Application of Hierarchical Clustering to Analyze Solvent-Accessible Surface Area Patterns in Amycolatopsis lipases
Abstract
:Simple Summary
Abstract
1. Introduction
2. Methods
Solvent Accessible Surface Area (SASA)
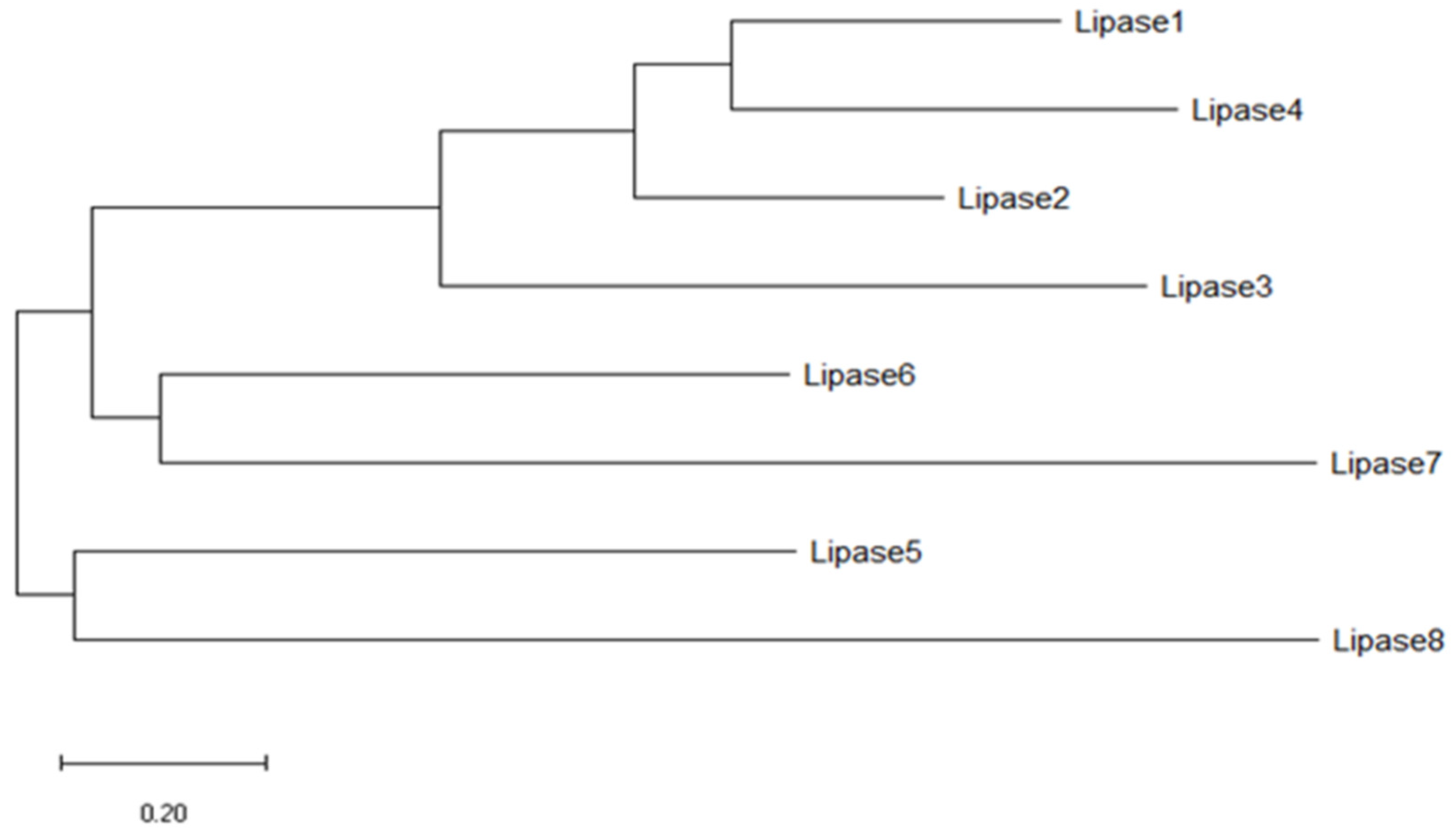
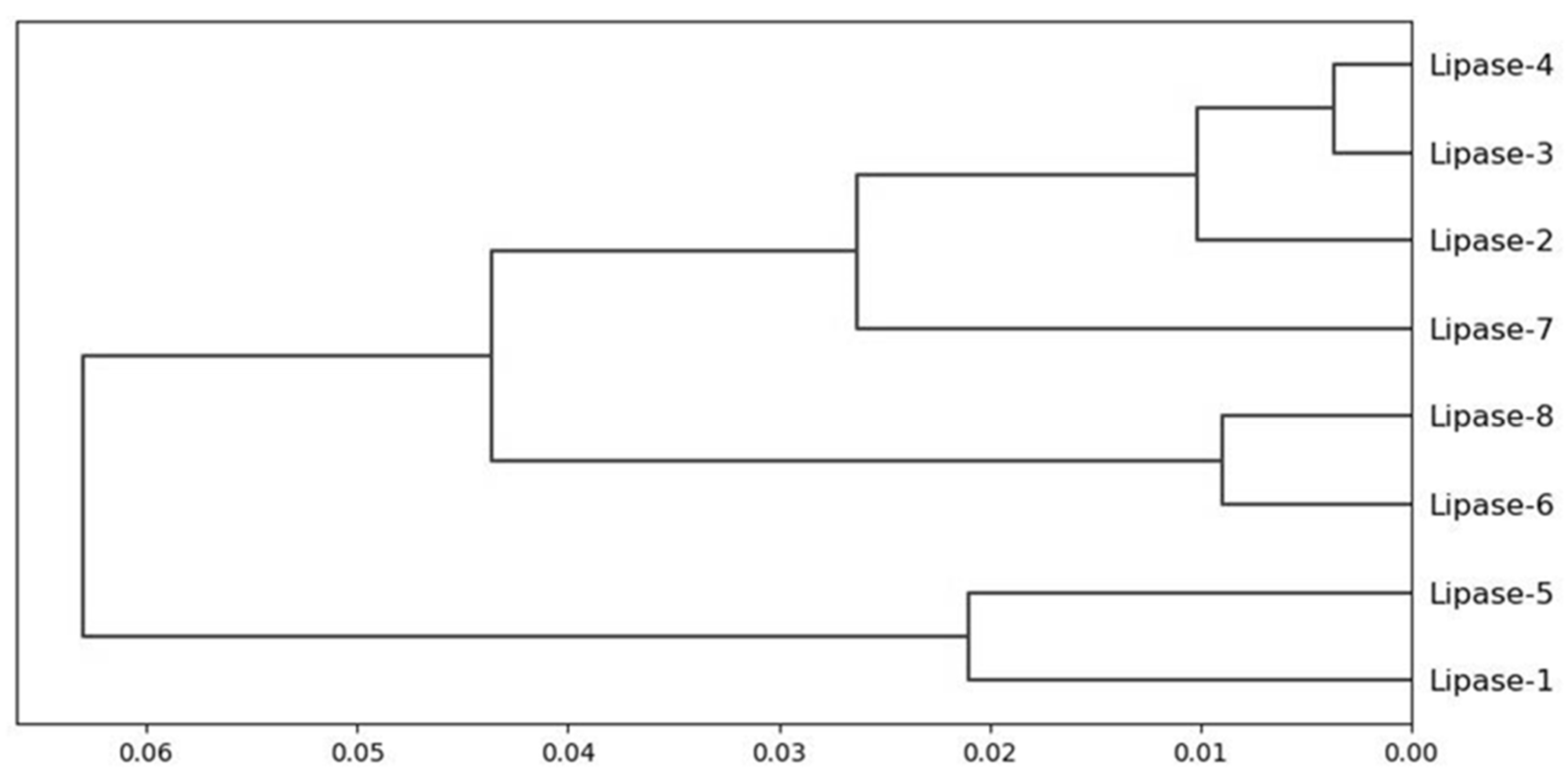
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. Recent advances in understanding catalysis of protein folding by molecular chaperones. FEBS Lett. 2020, 594, 2770–2781. [Google Scholar] [CrossRef] [PubMed]
- Dułak, D.; Gadzała, M.; Stapor, K.; Fabian, P.; Konieczny, L.; Roterman, I. Folding with active participation of water. In From Globular Proteins to Amyloids; Elsevier: Amsterdam, The Netherlands, 2020; pp. 13–26. [Google Scholar]
- Li, J.; Wang, J.; Zhao, Y.; Zhou, P.; Carter, J.; Li, Z.; Waigh, T.A.; Lu, J.R.; Xu, H. Surfactant-like peptides: From molecular design to controllable self-assembly with applications. Coord. Chem. Rev. 2020, 421, 213418. [Google Scholar] [CrossRef]
- Zhang, F.; Yu, L.; Zhang, W.; Liu, L.; Wang, C. A minireview on the perturbation effects of polar groups to direct nanoscale hydrophobic interaction and amphiphilic peptide assembly. RSC Adv. 2021, 11, 28667–28673. [Google Scholar] [CrossRef]
- Gao, J.; Zheng, S.; Yao, M.; Wu, P. Precise estimation of residue relative solvent accessible area from Cα atom distance matrix using a deep learning method. Bioinformatics 2021, 38, 94–98. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.; Karakasiliotis, I.; Anagnostopoulos, K.; Boulougouris, G.C. On the estimation of the molecular inaccessible volume and the molecular accessible surface of a ligand in protein–ligand systems. Mol. Syst. Des. Eng. 2021, 6, 946–963. [Google Scholar] [CrossRef]
- Gong, X.; Chiricotto, M.; Liu, X.; Nordquist, E.; Feig, M.; Brooks, C.L., III; Chen, J. Accelerating the generalized born with molecular volume and solvent accessible surface area implicit solvent model using graphics processing units. J. Comput. Chem. 2020, 41, 830–838. [Google Scholar] [CrossRef]
- Durham, E.; Dorr, B.; Woetzel, N.; Staritzbichler, R.; Meiler, J. Solvent accessible surface area approximations for rapid and accurate protein structure prediction. J. Mol. Model. 2009, 15, 1093–1108. [Google Scholar] [CrossRef] [Green Version]
- Pliego, J.; Mateos, J.C.; Rodriguez, J.; Valero, F.; Baeza, M.; Femat, R.; Camacho, R.; Sandoval, G.; Herrera-López, E.J. Monitoring lipase/esterase activity by stopped flow in a sequential injection analysis system using p-nitrophenyl butyrate. Sensors 2015, 15, 2798–2811. [Google Scholar] [CrossRef]
- Ananthi, S.; Ramasubburayan, R.; Palavesam, A.; Immanuel, G. Optimization and purification of lipase through solid state fermentation by bacillus cereus MSU as isolated from the gut of a marine fish Sardinella longiceps. Int. J. Pharm. Pharm. Sci. 2014, 6, 291–298. [Google Scholar]
- Iftikhar, T.; Niaz, M.; Ali, E.A.; Jabeen, R.; Abdullah, R. Production process of extracellular lipases by Fusarium sp. using agricultural by products. Pak. J. Bot. 2012, 44, 335–339. [Google Scholar]
- Kumar, A.; Parihar, S.S.; Batra, N. Enrichment, isolation and optimization of lipase-producing Staphylococcus sp. from oil mill waste (Oil cake). J. Exp. Sci. 2012, 3, 26–30. [Google Scholar]
- Ülker, S.; Özel, A.; Çolak, A.; Karaoğlu, Ş.A. Isolation, production, and characterization of an extracellular lipase from Trichoderma harzianum isolated from soil. Turk. J. Biol. 2011, 35, 543–550. [Google Scholar] [CrossRef]
- Laachari, F.; El Bergad, F.; Sadiki, M.; Sayari, A.; Bahafid, W.; Elabed, S.; Mohammed, I.; Ibnsouda, S.K. Higher tolerance of a novel lipase from Aspergillus flavus to the presence of free fatty acids at lipid/water interface. Afr. J. Biochem. Res. 2015, 9, 9–17. [Google Scholar] [CrossRef] [Green Version]
- Priji, P.; Unni, K.N.; Sajith, S.; Binod, P.; Benjamin, S. Production, optimization, and partial purification of lipase from Pseudomonas sp. strain BUP 6, a novel rumen bacterium characterized from Malabari goat. Biotechnol. Appl. Biochem. 2015, 62, 71–78. [Google Scholar] [CrossRef]
- Guo, J.; Chen, C.-P.; Wang, S.-G.; Huang, X.-J. A convenient test for lipase activity in aqueous-based solutions. Enzyme Microb. Technol. 2015, 71, 8–12. [Google Scholar] [CrossRef]
- Kapoor, M.; Gupta, M.N. Lipase promiscuity and its biochemical applications. Process Biochem. 2012, 47, 555–569. [Google Scholar] [CrossRef]
- Farrokh, P.; Yakhchali, B.; Asghar Karkhane, A. Cloning and characterization of newly isolated lipase from Enterobacter sp. Bn12. Braz. J. Microbiol. 2014, 45, 677–687. [Google Scholar] [CrossRef] [Green Version]
- Lee, L.P.; Karbul, H.M.; Citartan, M.; Gopinath, S.C.; Lakshmipriya, T.; Tang, T.-H. Lipase-secreting Bacillus species in an oil-contaminated habitat: Promising strains to alleviate oil pollution. Biomed. Res. Int. 2015, 2015, 1–9. [Google Scholar] [CrossRef]
- Nouioui, I.; Carro, L.; García-López, M.; Meier-Kolthoff, J.P.; Woyke, T.; Kyrpides, N.C.; Pukall, R.; Klenk, H.-P.; Goodfellow, M.; Göker, M. Genome-based taxonomic classification of the phylum Actinobacteria. Front. Microbiol. 2018, 9, 2007. [Google Scholar] [CrossRef] [Green Version]
- Ventura, M.; Canchaya, C.; Tauch, A.; Chandra, G.; Fitzgerald, G.F.; Chater, K.F.; van Sinderen, D. Genomics of Actinobacteria: Tracing the evolutionary history of an ancient phylum. Microbiol. Mol. Biol. Rev. 2007, 71, 495–548. [Google Scholar] [CrossRef] [Green Version]
- Bandyopadhyay, D.; Das, K.; Sen, S. Exploration of extracellular phytase production by Amycolatopsis vancoresmycina S-12 in submerged fermentation. Int. J. Curr. Microbiol. Appl. Sci. 2016, 5, 478–487. [Google Scholar] [CrossRef] [Green Version]
- Kshirsagar, S.D.; Saratale, G.D.; Saratale, R.G.; Govindwar, S.P.; Oh, M.-K. An isolated Amycolatopsis sp. GDS for cellulase and xylanase production using agricultural waste biomass. J. Appl. Microbiol. 2016, 120, 112–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peano, C.; Damiano, F.; Forcato, M.; Pietrelli, A.; Palumbo, C.; Corti, G.; Siculella, L.; Fuligni, F.; Tagliazucchi, G.M.; De Benedetto, G.E. Comparative genomics revealed key molecular targets to rapidly convert a reference rifamycin-producing bacterial strain into an overproducer by genetic engineering. Metab. Eng. 2014, 26, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Sharma, M.; Dangi, P.; Choudhary, M. Actinomycetes: Source, identification, and their applications. Int. J. Curr. Microbiol. Appl. Sci. 2014, 3, 801–832. [Google Scholar]
- Kumari, R.; Singh, P.; Lal, R. Genetics and genomics of the genus Amycolatopsis. Indian J. Microbiol. 2016, 56, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Nett, M.; Ikeda, H.; Moore, B.S. Genomic basis for natural product biosynthetic diversity in the actinomycetes. Nat. Prod. Rep. 2009, 26, 1362–1384. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Zhao, W.; Zheng, H.; Zhuo, Y.; Zhang, L.; Zhao, G.-P. Complete genome sequence of Amycolatopsis mediterranei S699 based on de novo assembly via a combinatorial sequencing strategy. J. Bacteriol. 2012, 194, 5699–5700. [Google Scholar] [CrossRef] [Green Version]
- Verma, M.; Kaur, J.; Kumar, M.; Kumari, K.; Saxena, A.; Anand, S.; Nigam, A.; Ravi, V.; Raghuvanshi, S.; Khurana, P. Whole genome sequence of the rifamycin B-producing strain Amycolatopsis mediterranei S699. J. Bacteriol. 2011, 193, 5562–5563. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Zhong, Y.; Yuan, H.; Wang, J.; Zheng, H.; Wang, Y.; Cen, X.; Xu, F.; Bai, J.; Han, X. Complete genome sequence of the rifamycin SV-producing Amycolatopsis mediterranei U32 revealed its genetic characteristics in phylogeny and metabolism. Cell Res. 2010, 20, 1096–1108. [Google Scholar] [CrossRef] [Green Version]
- Damborsky, J.; Brezovsky, J. Computational tools for designing and engineering enzymes. Curr. Opin. Chem. Biol. 2014, 19, 8–16. [Google Scholar] [CrossRef]
- Sraphet, S.; Javadi, B. Computational characterizations of GDP-mannose 4,6-dehydratase (NoeL) Rhizobial proteins. Curr. Genet. 2021, 67, 769–784. [Google Scholar] [CrossRef] [PubMed]
- García-Guevara, F.; Avelar, M.; Ayala, M.; Segovia, L. Computational tools applied to enzyme design—A review. Biocatalysis 2016, 1, 109–117. [Google Scholar] [CrossRef] [Green Version]
- Akmoussi-Toumi, S.; Khemili-Talbi, S.; Ferioune, I.; Kebbouche-Gana, S. Purification and characterization of an organic solvent-tolerant and detergent-stable lipase from Haloferax mediterranei CNCMM 50101. Int. J. Biol. Macromol. 2018, 116, 817–830. [Google Scholar] [CrossRef] [PubMed]
- Andualema, B.; Gessesse, A. Microbial lipases and their industrial applications: Review. Biotechnology 2012, 11, 100–118. [Google Scholar] [CrossRef] [Green Version]
- Nema, A.; Patnala, S.H.; Mandari, V.; Kota, S.; Devarai, S.K. Production and optimization of lipase using Aspergillus niger MTCC 872 by solid-state fermentation. Bull Natl. Res. Cent. 2019, 43, 82. [Google Scholar] [CrossRef] [Green Version]
- Dutta, M.; Tareq, A.M.; Rakib, A.; Mahmud, S.; Sami, S.A.; Mallick, J.; Islam, M.N.; Majumder, M.; Uddin, M.Z.; Alsubaie, A.; et al. Phytochemicals from Leucas zeylanica targeting main protease of SARS-CoV-2: Chemical profiles, molecular docking, and molecular dynamics simulations. Biology 2021, 10, 789. [Google Scholar] [CrossRef]
- Gasteiger, E.; Hoogland, C.; Gattiker, A.; Duvaud, S.E.; Wilkins, M.R.; Appel, R.D.; Bairoch, A. Protein identification and analysis tools on the ExPASy Server. In The Proteomics Protocols Handbook; Walker, J.M., Ed.; Humana Press: Totowa, NJ, USA, 2005; pp. 571–607. [Google Scholar]
- Yang, J.; Anishchenko, I.; Park, H.; Peng, Z.; Ovchinnikov, S.; Baker, D. Improved protein structure prediction using predicted interresidue orientations. Proc. Natl. Acad. Sci. USA 2020, 117, 1496–1503. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Kumar, T.A. CFSSP: Chou and Fasman secondary structure prediction server. Wide Spectr. 2013, 1, 15–19. [Google Scholar]
- Fraczkiewicz, R.; Braun, W. Exact and efficient analytical calculation of the accessible surface areas and their gradients for macromolecules. J. Comput. Chem. 1998, 19, 319–333. [Google Scholar] [CrossRef]
- Rost, B.; Sander, C. Conservation and prediction of solvent accessibility in protein families. Proteins Struct. Funct. Bioinf. 1994, 20, 216–226. [Google Scholar] [CrossRef] [PubMed]
- Pettersen, E.F.; Goddard, T.D.; Huang, C.C.; Couch, G.S.; Greenblatt, D.M.; Meng, E.C.; Ferrin, T.E. UCSF Chimera—a visualization system for exploratory research and analysis. J. Comput. Chem. 2004, 25, 1605–1612. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Javadi, B. In silico characterization of lipase architectural structure in Rhizobium leguminosarum. Plant Cell Biotechnol. Mol. Biol. 2020, 21, 14–26. [Google Scholar]
- Nadeem, U.; Muhammad, D.; Muhammad, S.; Özkan, A.; Sami, U.; Muhammad, Q. Screening identification and characterization of lipase producing soil bacteria from Upper Dir and Mardan Khyber Pakhtunkhwa, Pakistan. Int. J. Biosci. 2015, 6, 49–55. [Google Scholar] [CrossRef]
- Priji, P.; Sajith, S.; Faisal, P.A.; Benjamin, S. Pseudomonas sp. BUP6 produces a thermotolerant alkaline lipase with trans-esterification efficiency in producing biodiesel. 3 Biotech 2017, 7, 1–10. [Google Scholar] [CrossRef]
- Ramos-Sánchez, L.B.; Cujilema-Quitio, M.C.; Julian-Ricardo, M.C.; Cordova, J.; Fickers, P. Fungal lipase production by solid-state fermentation. J. Bioprocess. Biotech. 2015, 5, 1. [Google Scholar] [CrossRef] [Green Version]
- Do, H.; Lee, J.H.; Kwon, M.H.; Song, H.E.; An, J.Y.; Eom, S.H.; Lee, S.G.; Kim, H.J. Purification, characterization and preliminary X-ray diffraction analysis of a cold-active lipase (CpsLip) from the psychrophilic bacterium Colwellia psychrerythraea 34H. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2013, 69, 920–924. [Google Scholar] [CrossRef] [Green Version]
- Yang, W.; He, Y.; Xu, L.; Zhang, H.; Yan, Y. A new extracellular thermo-solvent-stable lipase from Burkholderia ubonensis SL-4: Identification, characterization and application for biodiesel production. J. Mol. Catal. B Enzym. 2016, 126, 76–89. [Google Scholar] [CrossRef]
- Chandra, P.; Singh, R.; Arora, P.K. Microbial lipases and their industrial applications: A comprehensive review. Microbial. Cell Factories 2020, 19, 169. [Google Scholar] [CrossRef]
- Javed, S.; Azeem, F.; Hussain, S.; Rasul, I.; Siddique, M.H.; Riaz, M.; Afzal, M.; Kouser, A.; Nadeem, H. Bacterial lipases: A review on purification and characterization. Prog. Biophys. Mol. Biol. 2018, 132, 23–34. [Google Scholar] [CrossRef]
- Melani, N.B.; Tambourgi, E.B.; Silveira, E. Lipases: From production to applications. Sep. Purif. Rev. 2020, 49, 143–158. [Google Scholar] [CrossRef]
- Uttatree, S.; Winayanuwattikun, P.; Charoenpanich, J. Isolation and characterization of a novel thermophilic-organic solvent stable lipase from Acinetobacter baylyi. Appl. Biochem. Biotechnol. 2010, 162, 1362–1376. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Xu, T.; Wang, J.; Hou, Y.; Liu, C.; Liu, S.; Wu, S. Secondary metabolites of the genus Amycolatopsis: Structures, bioactivities and biosynthesis. Molecules 2021, 26, 1884. [Google Scholar] [CrossRef]
- Xing, K.; Liu, W.; Zhang, Y.-J.; Bian, G.-K.; Zhang, W.-D.; Tamura, T.; Lee, J.-S.; Qin, S.; Jiang, J.-H. Amycolatopsis jiangsuensis sp. nov., a novel endophytic actinomycete isolated from a coastal plant in Jiangsu, China. Antonie Van Leeuwenhoek 2013, 103, 433–439. [Google Scholar] [CrossRef]
- Bharathi, D.; Rajalakshmi, G.; Komathi, S. Optimization and production of lipase enzyme from bacterial strains isolated from petrol spilled soil. J. King Saud. Univ. Sci. 2019, 31, 898–901. [Google Scholar] [CrossRef]
- Fjerbaek, L.; Christensen, K.V.; Norddahl, B. A review of the current state of biodiesel production using enzymatic transesterification. Biotechnol. Bioeng. 2009, 102, 1298–1315. [Google Scholar] [CrossRef]
- Street, G. Handbook of Enzyme Biotechnology; Wiley Online Library, Ellis Horwood Ltd.: Chichester, UK, 1977. [Google Scholar]
- Bakir, Z.B.; Metin, K. Purification and characterization of an alkali-thermostable lipase from thermophilic Anoxybacillus flavithermus HBB 134. J. Microbiol. Biotechnol. 2016, 26, 1087–1097. [Google Scholar] [CrossRef]
- Nagano, N.; Orengo, C.A.; Thornton, J.M. One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 2002, 321, 741–765. [Google Scholar] [CrossRef]
- Todd, A.E.; Orengo, C.A.; Thornton, J.M. Evolution of function in protein superfamilies, from a structural perspective. J. Mol. Biol. 2001, 307, 1113–1143. [Google Scholar] [CrossRef] [Green Version]
- Wierenga, R. The TIM-barrel fold: A versatile framework for efficient enzymes. FEBS Lett. 2001, 492, 193–198. [Google Scholar] [CrossRef] [Green Version]
- Leathers, T.D.; Rich, J.O.; Anderson, A.M.; Manitchotpisit, P. Lipase production by diverse phylogenetic clades of Aureobasidium pullulans. Biotechnol. Lett. 2013, 35, 1701–1706. [Google Scholar] [CrossRef] [PubMed]
- Mahmud, S.; Biswas, S.; Paul, G.K.; Mita, M.A.; Promi, M.M.; Afrose, S.; Hasan, M.R.; Zaman, S.; Uddin, M.S.; Dhama, K.; et al. Plant-based phytochemical screening by targeting main protease of SARS-CoV-2 to design effective potent inhibitors. Biology 2021, 10, 589. [Google Scholar] [CrossRef] [PubMed]
- El-Fakharany, E.M.; Hassan, M.A.; Taha, T.H. Production and application of extracellular laccase produced by Fusarium oxysporum EMT. Int. J. Agric. Biol. 2016, 18, 939–947. [Google Scholar] [CrossRef]
- da Silva, M.A.C.; Cavalett, A.; Spinner, A.; Rosa, D.C.; Jasper, R.B.; Quecine, M.C.; Bonatelli, M.L.; Pizzirani-Kleiner, A.; Corção, G.; de Souza Lima, A.O. Phylogenetic identification of marine bacteria isolated from deep-sea sediments of the eastern South Atlantic Ocean. SpringerPlus 2013, 2, 127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hassan, M.A.; Taha, T.H.; Hamad, G.M.; Hashem, M.; Alamri, S.; Mostafa, Y.S. Biochemical characterisation and application of keratinase from Bacillus thuringiensis MT1 to enable valorisation of hair wastes through biosynthesis of vitamin B-complex. Int. J. Biol. Macromol. 2020, 153, 561–572. [Google Scholar] [CrossRef]
- Ramani, K.; Kennedy, L.J.; Ramakrishnan, M.; Sekaran, G. Purification, characterization and application of acidic lipase from Pseudomonas gessardii using beef tallow as a substrate for fats and oil hydrolysis. Process Biochem. 2010, 45, 1683–1691. [Google Scholar] [CrossRef]
- Ramakrishnan, V.; Goveas, L.C.; Suralikerimath, N.; Jampani, C.; Halami, P.M.; Narayan, B. Extraction and purification of lipase from Enterococcus faecium MTCC5695 by PEG/phosphate aqueous-two phase system (ATPS) and its biochemical characterization. Biocatal. Agric. Biotechnol. 2016, 6, 19–27. [Google Scholar] [CrossRef]
- Castilla, A.; Panizza, P.; Rodríguez, D.; Bonino, L.; Díaz, P.; Irazoqui, G.; Giordano, S.R. A novel thermophilic and halophilic esterase from Janibacter sp. R02, the first member of a new lipase family (Family XVII). Enzyme Microb. Technol. 2017, 98, 86–95. [Google Scholar] [CrossRef]
- Bornscheuer, U.T. Alteration of lipase properties by protein engineering methods. Oléagineux Corps Gras Lipides 2008, 15, 184–188. [Google Scholar] [CrossRef] [Green Version]
- Bornscheuer, U.T. Enzymes in lipid modification: From classical biocatalysis with commercial enzymes to advanced protein engineering tools. Oléagineux Corps Gras Lipides 2013, 20, 45–49. [Google Scholar] [CrossRef]
- Lotti, M.; Alberghina, L. Lipases: Molecular Structure and Function; Springer: Amsterdam, The Netherlands, 2007; pp. 263–281. [Google Scholar]
- Ollis, D.L.; Cheah, E.; Cygler, M.; Dijkstra, B.; Frolow, F.; Franken, S.M.; Harel, M.; Remington, S.J.; Silman, I.; Schrag, J. The α/β hydrolase fold. Protein Eng. Des. Sel. 1992, 5, 197–211. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, F.I.; Lan, D.; Durrani, R.; Huan, W.; Zhao, Z.; Wang, Y. The Lid Domain in Lipases: Structural and Functional Determinant of Enzymatic Properties. Front. Bioeng. Biotechnol. 2017, 5, 16. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Lipase | NAA | MW | pI | Asp + Glu | Arg + Lys | AI | GRAVY | TPS | TAS | TSA | SCS | BBS |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 388 | 40,097.38 | 5.75 | 32 | 28 | 85.90 | 0.096 | 4790.13 | 9644.99 | 14,435.12 | 1476 | 1187 |
| 2 | 394 | 41,410.78 | 5.29 | 35 | 29 | 80.84 | −0.028 | 5317.97 | 9602.71 | 14,920.67 | 1488 | 1247 |
| 3 | 436 | 44,666.35 | 5.27 | 34 | 27 | 87.82 | 0.120 | 5554.14 | 10,311.73 | 15,865.87 | 1582 | 1279 |
| 4 | 404 | 42,150.78 | 5.73 | 38 | 31 | 90.97 | 0.111 | 5406.49 | 10,157.69 | 15,564.18 | 1526 | 1286 |
| 5 | 288 | 29,214.18 | 6.13 | 19 | 16 | 89.90 | 0.280 | 3386.73 | 7358.97 | 10,745.70 | 1003 | 793 |
| 6 | 252 | 24,997.27 | 4.52 | 24 | 12 | 97.26 | 0.222 | 3612.33 | 6175.37 | 9787.69 | 874 | 666 |
| 7 | 419 | 44,089.24 | 6.23 | 29 | 26 | 73.89 | −0.097 | 4917.24 | 9462.03 | 14,379.28 | 1310 | 867 |
| 8 | 380 | 40,419.88 | 5.96 | 47 | 39 | 95.87 | −0.110 | 7165.27 | 12,160.81 | 19,326.07 | 1719 | 1104 |
| Lipase | Entry | Oligo State | Ligand | GMQE | QMEAN | Cβ | Solvation | Torsion | Seq Identity | Seq Similarity | Coverage | Range | QSQE | Template |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A0A3R9KNJ9 | Monomer | None | 0.64 | −4.02 0.69 * | −1.96 −0.48 * | −1.46 0.06 * | −3.32 0.82 * | 29.49% | 0.35 | 0.92 | 25–388 | 0.00 | 2veo.1.A |
| 2 | A0A3R9DUJ4 | Monomer | None | 0.63 | −3.77 0.20 * | −1.98 −0.73 * | −0.85 −0.53 * | −3.27 0.53 * | 27.22% | 0.34 | 0.91 | 29–394 | 0.16 | 3zpx.1.A |
| 3 | A0A427T6P4 | Monomer | None | 0.56 | −4.02 −0.06 * | −3.87 −2.60 * | −1.01 0.32 * | −3.14 0.36 * | 26.60% | 0.33 | 0.86 | 28–422 | 0.12 | 3zpx.1.A |
| 4 | A0A3R9KMI2 | Monomer | None | 0.63 | −3.74 1.04 * | −3.15 −1.18 * | −1.82 0.26 * | −2.73 1.25 * | 30.41% | 0.35 | 0.90 | 22–403 | 0.00 | 3guu.1.A |
| 5 | A0A3R9EQB2 | Monomer | None | 0.66 | −2.24 0.84 * | −1.75 −1.67 * | −2.49 −0.52 * | −1.12 1.46 * | 44.80% | 0.40 | 0.87 | 33–282 | 0.00 | 5h6g.1.A |
| 6 | A0A3R9F8T1 | Monomer | None | 0.50 | −2.54 1.63 * | −2.32 −1.73 * | −1.60 −0.69 * | −1.48 2.34 * | 26.39% | 0.32 | 0.86 | 34–251 | 0.00 | 5h6b.1.A |
| 7 | A0A3R9DV90 | Monomer | None | 0.31 | −5.78 −1.98 * | −3.28 −2.13 * | −3.35 −3.36 * | −4.24 −0.58 * | 20.95% | 0.31 | 0.60 | 99–390 | 0.00 | 4bvj.1.A |
| 8 | A0A427T2R3 | Monomer | None | 0.59 | −4.36 1.34 * | −3.28 −0.91 * | −2.69 −0.38 * | −3.03 1.74 * | 32.33% | 0.35 | 0.87 | 4–378 | 0.00 | 3skv.1.A |
| Lipase | Sequences | Number of Residues in Favored Region | Number of Residues in Outlier Region | ||
|---|---|---|---|---|---|
| HM (%) | DM (%) | HM (%) | DM (%) | ||
| 1 | A0A3R9KNJ9 | 90.61 | 95.85 | 3.31 | 1.30 |
| 2 | A0A3R9DUJ4 | 90.93 | 96.68 | 2.47 | 0.77 |
| 3 | A0A427T6P4 | 89.82 | 96.77 | 2.80 | 0.92 |
| 4 | A0A3R9KMI2 | 90.79 | 96.02 | 2.89 | 0.25 |
| 5 | A0A3R9EQB2 | 96.77 | 96.85 | 0.40 | 0.00 |
| 6 | A0A3R9F8T1 | 93.06 | 98.40 | 2.31 | 0.40 |
| 7 | A0A3R9DV90 | 88.28 | 91.13 | 4.48 | 1.92 |
| 8 | A0A427T2R3 | 87.40 | 96.03 | 4.29 | 0.00 |
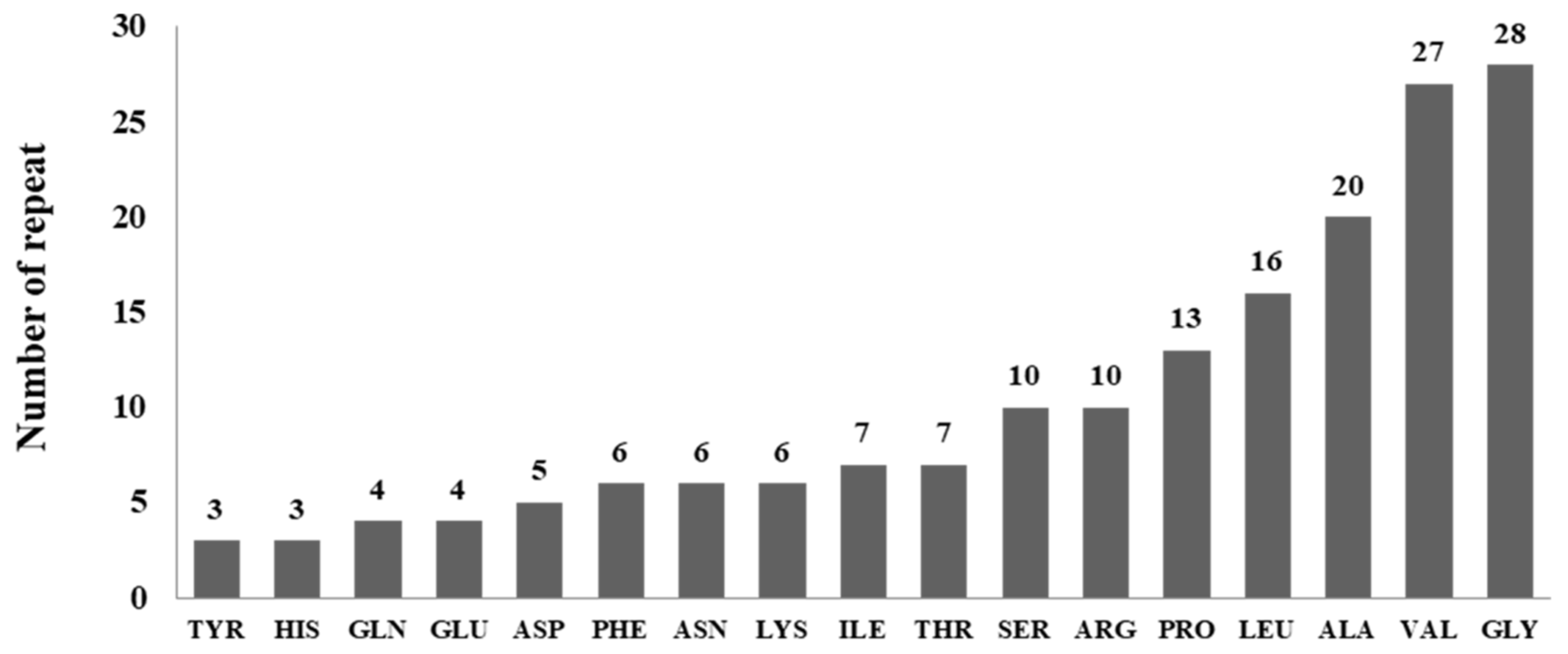
| Lipase | Entry | Length | Ala | Arg | Asn | Asp | Cys | Gln | Glu | Gly | His | Ile | Leu | Lys | Met | Phe | Pro | Ser | Thr | Trp | Tyr | Val |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | A0A3R9KNJ9 | 388 | 62 | 21 | 4 | 22 | 4 | 10 | 10 | 48 | 5 | 11 | 37 | 7 | 2 | 12 | 33 | 21 | 26 | 8 | 16 | 29 |
| 2 | A0A3R9DUJ4 | 394 | 58 | 13 | 10 | 24 | 4 | 18 | 11 | 39 | 4 | 9 | 34 | 16 | 4 | 16 | 30 | 20 | 29 | 5 | 18 | 32 |
| 3 | A0A427T6P4 | 436 | 65 | 18 | 13 | 22 | 2 | 9 | 12 | 50 | 5 | 14 | 40 | 9 | 5 | 11 | 33 | 33 | 34 | 5 | 19 | 37 |
| 4 | A0A3R9KMI2 | 404 | 69 | 20 | 8 | 20 | 4 | 11 | 18 | 39 | 9 | 14 | 38 | 11 | 3 | 14 | 28 | 24 | 21 | 3 | 17 | 33 |
| 5 | A0A3R9EQB2 | 288 | 45 | 8 | 7 | 14 | 6 | 7 | 5 | 37 | 7 | 9 | 28 | 8 | 3 | 9 | 17 | 18 | 22 | 3 | 11 | 24 |
| 6 | A0A3R9F8T1 | 252 | 44 | 9 | 3 | 10 | 4 | 13 | 14 | 31 | 3 | 4 | 26 | 3 | 2 | 0 | 20 | 11 | 20 | 3 | 3 | 29 |
| 7 | A0A3R9DV90 | 419 | 51 | 19 | 13 | 20 | 4 | 20 | 9 | 52 | 8 | 13 | 31 | 7 | 7 | 15 | 25 | 39 | 27 | 10 | 19 | 30 |
| 8 | A0A427T2R3 | 380 | 54 | 34 | 8 | 26 | 2 | 6 | 21 | 38 | 14 | 10 | 48 | 5 | 2 | 11 | 30 | 8 | 26 | 3 | 5 | 29 |
| Lipase | Entry | Helix (%) | Sheet (%) | Turn (%) |
|---|---|---|---|---|
| 1 | A0A3R9KNJ9 | 60.8 | 33.0 | 13.1 |
| 2 | A0A3R9DUJ4 | 62.7 | 37.8 | 13.7 |
| 3 | A0A427T6P4 | 51.4 | 34.4 | 11.0 |
| 4 | A0A3R9KMI2 | 68.3 | 50.5 | 12.4 |
| 5 | A0A3R9EQB2 | 56.2 | 60.8 | 9.7 |
| 6 | A0A3R9F8T1 | 59.9 | 51.6 | 10.7 |
| 7 | A0A3R9DV90 | 53.7 | 37.7 | 11.5 |
| 8 | A0A427T2R3 | 67.9 | 35.8 | 10.3 |
| Lipase | SASA | Total | Apolar | Backbone | Sidechain | Total Ave SASA |
|---|---|---|---|---|---|---|
| 1 | nucleus | 1612.05 | 1105.63 | 592.96 | 1019.10 | 39.65 |
| surface | 9087.30 | 6239.72 | 1796.53 | 7290.70 | ||
| 2 | nucleus | 1535.62 | 975.20 | 514.03 | 1021.70 | 40.76 |
| surface | 9201.62 | 5956.73 | 1673.27 | 7528.28 | ||
| 3 | nucleus | 1513.82 | 889.97 | 570.11 | 943.72 | 40.16 |
| surface | 10,211.71 | 6650.06 | 2225.90 | 7985.83 | ||
| 4 | nucleus | 1897.73 | 1227.46 | 601.65 | 1295.97 | 40.74 |
| surface | 10,134.94 | 6760.98 | 1846.70 | 8288.26 | ||
| 5 | nucleus | 839.39 | 498.14 | 390.68 | 448.79 | 42.98 |
| surface | 7280.09 | 5114.07 | 1544.76 | 5735.31 | ||
| 6 | nucleus | 941.66 | 630.22 | 411.88 | 529.76 | 44.89 |
| surface | 6561.57 | 4139.78 | 1521.13 | 5040.43 | ||
| 7 | nucleus | 1295.47 | 805.00 | 517.81 | 777.68 | 49.24 |
| surface | 9195.57 | 5912.66 | 2367.46 | 6828.11 | ||
| 8 | nucleus | 1686.72 | 1006.49 | 686.57 | 1000.18 | 51.53 |
| surface | 13,554.21 | 8567.14 | 2587.99 | 10,966.29 |
| Correlation Matrix | Lipase 1 | Lipase 2 | Lipase 3 | Lipase 4 | Lipase 5 | Lipase 6 | Lipase 7 | Lipase 8 |
|---|---|---|---|---|---|---|---|---|
| Lipase 1 | 1.00 | |||||||
| Lipase 2 | 0.91 | 1.00 | ||||||
| Lipase 3 | 0.54 | 0.72 | 1.00 | |||||
| Lipase 4 | 0.65 | 0.63 | 0.36 | 1.00 | ||||
| Lipase 5 | 0.64 | 0.78 | 0.88 | 0.33 | 1.00 | |||
| Lipase 6 | 0.55 | 0.69 | 0.85 | 0.48 | 0.92 | 1.00 | ||
| Lipase 7 | 0.60 | 0.72 | 0.60 | 0.36 | 0.80 | 0.83 | 1.00 | |
| Lipase 8 | 0.75 | 0.74 | 0.50 | 0.55 | 0.78 | 0.78 | 0.81 | 1.00 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sraphet, S.; Javadi, B. Application of Hierarchical Clustering to Analyze Solvent-Accessible Surface Area Patterns in Amycolatopsis lipases. Biology 2022, 11, 652. https://doi.org/10.3390/biology11050652
Sraphet S, Javadi B. Application of Hierarchical Clustering to Analyze Solvent-Accessible Surface Area Patterns in Amycolatopsis lipases. Biology. 2022; 11(5):652. https://doi.org/10.3390/biology11050652
Chicago/Turabian StyleSraphet, Supajit, and Bagher Javadi. 2022. "Application of Hierarchical Clustering to Analyze Solvent-Accessible Surface Area Patterns in Amycolatopsis lipases" Biology 11, no. 5: 652. https://doi.org/10.3390/biology11050652
APA StyleSraphet, S., & Javadi, B. (2022). Application of Hierarchical Clustering to Analyze Solvent-Accessible Surface Area Patterns in Amycolatopsis lipases. Biology, 11(5), 652. https://doi.org/10.3390/biology11050652





