Farnesol-Loaded Nanoliposomes Inhibit Inflammatory Gene Expression in Primary Human Skeletal Myoblasts
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Fatty Acids (FAs)
2.2. Nanoliposome Preparation and Quantification
2.3. Reverse-Phase High-Performance Liquid Chromatography (HPLC)
2.4. Cell Culture
2.5. Monitoring Mitochondrial Abundance and Function
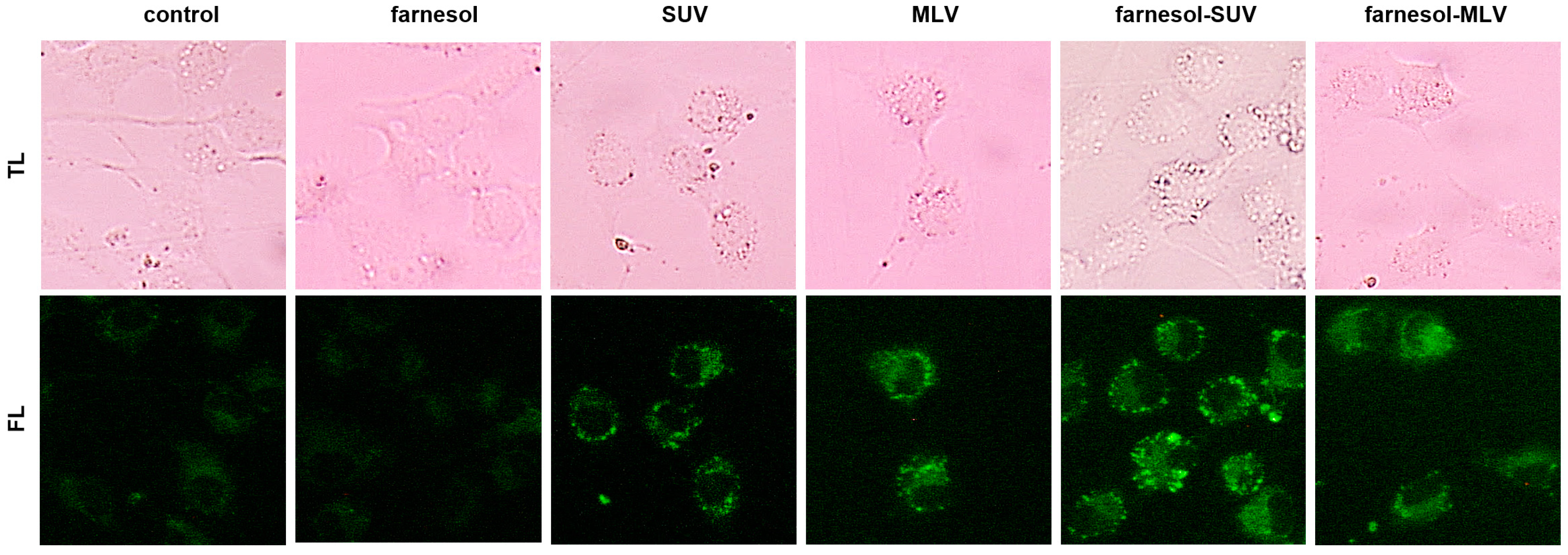
2.6. Lipid Vesicle Imaging
2.7. Multiplex Protein Quantification
2.8. Multiplex mRNA Quantification
2.9. Statistical Analysis
3. Results
3.1. Farnesol Can Be Efficiently Encapsulated in PC[soy]/DOTAP-Based SUV and MLV
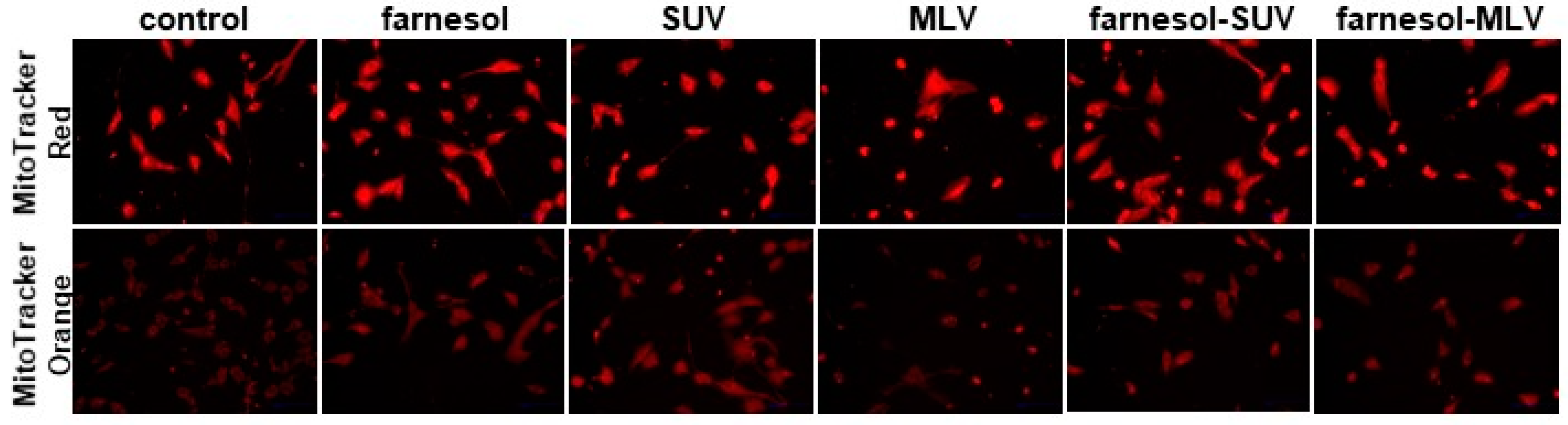
3.2. DOTAP-PC/Soy Vesicles Do Not Affect Mitochondrial Abundance or Oxidative Activity in SkMs
3.3. DOTAP-PC/Soy Vesicles Equally Attach to SkMs
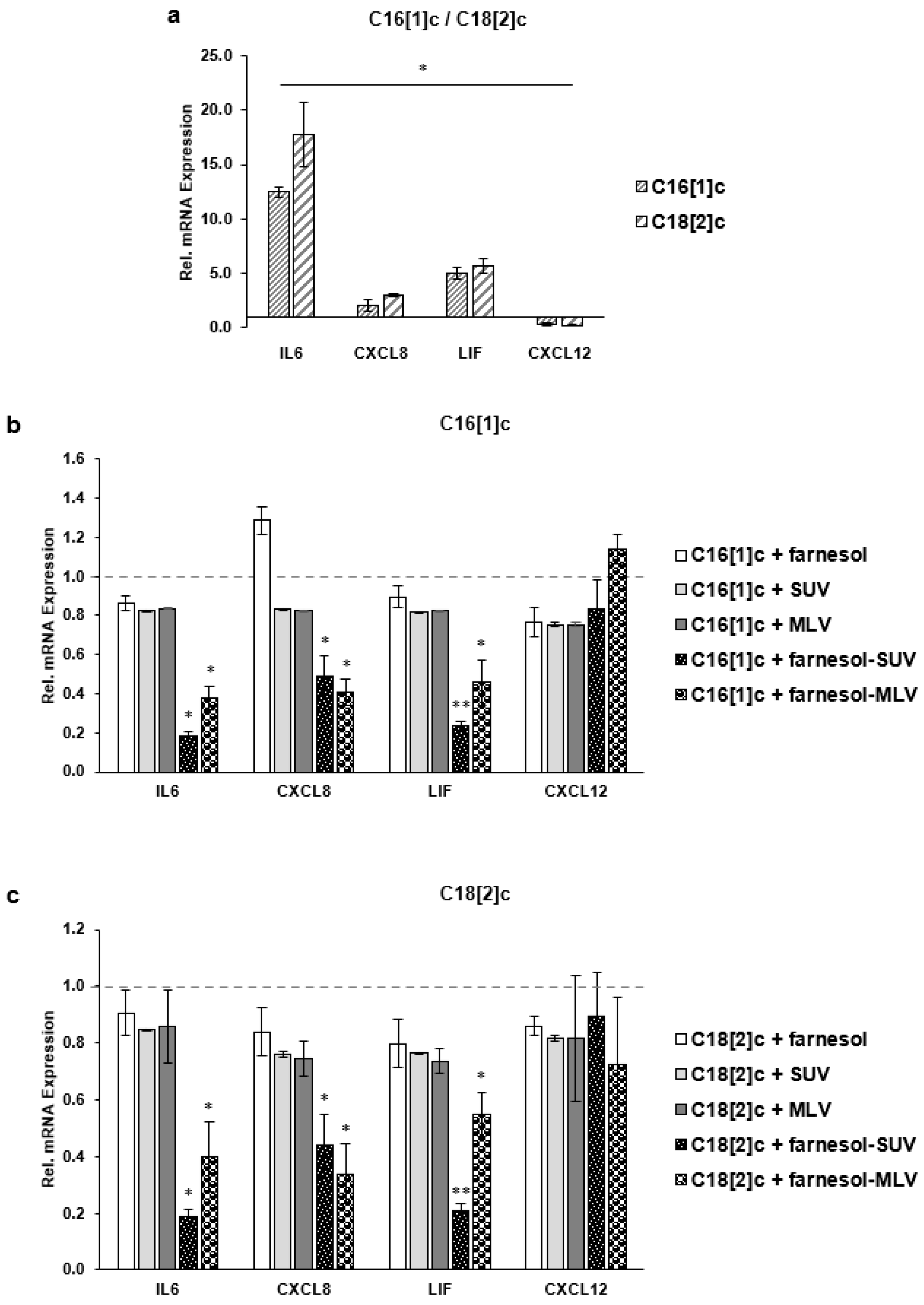
3.4. Unsaturated FFAs Modulate the Expression of Inflammatory Proteins in SkMs
3.5. Farnesol Inhibits the Expression of Inflammatory Proteins in Response to FFAs
3.6. Farnesol Inhibits the Transcription of IL6, CXCL8 and LIF Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stenholm, S.; Harris, T.B.; Rantanen, T.; Visser, M.; Kritchevsky, S.B.; Ferrucci, L. Sarcopenic obesity: Definition, cause and consequences. Curr. Opin. Clin. Nutr. Metab. Care 2008, 11, 693–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, W.-Q.; Xiao, G.-L.; Fan, Y.-B.; He, M.; Lv, S.; Li, Y.-S. Sarcopenic obesity: Research advances in pathogenesis and diagnostic criteria. Aging Clin. Exp. Res. 2019, 33, 247–252. [Google Scholar] [CrossRef] [PubMed]
- Puig, L.S.; Pillon, N.J.; Näslund, E.; Krook, A.; Zierath, J.R. Influence of obesity, weight loss, and free fatty acids on skeletal muscle clock gene expression. Am. J. Physiol. Endocrinol. Metab. 2020, 318, E1–E10. [Google Scholar] [CrossRef] [PubMed]
- Guo, A.; Li, K.; Xiao, Q. Sarcopenic obesity: Myokines as potential diagnostic biomarkers and therapeutic targets? Exp. Gerontol. 2020, 139, 111022. [Google Scholar] [CrossRef]
- Lynch, G.M.; Murphy, C.H.; Castro, E.D.M.; Roche, H.M. Inflammation and metabolism: The role of adiposity in sarcopenic obesity. Proc. Nutr. Soc. 2020, 79, 435–447. [Google Scholar] [CrossRef]
- Nelke, C.; Dziewas, R.; Minnerup, J.; Meuth, S.G.; Ruck, T. Skeletal muscle as potential central link between sarcopenia and immune senescence. eBioMedicine 2019, 49, 381–388. [Google Scholar] [CrossRef] [Green Version]
- Bano, G.; Trevisan, C.; Carraro, S.; Solmi, M.; Luchini, C.; Stubbs, B.; Manzato, E.; Sergi, G.; Veronese, N. Inflammation and sarcopenia: A systematic review and meta-analysis. Maturitas 2017, 96, 10–15. [Google Scholar] [CrossRef]
- Kob, R.; Fellner, C.; Bertsch, T.; Wittmann, A.; Mishura, D.; Sieber, C.C.; Fischer, B.E.; Stroszczynski, C.; Bollheimer, C.L. Gender-specific differences in the development of sarcopenia in the rodent model of the ageing high-fat rat. J. Cachex-Sarcopenia Muscle 2015, 6, 181–191. [Google Scholar] [CrossRef]
- Collins, K.H.; Paul, H.A.; Hart, D.A.; Reimer, H.A.P.R.A.; Smith, I.C.; Rios, J.L.; Seerattan, R.A.; Herzog, W. A High-Fat High-Sucrose Diet Rapidly Alters Muscle Integrity, Inflammation and Gut Microbiota in Male Rats. Sci. Rep. 2016, 6, 37278. [Google Scholar] [CrossRef]
- Laurentius, T.; Kob, R.; Fellner, C.; Nourbakhsh, M.; Bertsch, T.; Sieber, C.C.; Bollheimer, L.C. Long-Chain Fatty Acids and Inflammatory Markers Coaccumulate in the Skeletal Muscle of Sarcopenic Old Rats. Dis. Markers 2019, 2019, 9140789. [Google Scholar] [CrossRef] [Green Version]
- Honda, K.L.; Lamon-Fava, S.; Matthan, N.R.; Wu, D.; Lichtenstein, A.H. EPA and DHA Exposure Alters the Inflammatory Response but not the Surface Expression of Toll-like Receptor 4 in Macrophages. Lipids 2014, 50, 121–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frommer, K.W.; Schäffler, A.; Rehart, S.; Lehr, A.; Müller-Ladner, U.; Neumann, E. Free fatty acids: Potential proinflammatory mediators in rheumatic diseases. Ann. Rheum. Dis. 2013, 74, 303–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schaeffler, A.; Gross, P.; Buettner, R.; Bollheimer, C.; Buechler, C.; Neumeier, M.; Kopp, A.; Schoelmerich, J.; Falk, W. Fatty acid-induced induction of Toll-like receptor-4/nuclear factor-kappaB pathway in adipocytes links nutritional signalling with innate immunity. Immunology 2009, 126, 233–245. [Google Scholar] [CrossRef] [PubMed]
- Håversen, L.; Danielsson, K.N.; Fogelstrand, L.; Wiklund, O. Induction of proinflammatory cytokines by long-chain saturated fatty acids in human macrophages. Atherosclerosis 2009, 202, 382–393. [Google Scholar] [CrossRef] [PubMed]
- Rauen, M.; Hao, D.; Müller, A.; Mückter, E.; Bollheimer, L.C.; Nourbakhsh, M. Free Fatty Acid Species Differentially Modulate the Inflammatory Gene Response in Primary Human Skeletal Myoblasts. Biology 2021, 10, 1318. [Google Scholar] [CrossRef]
- Goto, T.; Kim, Y.I.; Funakoshi, K.; Teraminami, A.; Uemura, T.; Hirai, S.; Lee, J.Y.; Makishima, M.; Nakata, R.; Inoue, H.; et al. Farnesol, an isoprenoid, improves metabolic abnormalities in mice via both PPARalpha-dependent and -independent pathways. Am. J. Physiol.-Endocrinol. Metab. 2011, 301, E1022–E1032. [Google Scholar] [CrossRef] [Green Version]
- Ku, C.-M.; Lin, J.-Y. Farnesol, a Sesquiterpene Alcohol in Herbal Plants, Exerts Anti-Inflammatory and Antiallergic Effects on Ovalbumin-Sensitized and -Challenged Asthmatic Mice. Evid.-Based Complement. Altern. Med. 2015, 2015, 387357. [Google Scholar] [CrossRef] [Green Version]
- Jung, Y.Y.; Hwang, S.T.; Sethi, G.; Fan, L.; Arfuso, F.; Ahn, K.S. Potential Anti-Inflammatory and Anti-Cancer Properties of Farnesol. Molecules 2018, 23, 2827. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, S.; Howe, N.; Volk, K.; Tati, S.; Nickerson, K.W.; Petro, T.M. Candida albicans cell wall components and farnesol stimulate the expression of both inflammatory and regulatory cytokines in the murine RAW264.7 macrophage cell line. FEMS Immunol. Med. Microbiol. 2010, 60, 63–73. [Google Scholar] [CrossRef]
- Joo, J.H.; Jetten, A.M. NF-kappaB-dependent transcriptional activation in lung carcinoma cells by farnesol involves p65/RelA(Ser276) phosphorylation via the MEK-MSK1 signaling pathway. J. Biol. Chem. 2008, 283, 16391–16399. [Google Scholar] [CrossRef] [Green Version]
- Navarathna, D.H.M.L.P.; Nickerson, K.W.; Duhamel, G.E.; Jerrels, T.R.; Petro, T.M. Exogenous Farnesol Interferes with the Normal Progression of Cytokine Expression during Candidiasis in a Mouse Model. Infect. Immun. 2007, 75, 4006–4011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, R.; Sultana, S. Farnesol attenuates 1,2-dimethylhydrazine induced oxidative stress, inflammation and apoptotic responses in the colon of Wistar rats. Chem. Interact. 2011, 192, 193–200. [Google Scholar] [CrossRef] [PubMed]
- Santhanasabapathy, R.; Sudhandiran, G. Farnesol attenuates lipopolysaccharide-induced neurodegeneration in Swiss albino mice by regulating intrinsic apoptotic cascade. Brain Res. 2015, 1620, 42–56. [Google Scholar] [CrossRef] [PubMed]
- Abukhalil, M.H.; Hussein, O.E.; Bin-Jumah, M.; Saghir, S.A.M.; Germoush, M.O.; Elgebaly, H.A.; Mosa, N.M.; Hamad, I.; Qarmush, M.M.; Hassanein, E.M.; et al. Farnesol attenuates oxidative stress and liver injury and modulates fatty acid synthase and acetyl-CoA carboxylase in high cholesterol-fed rats. Environ. Sci. Pollut. Res. Int. 2020, 27, 30118–30132. [Google Scholar] [CrossRef]
- Sercombe, L.; Veerati, T.; Moheimani, F.; Wu, S.Y.; Sood, A.K.; Hua, S. Advances and Challenges of Liposome Assisted Drug Delivery. Front. Pharmacol. 2015, 6, 286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alsabeeh, N.; Chausse, B.; Kakimoto, P.A.; Kowaltowski, A.J.; Shirihai, O. Cell culture models of fatty acid overload: Problems and solutions. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2017, 1863, 143–151. [Google Scholar] [CrossRef] [PubMed]
- Weissig, V. (Ed.) Liposomes: Methods and Protocols; Methods in Molecular Biology; Humana Press: Totowa, NJ, USA, 2010; Volume 1. [Google Scholar]
- Hall, R.; Alasmari, A.; Mozaffari, S.; Mahdipoor, P.; Parang, K.; Aliabadi, H.M. Peptide/Lipid-Associated Nucleic Acids (PLANAs) as a Multicomponent siRNA Delivery System. Mol. Pharm. 2021, 18, 986–1002. [Google Scholar] [CrossRef]
- Chen, F.; Liu, X.-M.; Rice, K.C.; Li, X.; Yu, F.; Reinhardt, R.A.; Bayles, K.W.; Wang, D. Tooth-Binding Micelles for Dental Caries Prevention. Antimicrob. Agents Chemother. 2009, 53, 4898–4902. [Google Scholar] [CrossRef] [Green Version]
- Bandara, H.M.H.N.; Herpin, M.J.; Kolacny, D., Jr.; Harb, A.; Romanovicz, D.; Smyth, H.D.C. Incorporation of Farnesol Significantly Increases the Efficacy of Liposomal Ciprofloxacin against Pseudomonas aeruginosa Biofilms in Vitro. Mol. Pharm. 2016, 13, 2760–2770. [Google Scholar] [CrossRef] [Green Version]
- de Mello, A.H.; Costa, A.B.; Engel, J.D.G.; Rezin, G.T. Mitochondrial dysfunction in obesity. Life Sci. 2018, 192, 26–32. [Google Scholar] [CrossRef] [PubMed]
- Weissig, V.; Torchilin, V. (Eds.) Liposomes: A Practical Approach; Practical Approach Series; Oxford University Press: Oxford, UK, 2003; pp. 1–396. [Google Scholar]
- Axelrod, C.L.; Fealy, C.E.; Erickson, M.L.; Davuluri, G.; Fujioka, H.; Dantas, W.S.; Huang, E.; Pergola, K.; Mey, J.T.; King, W.T.; et al. Lipids activate skeletal muscle mitochondrial fission and quality control networks to induce insulin resistance in humans. Metabolism 2021, 121, 154803. [Google Scholar] [CrossRef] [PubMed]
- Løvsletten, N.G.; Vu, H.; Skagen, C.; Lund, J.; Kase, E.T.; Thoresen, G.H.; Zammit, V.A.; Rustan, A.C. Treatment of human skeletal muscle cells with inhibitors of diacylglycerol acyltransferases 1 and 2 to explore isozyme-specific roles on lipid metabolism. Sci. Rep. 2020, 10, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
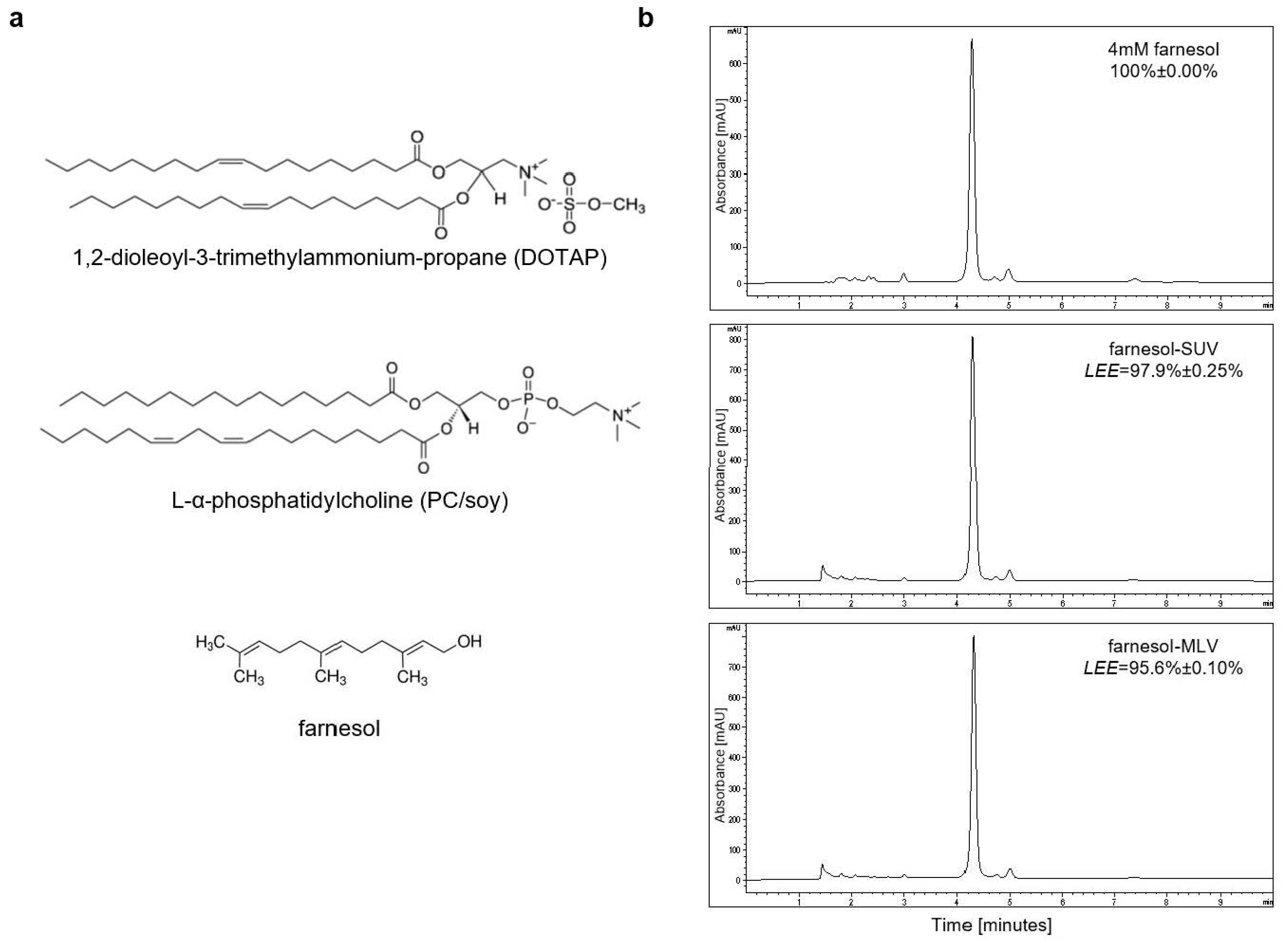


| Liposomes | Composition | Size by Volume ± SD (nm) | Polydispersity Index |
|---|---|---|---|
| SUV | PCsoy/TAP (98:2) | 27.3 ± 5.7 | 0.384 |
| farnesol-SUV | PCsoy/TAP:4.0 mM Farnesol | 33.77 ± 0.76 | 0.396 |
| MLV | PCsoy/TAP (98:2) | 322.6 ± 28.48 | 0.247 |
| farnesol-MLV | PCsoy/TAP/4.0 mM Farnesol | 195.7 ± 1.58 | 0.163 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mückter, E.; Lozoya, M.; Müller, A.; Weissig, V.; Nourbakhsh, M. Farnesol-Loaded Nanoliposomes Inhibit Inflammatory Gene Expression in Primary Human Skeletal Myoblasts. Biology 2022, 11, 701. https://doi.org/10.3390/biology11050701
Mückter E, Lozoya M, Müller A, Weissig V, Nourbakhsh M. Farnesol-Loaded Nanoliposomes Inhibit Inflammatory Gene Expression in Primary Human Skeletal Myoblasts. Biology. 2022; 11(5):701. https://doi.org/10.3390/biology11050701
Chicago/Turabian StyleMückter, Eva, Maria Lozoya, Aline Müller, Volkmar Weissig, and Mahtab Nourbakhsh. 2022. "Farnesol-Loaded Nanoliposomes Inhibit Inflammatory Gene Expression in Primary Human Skeletal Myoblasts" Biology 11, no. 5: 701. https://doi.org/10.3390/biology11050701
APA StyleMückter, E., Lozoya, M., Müller, A., Weissig, V., & Nourbakhsh, M. (2022). Farnesol-Loaded Nanoliposomes Inhibit Inflammatory Gene Expression in Primary Human Skeletal Myoblasts. Biology, 11(5), 701. https://doi.org/10.3390/biology11050701







