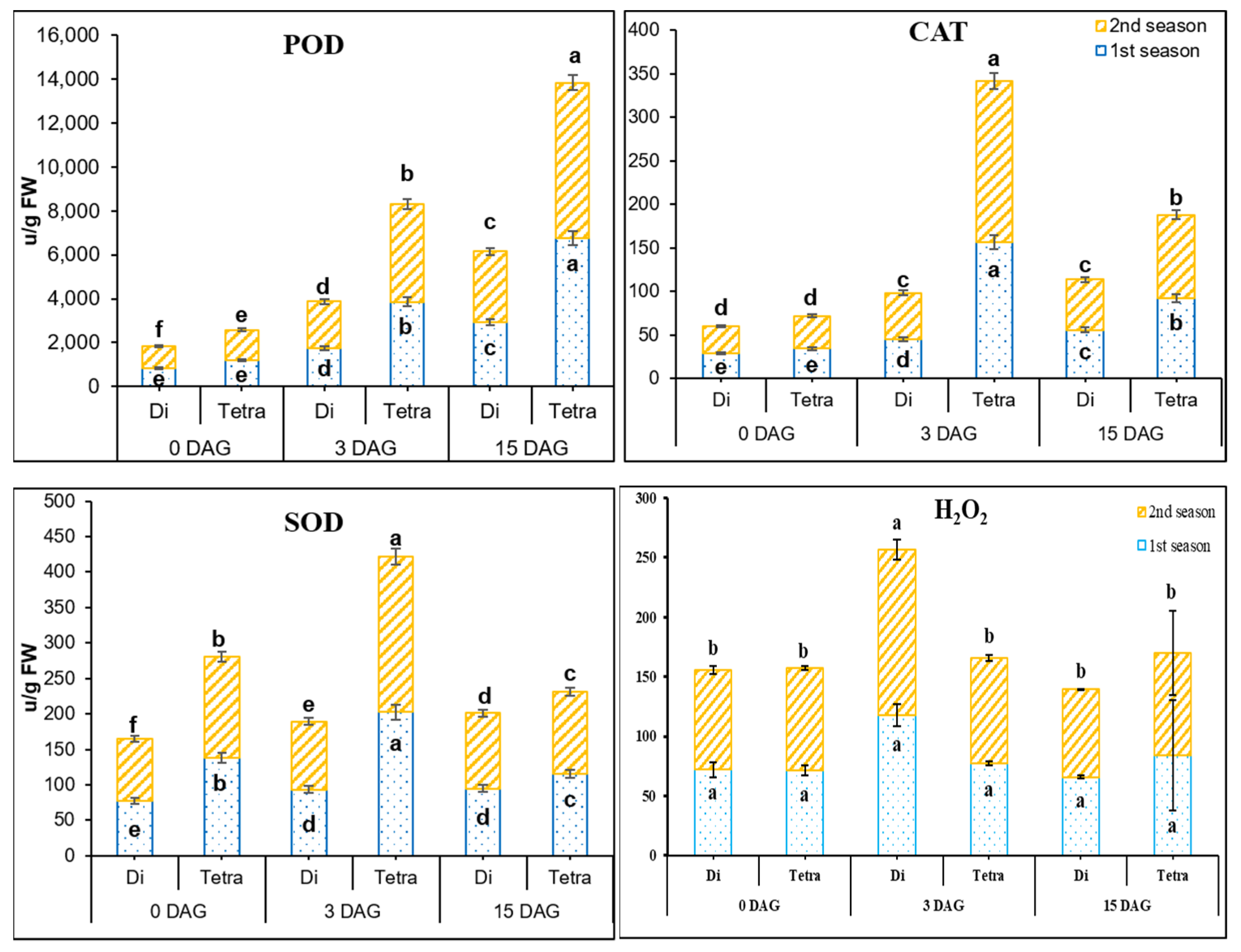
2.5. Assay for Antioxidant Enzymes’ Activity and H2O2 Contents
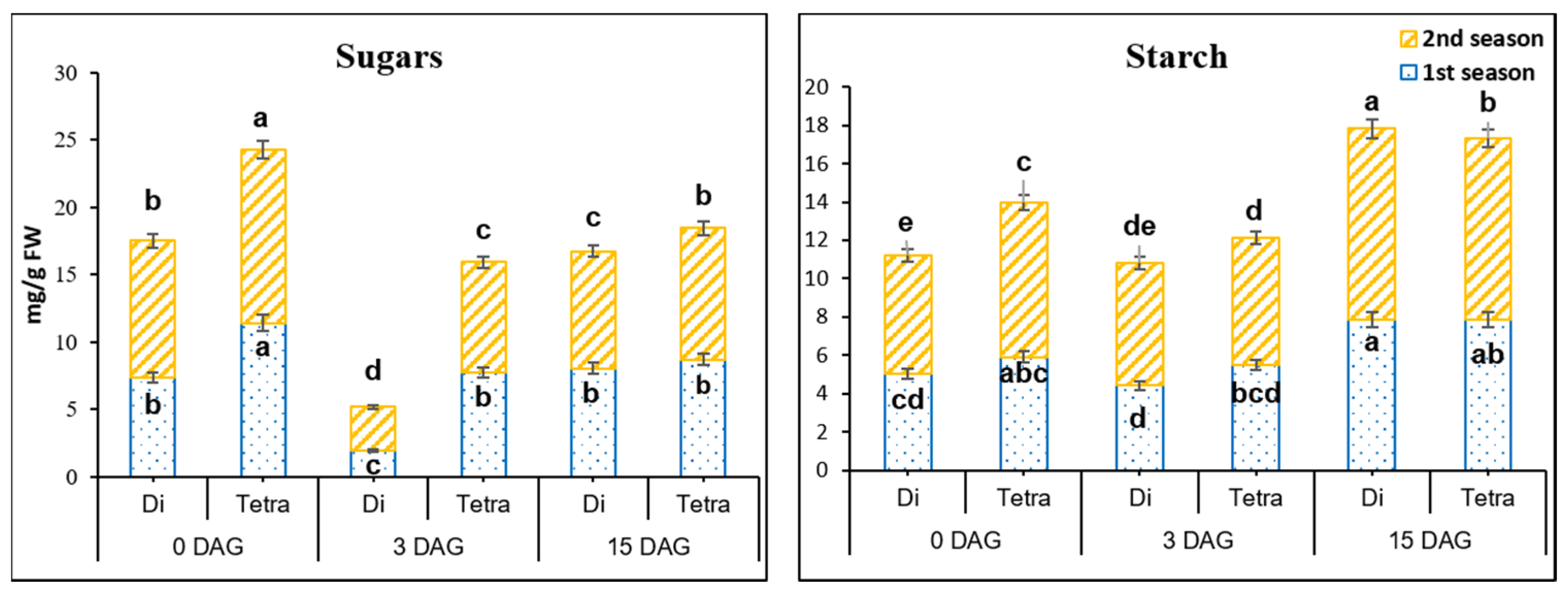
2.6. Quantification of Sugars and Starch
2.7. Construction and Sequencing of the RNA-Sequencing Library
2.8. Mapping to Watermelon Genome V2 and Quantification of Gene Expression
2.9. Enrichment Analysis of DEGs and Weighted Gene Coexpression Network Analysis (WGCNA) for Identifying Correlated Gene Networks
2.10. Validation of Intramodular Candidates through RT-qPCR Analysis
2.11. Statistical Analysis
3. Results
3.1. Survival Rates of Diploid and Tetraploid Watermelon Grafted by Splice Method
3.2. Measurement of IAA and ZR in the Grafting Union among Diploid and Tetraploid Plants of Watermelon during the Grafting Process
3.3. Measurement of POD, SOD, CAT, and H2O2 Contents in the Grafting Union among Diploid and Tetraploid Plants of Watermelon
3.4. Sugars’ and Starch Contents in the Grafting Union among Diploid and Tetraploid Plants of Watermelon during the Grafting Process
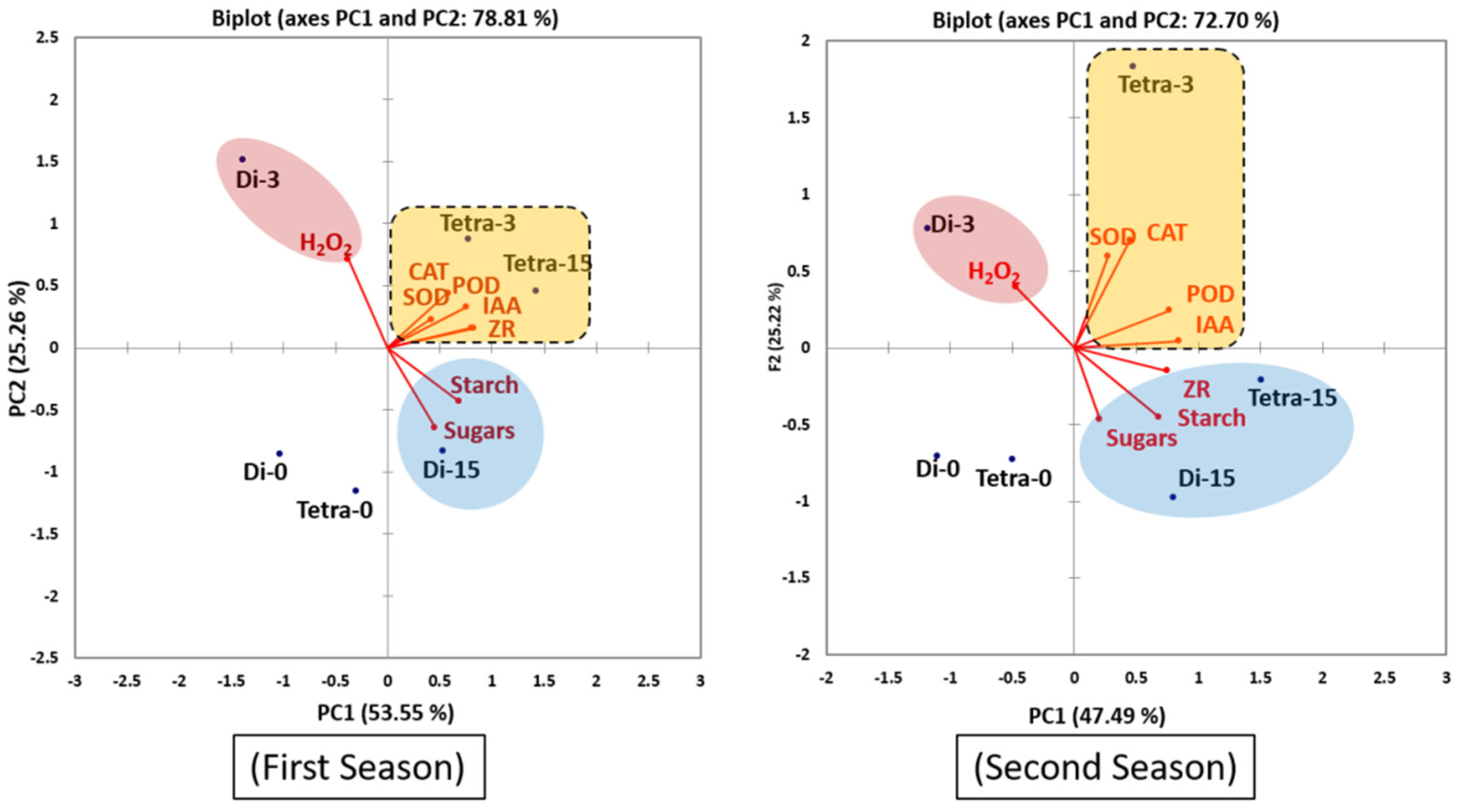
3.5. Principal Component Analysis (PCA)
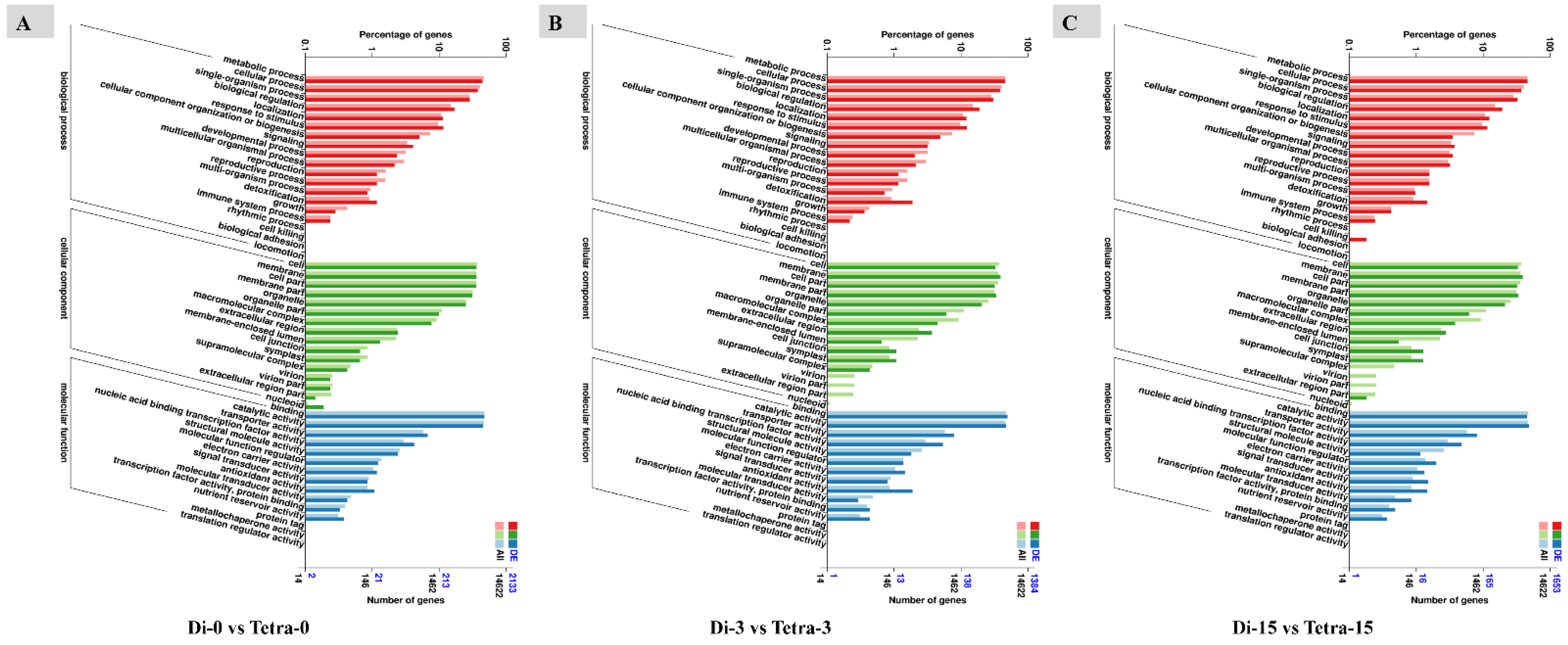
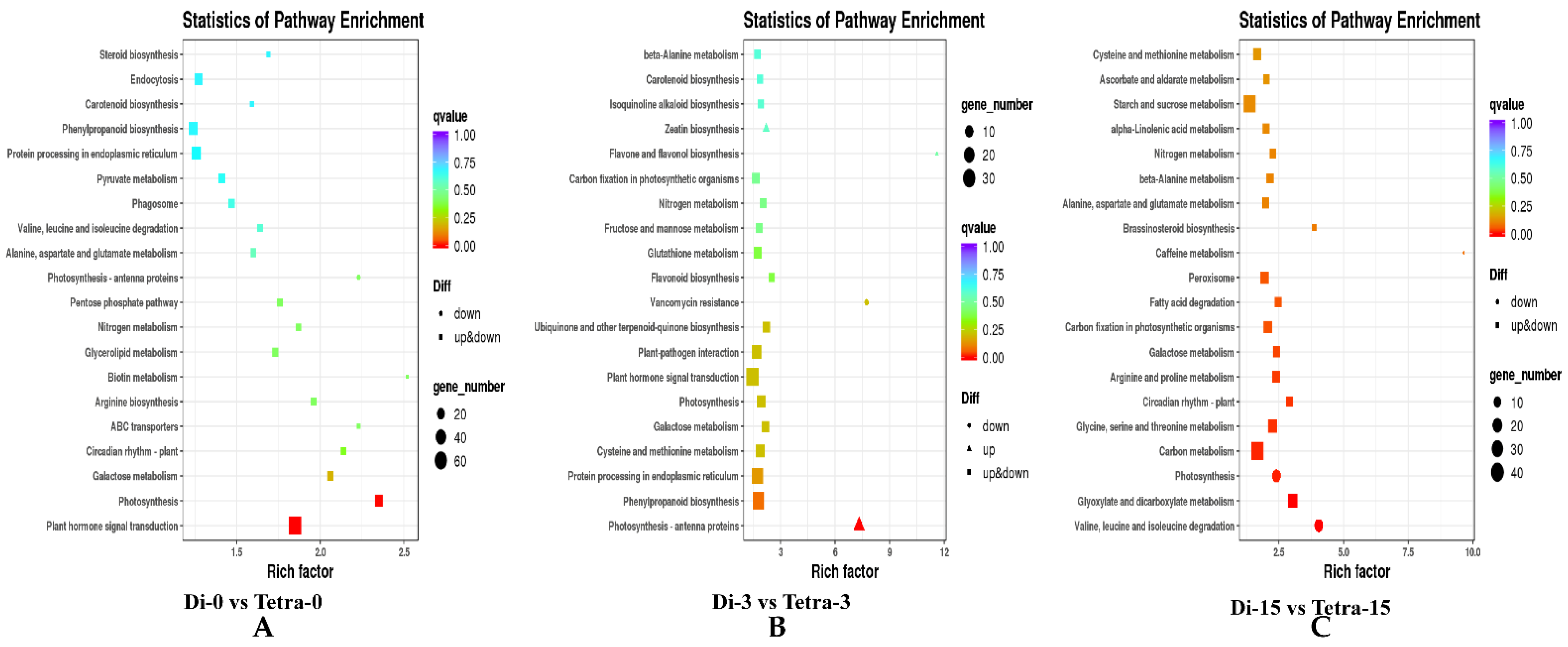
3.6. Genome-Wide Transcriptomic Analyses of Diploid and Tetraploid Plants of Watermelon
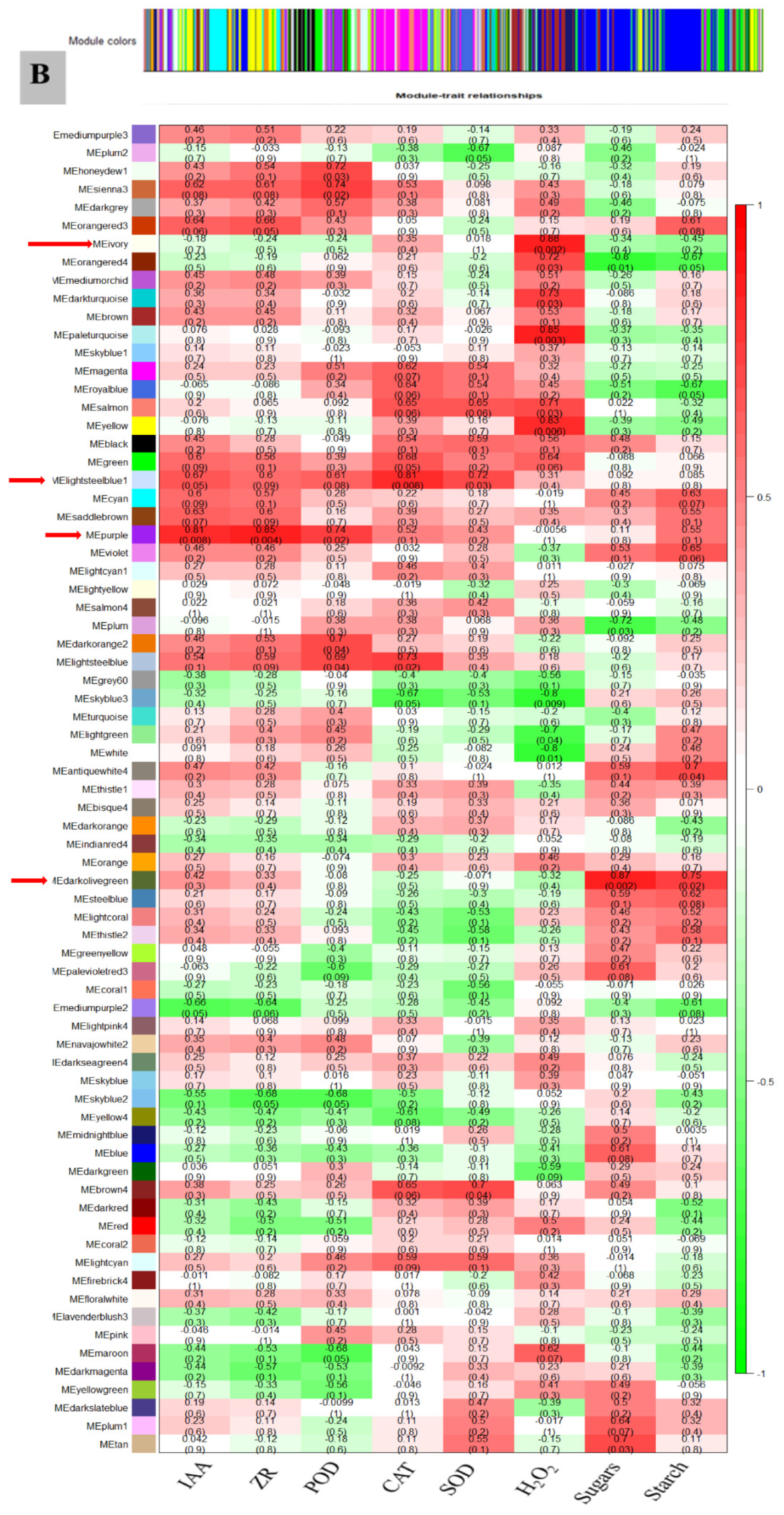
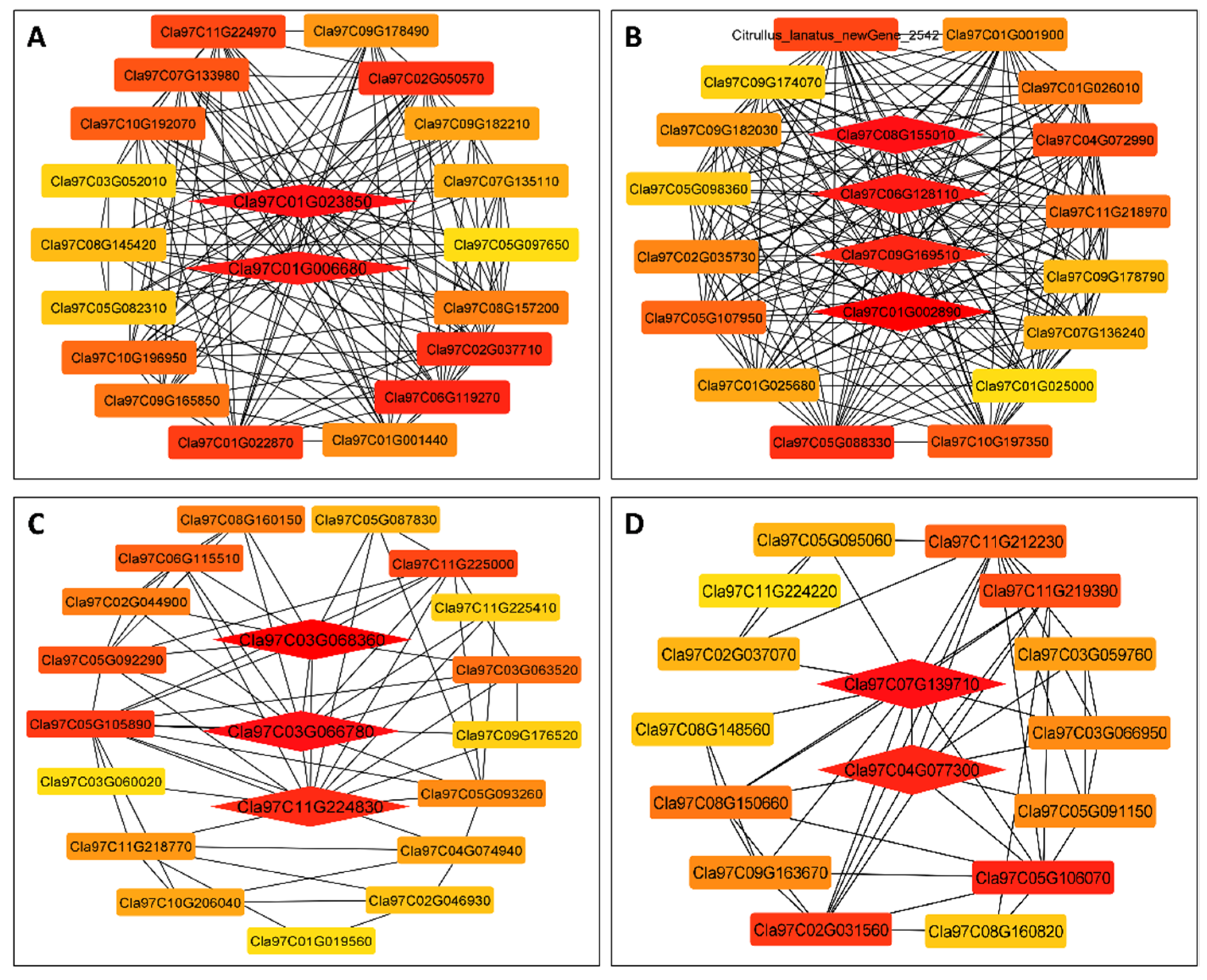
3.7. WGCNA for the Identification of Highly Connected “Hub Genes” and Correlated Gene Modules
3.8. Module Analysis Based on WGCNA
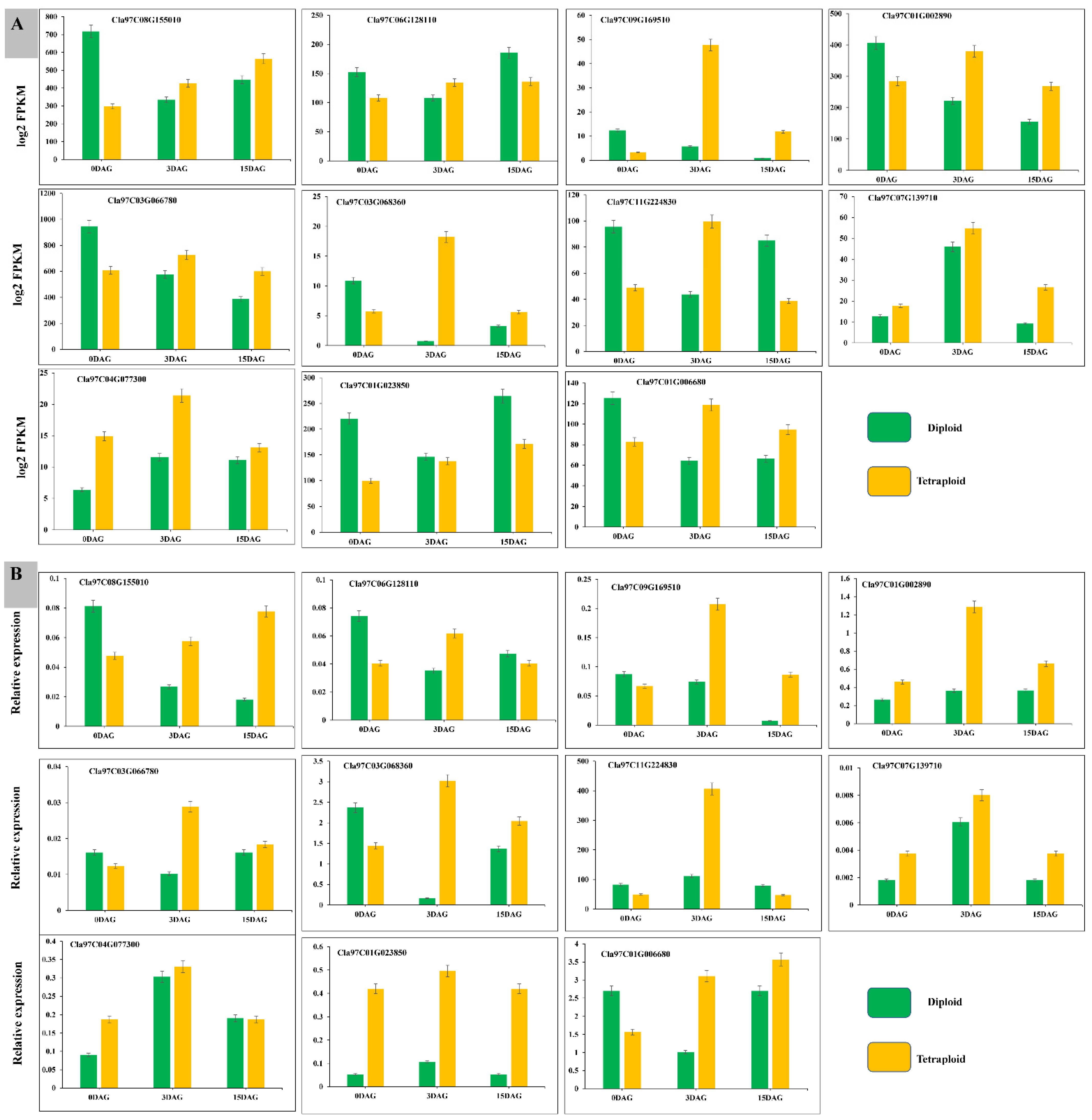
3.9. RT-qPCR to Verify the Accuracy of Transcriptome Data
4. Discussion
4.1. Role of Hormones, AOX and Carbohydrates in the Graft Union during the Grafting Seedling Process in Tetraploid Compared to Diploid Watermelon
4.2. Transcriptome Sequencing and DEG Screening in Diploid and Tetraploid Plants of Watermelon
4.3. Role of Genes Related to Hormonal Signaling in Rootstock–Scion Interaction
4.4. Expression of Antioxidant Enzymes and Growth-Regulating Factors
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Meng, J.; Wu, S.; Wang, X.; Yu, X.; Jiang, R. Effects of Different Rootstocks on Plant Growth and Fruit Quality of Watermelon. Agric. Biotechnol. 2019, 8, 64–68. [Google Scholar] [CrossRef]
- Marr, C.W.; Gast, K.L. Reactions by consumers in a farmers’ market to prices for seedless watermelon and ratings of eating quality. HortTechnology 1991, 1, 105–106. [Google Scholar] [CrossRef] [Green Version]
- Jaskani, M.J.; Kwon, S.W.; Kim, D.H. Comparative study on vegetative, reproductive and qualitative traits of seven diploid and tetraploid watermelon lines. Euphytica 2005, 145, 259–268. [Google Scholar] [CrossRef]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The polyploidy and its key role in plant breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.; Sheikh, S.; Chon, H.G.; Seong, M.H.; Lim, J.H.; Lee, S.G.; Jung, G.T.; Kim, J.M.; Ju, H.-J.; Huh, Y.C. Screening different methods of tetraploid induction in watermelon [Citrullus lanatus (thunb.) Manst. and Nakai]. Hortic. Environ. Biotechnol. 2012, 53, 521–529. [Google Scholar] [CrossRef]
- Zhang, N.; Bao, Y.; Xie, Z.; Huang, X.; Sun, Y.; Feng, G.; Zeng, H.; Ren, J.; Li, Y.; Xiong, J. Efficient Characterization of Tetraploid Watermelon. Plants 2019, 8, 419. [Google Scholar] [CrossRef] [Green Version]
- Hassan, J.; Miyajima, I.; Ozaki, Y.; Mizunoe, Y.; Sakai, K.; Zaland, W. Tetraploid Induction by Colchicine Treatment and Crossing with a Diploid Reveals Less-Seeded Fruit Production in Pointed Gourd (Trichosanthes dioica Roxb.). Plants 2020, 9, 370. [Google Scholar] [CrossRef] [Green Version]
- Doyle, J.J.; Coate, J.E. Polyploidy, the nucleotype, and novelty: The impact of genome doubling on the biology of the cell. Int. J. Plant Sci. 2019, 180, 1–52. [Google Scholar] [CrossRef]
- Budhani, A.; Bhanderi, D.R.; Saravaiya, S.N.; Patel, A.I.; Patel, K. Polyploidy breeding in vegetable and tuber crops—A review. Int. J. Sci. Environ. 2018, 7, 968–972. [Google Scholar]
- Comai, L. The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Soltis, P.S.; Marchant, D.B.; Van de Peer, Y.; Soltis, D.E. Polyploidy and genome evolution in plants. Curr. Opin. Genet. Dev. 2015, 35, 119–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, K.L.; Wendel, J.F. Polyploidy and genome evolution in plants. Curr. Opin. Plant Biol. 2005, 8, 135–141. [Google Scholar] [CrossRef] [PubMed]
- Soltis, D.E.; Misra, B.B.; Shan, S.; Chen, S.; Soltis, P.S. Polyploidy and the proteome. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2016, 1864, 896–907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Zhao, S.; Cheng, Z.; Wan, X.; Yan, Z.; King, S. Lycopene and citrulline contents in watermelon (Citrullus lanatus) fruit with different ploidy and changes during fruit development. Acta Hortic. 2010, 871, 543–550. [Google Scholar] [CrossRef] [Green Version]
- Duval, J.R.; NeSmith, D.S. Emergence ofGenesis’ triploid watermelon following mechanical scarification. J. Am. Soc. Hortic. Sci. 1999, 124, 430–432. [Google Scholar] [CrossRef] [Green Version]
- Raza, H.; Jaskani, M.J.; Khan, M.M.; Malik, T.A. In vitro induction of polyploids in watermelon and estimation based on DNA content. Int. J. Agri. Biol 2003, 5, 298–302. [Google Scholar]
- Estilai, A.; Shannon, M.C. Salt tolerance in relation to ploidy level in guayule. In New Crops; Janick, J., Simon, J.E., Eds.; Wiley: New York, NY, USA, 1993; pp. 349–351. [Google Scholar]
- Zhu, H.; Zhao, S.; Lu, X.; He, N.; Gao, L.; Dou, J.; Bie, Z.; Liu, W. Genome duplication improves the resistance of watermelon root to salt stress. Plant Physiol. Biochem. 2018, 133, 11–21. [Google Scholar] [CrossRef]
- Jiang, A.; Gan, L.; Tu, Y.; Ma, H.; Zhang, J.; Song, Z.; He, Y.; Cai, D.; Xue, X. The effect of genome duplication on seed germination and seedling growth of rice under salt stress. Aust. J. Crop Sci. 2013, 7, 1814. [Google Scholar]
- Tu, Y.; Jiang, A.; Gan, L.; Hossain, M.; Zhang, J.; Peng, B.; Xiong, Y.; Song, Z. Genome duplication improves rice root resistance to salt stress. Rice 2014, 7, 15. [Google Scholar] [CrossRef] [Green Version]
- Ruiz, M.; Quiñones, A.; Martínez-Alcántara, B.; Aleza, P.; Morillon, R.; Navarro, L.; Primo-Millo, E.; Martínez-Cuenca, M.-R. Effects of salinity on diploid (2x) and doubled diploid (4x) Citrus macrophylla genotypes. Sci. Hortic. 2016, 207, 33–40. [Google Scholar] [CrossRef]
- Meng, F.; Luo, Q.; Wang, Q.; Zhang, X.; Qi, Z.; Xu, F.; Lei, X.; Cao, Y.; Chow, W.S.; Sun, G. Physiological and proteomic responses to salt stress in chloroplasts of diploid and tetraploid black locust (Robinia pseudoacacia L.). Sci. Rep. 2016, 6, 23098. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, K.; Wu, C.; Zhang, L.; Chen, X. Contrasting photosynthesis and photoinhibition in tetraploid and its autodiploid honeysuckle (Lonicera japonica Thunb.) under salt stress. Front. Plant Sci. 2015, 6, 227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balal, R.M.; Shahid, M.A.; Vincent, C.; Zotarelli, L.; Liu, G.; Mattson, N.S.; Rathinasabapathi, B.; Martínez-Nicolas, J.J.; Garcia-Sanchez, F. Kinnow mandarin plants grafted on tetraploid rootstocks are more tolerant to Cr-toxicity than those grafted on its diploids one. Environ. Exp. Bot. 2017, 140, 8–18. [Google Scholar] [CrossRef]
- Said, J.I.; Lin, Z.; Zhang, X.; Song, M.; Zhang, J. A comprehensive meta QTL analysis for fiber quality, yield, yield related and morphological traits, drought tolerance, and disease resistance in tetraploid cotton. BMC Genom. 2013, 14, 776. [Google Scholar] [CrossRef] [Green Version]
- Allario, T.; Brumos, J.; Colmenero-Flores, J.M.; Iglesias, D.J.; Pina, J.A.; Navarro, L.; Talon, M.; Ollitrault, P.; Morillon, R. Tetraploid Rangpur lime rootstock increases drought tolerance via enhanced constitutive root abscisic acid production. Plant Cell Env. 2013, 36, 856–868. [Google Scholar] [CrossRef] [Green Version]
- Savvas, D.; Colla, G.; Rouphael, Y.; Schwarz, D. Amelioration of heavy metal and nutrient stress in fruit vegetables by grafting. Sci. Hortic. 2010, 127, 156–161. [Google Scholar] [CrossRef]
- Bantis, F.; Dangitsis, C.; Koukounaras, A. Influence of Light Spectra from LEDs and Scion× Rootstock Genotype Combinations on the Quality of Grafted Watermelon Seedlings. Plants 2021, 10, 353. [Google Scholar] [CrossRef]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [Green Version]
- Li, H.L.; Zhang, H.; Yu, C.; Ma, L.; Wang, Y.; Zhang, X.Z.; Han, Z.H. Possible roles of auxin and zeatin for initiating the dwarfing effect of M9 used as apple rootstock or interstock. Acta Physiol. Plant. 2012, 34, 235–244. [Google Scholar] [CrossRef]
- Liu, X.-Y.; Li, J.; Liu, M.-M.; Yao, Q.; Chen, J.-Z. Transcriptome profiling to understand the effect of citrus rootstocks on the growth of ‘Shatangju’Mandarin. PLoS ONE 2017, 12, e0169897. [Google Scholar] [CrossRef] [Green Version]
- Prassinos, C.; Ko, J.-H.; Lang, G.; Iezzoni, A.F.; Han, K.-H. Rootstock-induced dwarfing in cherries is caused by differential cessation of terminal meristem growth and is triggered by rootstock-specific gene regulation. Tree Physiol. 2009, 29, 927–936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cookson, S.J.; Ollat, N. Grafting with rootstocks induces extensive transcriptional re-programming in the shoot apical meristem of grapevine. BMC Plant Biol. 2013, 13, 147. [Google Scholar] [CrossRef] [PubMed]
- Kawaguchi, M.; Taji, A.; Backhouse, D.; Oda, M. Anatomy and physiology of graft incompatibility in solanaceous plants. J. Hortic. Sci. Biotechnol. 2008, 83, 581–588. [Google Scholar] [CrossRef]
- El-Gazzar, T.; Dawa, K.K.; Ibrahim, E.; El-Banna, M.; Mohamed, A. Anatomical Study on Watermelon Grafting. J. Plant Prod. 2017, 8, 999–1009. [Google Scholar] [CrossRef] [Green Version]
- Assunção, M.; Canas, S.; Cruz, S.; Brazão, J.; Zanol, G.C.; Eiras-Dias, J.E. Graft compatibility of Vitis spp.: The role of phenolic acids and flavanols. Sci. Hortic. 2016, 207, 140–145. [Google Scholar] [CrossRef]
- Pina, A.; Cookson, S.J.; Calatayud, A.; Trinchera, A.; Errea, P. Physiological and molecular mechanisms underlying graft compatibility. In Vegetable Grafting: Principles and Practices; CABI: London, UK, 2017; pp. 132–154. [Google Scholar] [CrossRef]
- Dogra, K.; Kour, K.; Kumar, R.; Bakshi, P.; Kumar, V. Graft-Incompatibility in Horticultural Crops Graft-Incompatibility in Horticultural Crops. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1805–1820. [Google Scholar] [CrossRef]
- Melnyk, C.W. Plant grafting: Insights into tissue regeneration. Regeneration 2017, 4, 3–14. [Google Scholar] [CrossRef]
- Mo, Z.; Feng, G.; Su, W.; Liu, Z.; Peng, F. Transcriptomic Analysis Provides Insights into Grafting Union Development in Pecan (Carya illinoinensis). Genes 2018, 9, 71. [Google Scholar] [CrossRef] [Green Version]
- Davis, A.R.; Perkins-Veazie, P.; Sakata, Y.; López-Galarza, S.; Maroto, J.V.; Lee, S.-G.; Huh, Y.-C.; Sun, Z.; Miguel, A.; King, S.R.; et al. Cucurbit Grafting. Crit. Rev. Plant Sci. 2008, 27, 50–74. [Google Scholar] [CrossRef]
- Lu, S.; Song, Y. Relation between phytohormone level and vascular bridge differentiation in graft union of explanted internode autografting. Chin. Sci. Bull. 1999, 44, 1874–1878. [Google Scholar] [CrossRef]
- Ren, Y.; Guo, S.R.; Shu, S.; Xu, Y.; Sun, J. Isolation and expression pattern analysis of CmRNF5 and CmNPH3L potentially involved in graft compatibility in cucumber/pumpkin graft combinations. Sci. Hortic. 2018, 227, 92–101. [Google Scholar] [CrossRef]
- Ren, Y.; Xu, Q.; Wang, L.; Guo, S.; Shu, S.; Lu, N.; Sun, J. Involvement of metabolic, physiological and hormonal responses in the graft-compatible process of cucumber/pumpkin combinations was revealed through the integrative analysis of mRNA and miRNA expression. Plant Physiol. Biochem. 2018, 129, 368–380. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zhao, J.; Hu, F.; Qin, Y.; Wang, X.; Hu, G. Transcriptome changes between compatible and incompatible graft combination of Litchi chinensis by digital gene expression profile. Sci. Rep. 2017, 7, 3954. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnyk, C.W.; Gabel, A.; Hardcastle, T.J.; Robinson, S.; Miyashima, S.; Grosse, I.; Meyerowitz, E.M. Transcriptome dynamics at Arabidopsis graft junctions reveal an intertissue recognition mechanism that activates vascular regeneration. Proc. Natl. Acad. Sci. USA 2018, 115, E2447–E2456. [Google Scholar] [CrossRef] [Green Version]
- Tanimoto, E. Regulation of root growth by plant hormones—roles for auxin and gibberellin. Crit. Rev. Plant Sci. 2005, 24, 249–265. [Google Scholar] [CrossRef]
- Taiz, L.; Zeiger, E.; Møller, I.M.; Murphy, A. Plant Physiology and Development; Sinauer Associates Incorporated, Inc.: Sunderland, MA, USA, 2015. [Google Scholar]
- Pilet, P.-E.; Saugy, M. Effect on root growth of endogenous and applied IAA and ABA: A critical reexamination. Plant Physiol. 1987, 83, 33–38. [Google Scholar] [CrossRef]
- Zafar, S.A.; Patil, S.B.; Uzair, M.; Fang, J.; Zhao, J.; Guo, T.; Yuan, S.; Uzair, M.; Luo, Q.; Shi, J.; et al. DEGENERATED PANICLE AND PARTIAL STERILITY 1 (DPS1) encodes a cystathionine β-synthase domain containing protein required for anther cuticle and panicle development in rice. New Phytol. 2020, 225, 356–375. [Google Scholar] [CrossRef] [Green Version]
- Hussain, H.A.; Men, S.; Hussain, S.; Chen, Y.; Ali, S.; Zhang, S.; Zhang, K.; Li, Y.; Xu, Q.; Liao, C.; et al. Interactive effects of drought and heat stresses on morpho-physiological attributes, yield, nutrient uptake and oxidative status in maize hybrids. Sci. Rep. 2019, 9, 3890. [Google Scholar] [CrossRef] [Green Version]
- Noor, R.S.; Wang, Z.; Umair, M.; Yaseen, M.; Ameen, M.; Rehman, S.-U.; Khan, M.U.; Imran, M.; Ahmed, W.; Sun, Y. Interactive Effects of Grafting Techniques and Scion-Rootstocks Combinations on Vegetative Growth, Yield and Quality of Cucumber (Cucumis sativus L.). Agronomy 2019, 9, 288. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, P.; Wani, M.R. Physiological Mechanisms and Adaptation Strategies in Plants under Changing Environment; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2014; Volume 1. [Google Scholar]
- Chen, Z.; Zhao, J.; Qin, Y.; Hu, G. Study on the graft compatibility between ‘Jingganghongnuo’and other litchi cultivars. Sci. Hortic. 2016, 199, 56–62. [Google Scholar] [CrossRef]
- Fernandez-Garcia, N.; Carvajal, M.; Olmos, E. Graft union formation in tomato plants: Peroxidase and catalase involvement. Ann. Bot. 2004, 93, 53–60. [Google Scholar] [CrossRef] [PubMed]
- Aloni, B.; Karni, L.; Deventurero, G.; Levin, Z.; Cohen, R.; Katzir, N.; Lotan-Pompan, M.; Edelstein, M.; Aktas, H.; Turhan, E.; et al. Physiological and biochemical changes at the rootstock-scion interface in graft combinations between Cucurbita rootstocks and a melon scion. J. Hortic. Sci. Biotechnol. 2008, 83, 777–783. [Google Scholar] [CrossRef]
- Nocito, F.F.; Espen, L.; Fedeli, C.; Lancilli, C.; Musacchi, S.; Serra, S.; Sansavini, S.; Cocucci, M.; Sacchi, G.A. Oxidative stress and senescence-like status of pear calli co-cultured on suspensions of incompatible quince microcalli. Tree Physiol. 2010, 30, 450–458. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rapaka, V.K.; Faust, J.E.; Dole, J.M.; Runkle, E.S. Diurnal carbohydrate dynamics affect postharvest ethylene responsiveness in portulaca (Portulaca grandiflora ‘Yubi Deep Rose’) unrooted cuttings. Postharvest Biol. Technol. 2007, 44, 293–299. [Google Scholar] [CrossRef]
- Dabirian, S.; Miles, C.A. Increasing Survival of Splice-grafted Watermelon Seedlings Using a Sucrose Application. HortScience 2017, 52, 579–583. [Google Scholar] [CrossRef]
- Hunter, J.; Volschenk, C.; Le Roux, D.; Fouché, G.; Adams, L. Plant Material Quality, a Compilation of Research; Research Reports; ARC Infruitec-Nietvoorbij: Stellenbosch, South Africa, 2004. [Google Scholar]
- Xu, Q.; Guo, S.-r.; Li, H.; Du, N.-s.; Shu, S. Physiological Aspects of Compatibility and Incompatibility in Grafted Cucumber Seedlings. J. Am. Soc. Hortic. Sci. 2015, 140, 299–307. [Google Scholar] [CrossRef] [Green Version]
- Kaseb, M.O.; Umer, M.J.; Mahmud, E.; Anees, M.; Diao, W.; Yuan, P.; Zhu, H.; Zhao, S.; Lu, X.; He, N.; et al. Physio-Anatomical Study of Polyploid Watermelon Grafted by Different Methods. Agronomy 2021, 11, 913. [Google Scholar] [CrossRef]
- Gainza, F.; Opazo, I.; Muñoz, C. Graft incompatibility in plants: Metabolic changes during formation and establishment of the rootstock/scion union with emphasis on Prunus species. Chil. J. Agric. Res. 2015, 75, 28–34. [Google Scholar] [CrossRef] [Green Version]
- El-Eslamboly, A. Effect of watermelon propagation by cuttings on vegetative growth, yield and fruit quality. Egypt. J. Agric. Res. 2014, 92, 553–579. [Google Scholar] [CrossRef]
- Wang, N.; Liu, W.; Zhang, T.; Jiang, S.; Xu, H.; Wang, Y.; Zhang, Z.; Wang, C.; Chen, X. Transcriptomic analysis of red-fleshed apples reveals the novel role of MdWRKY11 in flavonoid and anthocyanin biosynthesis. J. Agric. Food Chem. 2018, 66, 7076–7086. [Google Scholar] [CrossRef]
- Mortazavi, A.; Williams, B.A.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Andino, G.K.; Gribskov, M.; Anderson, D.L.; Evans, J.D.; Hunt, G.J. Differential gene expression in Varroa jacobsoni mites following a host shift to European honey bees (Apis mellifera). BMC Genom. 2016, 17, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Nat. Preced. 2010, 11, R106. [Google Scholar] [CrossRef] [Green Version]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Langfelder, P.; Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 2008, 9, 559. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Horvath, S. A general framework for weighted gene co-expression network analysis. Stat. Appl. Genet. Mol. Biol. 2005, 4, 1–45. [Google Scholar] [CrossRef]
- Kariyanna, B.; Prabhuraj, A.; Asokan, R.; Babu, P.; Jalali, S.K.; Venkatesan, T.; Gracy, R.G.; Mohan, M. Identification of suitable reference genes for normalization of RT-qPCR data in eggplant fruit and shoot borer (Leucinodes orbonalis Guenée). Biologia 2020, 75, 289–297. [Google Scholar] [CrossRef]
- Jawad, U.M.; Gao, L.; Gebremeskel, H.; Safdar, L.B.; Yuan, P.; Zhao, S.; Xuqiang, L.; Nan, H.; Hongju, Z.; Liu, W. Expression pattern of sugars and organic acids regulatory genes during watermelon fruit development. Sci. Hortic. 2020, 265, 109102. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Gao, L.; Zhao, S.; Jiang, W.; Huang, Y.; Bie, Z. Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE 2014, 9, e90612. [Google Scholar] [CrossRef]
- Gao, L.; Zhao, S.; Lu, X.; He, N.; Zhu, H.; Dou, J.; Liu, W. Comparative transcriptome analysis reveals key genes potentially related to soluble sugar and organic acid accumulation in watermelon. PLoS ONE 2018, 13, e0190096. [Google Scholar] [CrossRef] [PubMed]
- Zheng, B.S.; Huai, L.C.; Song, H.J.; Huang, Y.J.; Zheng, J.W.; Chen, M.; Huang, J.Q. cDNA-AFLP analysis of gene expression in hickory (Carya cathayensis) during graft process. Tree Physiol. 2009, 30, 297–303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnyk, C.W.; Schuster, C.; Leyser, O.; Meyerowitz, E.M. A Developmental Framework for Graft Formation and Vascular Reconnection in Arabidopsis thaliana. Curr. Biol. 2015, 25, 1306–1318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aloni, B.; Cohen, R.; Karni, L.; Aktas, H.; Edelstein, M. Hormonal signaling in rootstock-scion interactions. Sci. Hortic. 2010, 127, 119–126. [Google Scholar] [CrossRef]
- Pina, A.; Errea, P. Influence of graft incompatibility on gene expression and enzymatic activity of UDP-glucose pyrophosphorylase. Plant Sci. 2008, 174, 502–509. [Google Scholar] [CrossRef]
- Cookson, S.J.; Clemente Moreno, M.J.; Hevin, C.; Nyamba Mendome, L.Z.; Delrot, S.; Trossat-Magnin, C.; Ollat, N. Graft union formation in grapevine induces transcriptional changes related to cell wall modification, wounding, hormone signalling, and secondary metabolism. J. Exp. Bot. 2013, 64, 2997–3008. [Google Scholar] [CrossRef]
- Schaller, G.E.; Bishopp, A.; Kieber, J.J. The yin-yang of hormones: Cytokinin and auxin interactions in plant development. Plant Cell 2015, 27, 44–63. [Google Scholar] [CrossRef] [Green Version]
- Ganie, S.A.; Molla, K.A.; Henry, R.J.; Bhat, K.V.; Mondal, T.K. Advances in understanding salt tolerance in rice. Theor. Appl. Genet. 2019, 132, 851–870. [Google Scholar] [CrossRef]
- Schwarz, D.; Rouphael, Y.; Colla, G.; Venema, J.H. Grafting as a tool to improve tolerance of vegetables to abiotic stresses: Thermal stress, water stress and organic pollutants. Sci. Hortic. 2010, 127, 162–171. [Google Scholar] [CrossRef]
- Bartolini, G.; Pestelli, P.; Toponi, M.; Di Monte, G. Rooting and carbohydrate availability in Vitis 140 Ruggeri stem cuttings. Vitis 1996, 35, 11–14. [Google Scholar]
- Beaumont, J.H.; Egede, M.R.H. Nursery Propagation and Topworking of the Macadamia; Hawaii Agricultural Experiment Station, University of Hawaii: Hawaii, HI, USA, 1937. [Google Scholar]
- Rodrigues, J.; Ryan, G.; Frolich, E. Some factors influencing grafting success with avocados. Calif. Avocado Soc. Yearb. 1960, 44, 89–92. [Google Scholar]
- Lu, J.; Cheng, F.; Huang, Y.; Bie, Z. Grafting Watermelon Onto Pumpkin Increases Chilling Tolerance by Up Regulating Arginine Decarboxylase to Increase Putrescine Biosynthesis. Front. Plant Sci. 2022, 12, 812396. [Google Scholar] [CrossRef] [PubMed]
- Xie, L.; Dong, C.; Shang, Q. Gene co-expression network analysis reveals pathways associated with graft healing by asymmetric profiling in tomato. BMC Plant Biol. 2019, 19, 373. [Google Scholar] [CrossRef] [PubMed]
- Guilfoyle, T. Sticking with auxin. Nature 2007, 446, 621–622. [Google Scholar] [CrossRef] [PubMed]
- Zenser, N.; Ellsmore, A.; Leasure, C.; Callis, J. Auxin modulates the degradation rate of Aux/IAA proteins. Proc. Natl. Acad. Sci. USA 2001, 98, 11795–11800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dreher, K.A.; Brown, J.; Saw, R.E.; Callis, J. The Arabidopsis Aux/IAA Protein Family Has Diversified in Degradation and Auxin Responsiveness. Plant Cell 2006, 18, 699–714. [Google Scholar] [CrossRef] [Green Version]
- Passardi, F.; Penel, C.; Dunand, C. Performing the paradoxical: How plant peroxidases modify the cell wall. Trends Plant Sci. 2004, 9, 534–540. [Google Scholar] [CrossRef]
- Rhee, S.G.; Woo, H.A. Multiple functions of peroxiredoxins: Peroxidases, sensors and regulators of the intracellular messenger H2O2, and protein chaperones. Antioxid. Redox Signal. 2011, 15, 781–794. [Google Scholar] [CrossRef]
| Samples | Di-0DAG | Di-3DAG | Di-15DAG | Tetra-0DAG | Tetra-3DAG | Tetra-15DAG |
|---|---|---|---|---|---|---|
| Clean reads | 21,931,767 | 24,794,003 | 21,124,267 | 22,596,008 | 21,227,314 | 21,498,313 |
| Clean bases | 6,551,749,351 | 7,401,188,622 | 6,316,423,223 | 6,755,177,632 | 6,352,106,792 | 6,428,408,569 |
| GC Content | 45.54% | 44.51% | 45.73% | 44.85% | 44.98% | 44.73% |
| % ≥ Q30 | 94.15% | 93.89% | 94.61% | 94.93% | 94.26% | 94.27% |
| Total Reads | 43,863,533 | 49,588,005 | 42,248,535 | 45,192,017 | 42,454,628 | 42,996,625 |
| Mapped Reads | 18,004,952 | 23,863,421 | 12,594,273 | 34,743,569 | 22,683,237 | 22,038,804 |
| Mapped Reads ratio | 41.35% | 47.58% | 29.72% | 76.69% | 53.17% | 52.67% |
| Uniq Mapped Reads | 17,456,213 | 23,213,051 | 12,223,916 | 33,769,354 | 22,049,895 | 21,546,888 |
| Uniq Mapped ratio | 40.09% | 46.28% | 28.85% | 74.53% | 51.69% | 51.51% |
| Multiple Map Reads | 548,739 | 650,370 | 370,356 | 974,215 | 633,342 | 491,917 |
| Multiple Map ratio | 1.26% | 1.30% | 0.87% | 2.16% | 1.48% | 1.16% |
| SNP Number | 21,441 | 33,225 | 27,477 | 26,737 | 43,712 | 45,905 |
| DEG Set | DEG Number | Upregulated | Downregulated |
|---|---|---|---|
| Diploid-0_vs_Tetraploid-0 | 3178 | 1746 | 1432 |
| Diploid-3_vs_Tetraploid-3 | 2002 | 967 | 1035 |
| Diploid-15_vs_Tetraploid-15 | 2457 | 1232 | 1225 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaseb, M.O.; Umer, M.J.; Anees, M.; Zhu, H.; Zhao, S.; Lu, X.; He, N.; El-Remaly, E.; El-Eslamboly, A.; Yousef, A.F.; et al. Transcriptome Profiling to Dissect the Role of Genome Duplication on Graft Compatibility Mechanisms in Watermelon. Biology 2022, 11, 575. https://doi.org/10.3390/biology11040575
Kaseb MO, Umer MJ, Anees M, Zhu H, Zhao S, Lu X, He N, El-Remaly E, El-Eslamboly A, Yousef AF, et al. Transcriptome Profiling to Dissect the Role of Genome Duplication on Graft Compatibility Mechanisms in Watermelon. Biology. 2022; 11(4):575. https://doi.org/10.3390/biology11040575
Chicago/Turabian StyleKaseb, Mohamed Omar, Muhammad Jawad Umer, Muhammad Anees, Hongju Zhu, Shengjie Zhao, Xuqiang Lu, Nan He, Eman El-Remaly, Ahmed El-Eslamboly, Ahmed F. Yousef, and et al. 2022. "Transcriptome Profiling to Dissect the Role of Genome Duplication on Graft Compatibility Mechanisms in Watermelon" Biology 11, no. 4: 575. https://doi.org/10.3390/biology11040575
APA StyleKaseb, M. O., Umer, M. J., Anees, M., Zhu, H., Zhao, S., Lu, X., He, N., El-Remaly, E., El-Eslamboly, A., Yousef, A. F., Salama, E. A. A., Alrefaei, A. F., Kalaji, H. M., & Liu, W. (2022). Transcriptome Profiling to Dissect the Role of Genome Duplication on Graft Compatibility Mechanisms in Watermelon. Biology, 11(4), 575. https://doi.org/10.3390/biology11040575











