Deciphering the Molecular Mechanism Underlying African Animal Trypanosomiasis by Means of the 1000 Bull Genomes Project Genomic Dataset
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Monotonically Expressed Genes
2.2. Genotype Data
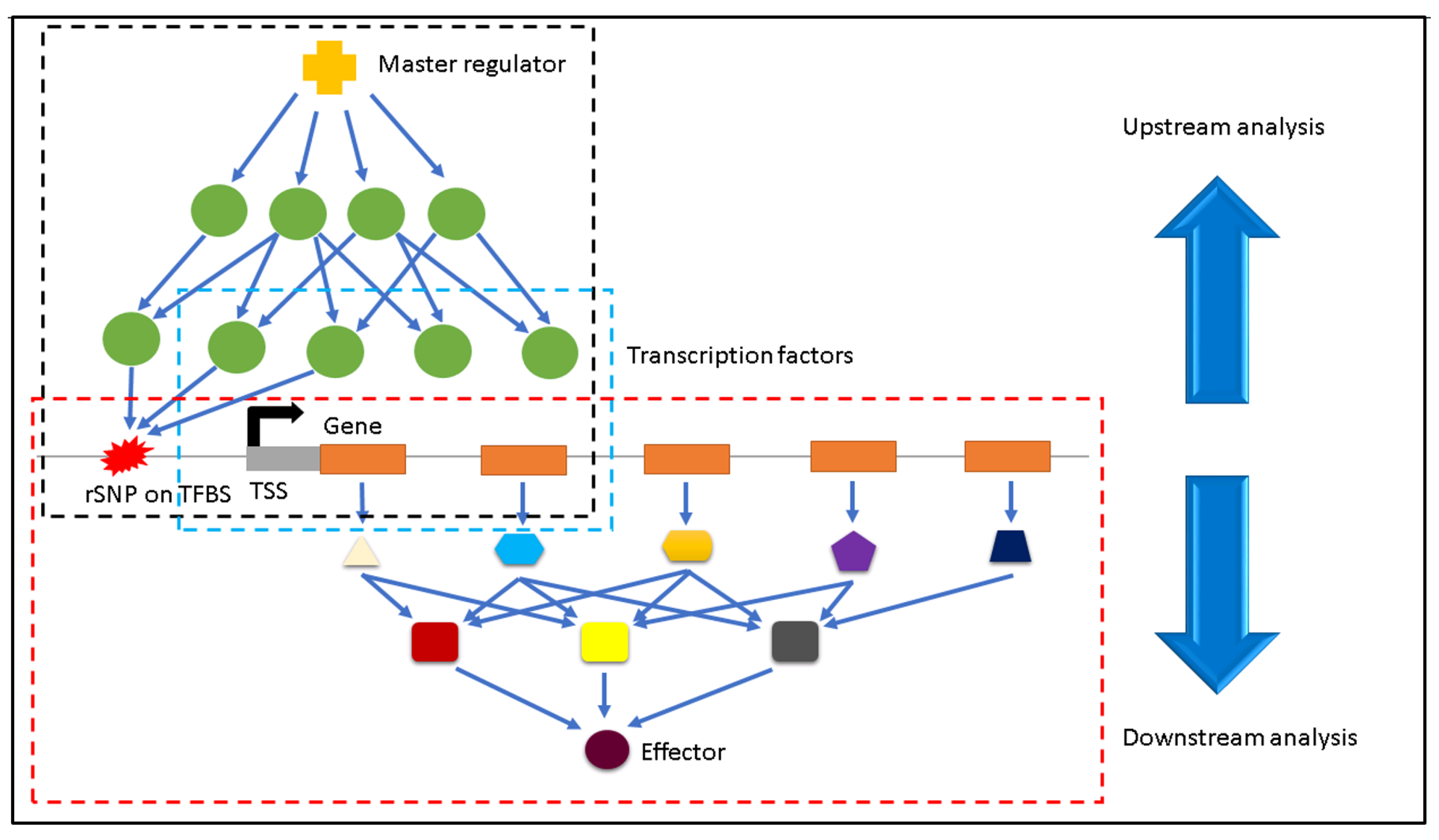
2.3. Identification of Regulatory SNPs
2.4. Finding the Effectors
3. Results and Discussion
3.1. Identification of Downstream Effectors
3.2. Downstream Effectors for Liver Tissue
3.3. Downstream Effectors for Spleen
3.4. Downstream Effectors for Lymph Node
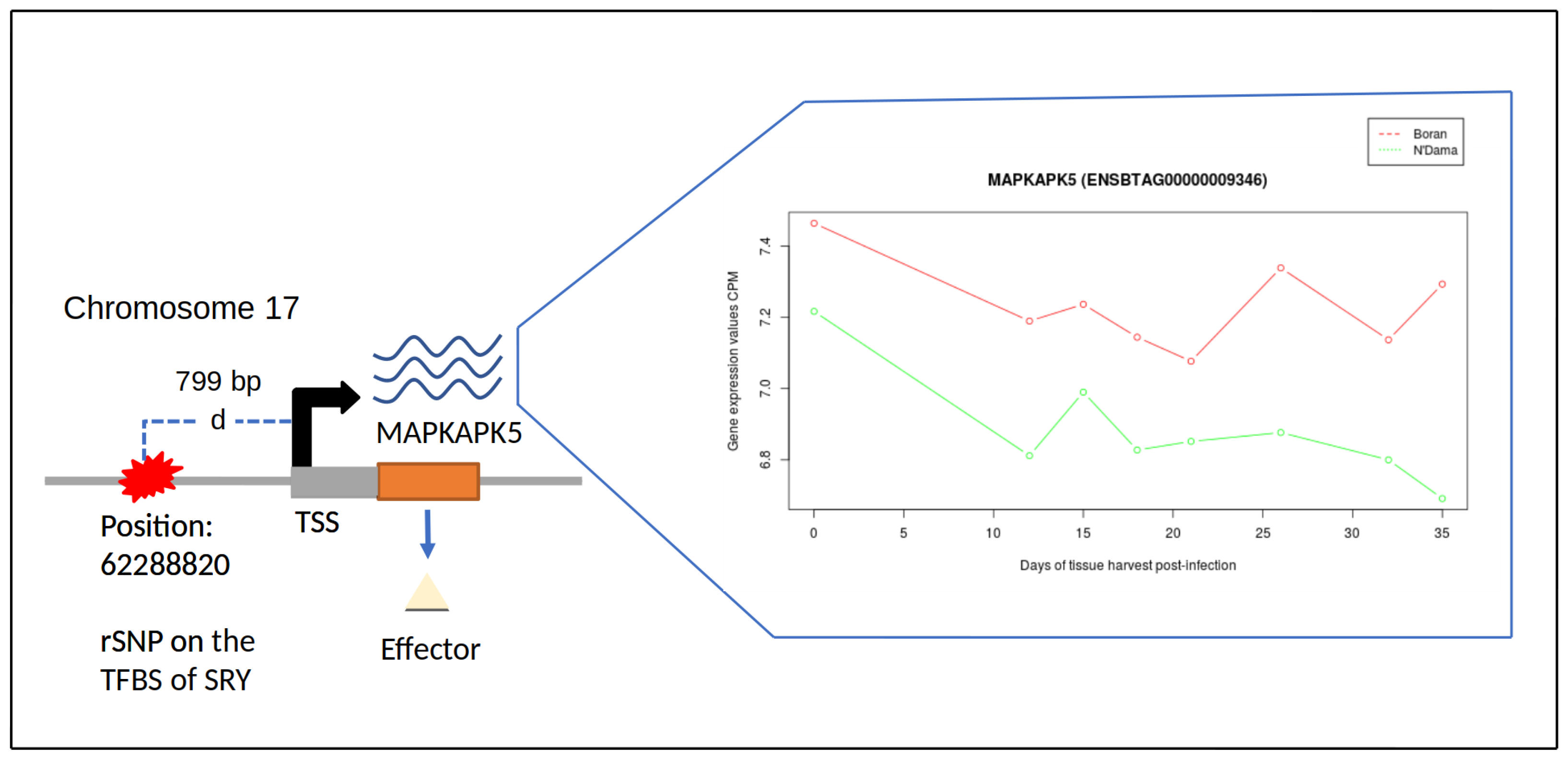
3.5. Gene Expression Profile Analysis of MEGs Harbouring rSNPs
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Brun, R.; Blum, J. Human African trypanosomiasis. Infect. Dis. Clin. N. Am. 2012, 26, 261–273. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barrett, M.P.; Boykin, D.W.; Brun, R.; Tidwell, R.R. Human African trypanosomiasis: Pharmacological re-engagement with a neglected disease. Br. J. Pharmacol. 2007, 152, 1155–1171. [Google Scholar] [CrossRef] [PubMed]
- Kappmeier, K.; Nevill, E.; Bagnall, R. Review of Tsetse Flies and Trypanosomosis in South Africa. Onderstepoort J. Vet. Res. 1998, 65, 195–203. [Google Scholar] [PubMed]
- Van den Bossche, P. Some general aspects of the distribution and epidemiology of bovine trypanosomosis in southern Africa. Int. J. Parasitol. 2001, 31, 592–598. [Google Scholar] [CrossRef]
- Firesbhat, A.; Desalegn, C. Epidemiology and impacts of trypanosomosis in cattle. Eur. J. Appl. Sci. 2015, 7, 220–225. [Google Scholar]
- Radostits, O.M.; Gay, C.C.; Hinchcliff, K.W.; Constable, P.D. Veterinary Medicine E-Book: A Textbook of the Diseases of Cattle, Horses, Sheep, Pigs and Goats; Elsevier Health Sciences: Amsterdam, The Netherlands, 2006. [Google Scholar]
- Losos, G.J.; Ikede, B. Review of pathology of diseases in domestic and laboratory animals caused by Trypanosoma congolense, T. vivax, T. brucei, T. rhodesiense and T. gambiense. Vet. Pathol. 1972, 9, 1–79. [Google Scholar] [CrossRef] [Green Version]
- Buguet, A.; Bert, J.; Tapie, P.; Tabaraud, F.; Doua, F.; Lonsdorfer, J.; Bogui, P.; Dumas, M. Sleep-wake cycle in human African trypanosomiasis. J. Clin. Neurophysiol. 1993, 10, 190–196. [Google Scholar] [CrossRef]
- Rijo-Ferreira, F.; Carvalho, T.; Afonso, C.; Sanches-Vaz, M.; Costa, R.M.; Figueiredo, L.M.; Takahashi, J.S. Sleeping sickness is a circadian disorder. Nat. Commun. 2018, 9, 62. [Google Scholar] [CrossRef] [Green Version]
- Rassi, A., Jr.; Rassi, A.; Marin-Neto, J.A. Chagas disease. Lancet 2010, 375, 1388–1402. [Google Scholar] [CrossRef]
- Rassi, A.; de Rezende, J.M. American trypanosomiasis (Chagas disease). Infect. Dis. Clin. 2012, 26, 275–291. [Google Scholar] [CrossRef]
- D’Ieteren, G.; Authié, E.; Wissocq, N.; Murray, M. Trypanotolerance, an option for sustainable livestock production in areas at risk from trypanosomosis. Rev. Sci. Tech. 1998, 17, 154–175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roelants, G. Natural resistance to African trypanosomiasis. Parasite Immunol. 1986, 8, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Authie, É. Contribution à L’étude des Mécanismes Immunologiques Impliqués dans la Trypanotolérance des Taurins d’Afrique. Ph.D. Thesis, Université de Bordeaux 2, Bordeaux, France, 1993. [Google Scholar]
- Ellis, J.; Scott, J.; MacHugh, N.D.; Gettinby, G.; Davis, W. Peripheral blood leucocytes subpopulation dynamics during Trypanosoma congolense infection in Boran and N’Dama cattle: An analysis using monoclonal antibodies and flow cytometry. Parasite Immunol. 1987, 9, 363–378. [Google Scholar] [CrossRef]
- Mulligan, H.W.; Porrs, W. (Eds.) The African Trypanosomiases; Allen and Unwin: Crows Nest, Australia, 1970; p. 950. [Google Scholar]
- Luckins, A.; Gray, A. An extravascular site of development of Trypanosoma congolense. Nature 1978, 272, 613–614. [Google Scholar] [CrossRef] [PubMed]
- Emery, D.; Moloo, S. The dynamics of the cellular reactions elicited in the skin of goats by Glossina morsitans morsitans infected with Trypanosoma (Nannomonas) congolense or T. (Duttonella) vivax. Acta Trop. 1981, 38, 15–28. [Google Scholar] [PubMed]
- Akol, G.; Murray, M.; Hirumi, H.; Hirumi, K.; Moloo, S. Infectivity to cattle of metacyclic forms of Trypanosoma (Nannomonas) congolense propagated in vitro. I. Development of localized skin reactions following intradermal inoculation. Acta Trop. 1986, 43, 207–214. [Google Scholar]
- Wellde, B.T.; Kovatch, R.M.; Chumo, D.A.; Wykoff, D.E. Trypanosoma congolense: Thrombocytopenia in experimentally infected cattle. Exp. Parasitol. 1978, 45, 26–33. [Google Scholar] [CrossRef]
- Rurangirwa, F.; Tabel, H.; Losos, G.; Masiga, W.; Mwambu, P. Immunosuppressive effect of Trypanosoma congolense and Trypanosoma vivax on the secondary immune response of cattle to Mycoplasma mycoides subsp mycoides. Res. Vet. Sci. 1978, 25, 395–397. [Google Scholar] [CrossRef]
- Rurangirwa, F.; Tabel, H.; Losos, G.; Tizard, I. Suppression of antibody response to Leptospira biflexa and Brucella abortus and recovery from immunosuppression after Berenil treatment. Infect. Immun. 1979, 26, 822–826. [Google Scholar] [CrossRef] [Green Version]
- Rurangirwa, F.; Musoke, A.; Nantulya, V.; Tabel, H. Immune depression in bovine trypanosomiasis: Effects of acute and chronic Trypanosoma congolense and chronic Trypanosoma vivax infections on antibody response to Brucella abortus vaccine. Parasite Immunol. 1983, 5, 267–276. [Google Scholar] [CrossRef]
- Tabel, H.; Losos, G.; Maxie, M. Experimental bovine trypanosomiasis (Trypanosoma vivax and T. congolense). II. Serum levels of total protein, albumin, hemolytic complement, and complement component C3. Tropenmedizin Und Parasitol. 1980, 31, 99–104. [Google Scholar]
- Starkey, P.H. N’Dama cattle—A productive trypanotolerant breed. World Anim. Rev. 1984, 50, 2–15. [Google Scholar]
- Courtin, D.; Berthier, D.; Thevenon, S.; Dayo, G.K.; Garcia, A.; Bucheton, B. Host genetics in African trypanosomiasis. Infect. Genet. Evol. 2008, 8, 229–238. [Google Scholar] [CrossRef]
- Bahbahani, H.; Salim, B.; Almathen, F.; Al Enezi, F.; Mwacharo, J.M.; Hanotte, O. Signatures of positive selection in African Butana and Kenana dairy zebu cattle. PLoS ONE 2018, 13, e0190446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tijjani, A.; Utsunomiya, Y.T.; Ezekwe, A.G.; Nashiru, O.; Hanotte, O. Genome sequence analysis reveals selection signatures in endangered trypanotolerant West African Muturu cattle. Front. Genet. 2019, 10, 442. [Google Scholar] [CrossRef]
- Mekonnen, Y.A.; Gültas, M.; Effa, K.; Hanotte, O.; Schmitt, A.O. Identification of candidate signature genes and key regulators associated with Trypanotolerance in the Sheko Breed. Front. Genet. 2019, 1095. [Google Scholar] [CrossRef] [Green Version]
- Hanotte, O.; Ronin, Y.; Agaba, M.; Nilsson, P.; Gelhaus, A.; Horstmann, R.; Sugimoto, Y.; Kemp, S.; Gibson, J.; Korol, A.; et al. Mapping of quantitative trait loci controlling trypanotolerance in a cross of tolerant West African N’Dama and susceptible East African Boran cattle. Proc. Natl. Acad. Sci. USA 2003, 100, 7443–7448. [Google Scholar] [CrossRef] [Green Version]
- Hill, E.W.; O’Gorman, G.M.; Agaba, M.; Gibson, J.P.; Hanotte, O.; Kemp, S.J.; Naessens, J.; Coussens, P.M.; MacHugh, D.E. Understanding bovine trypanosomiasis and trypanotolerance: The promise of functional genomics. Vet. Immunol. Immunopathol. 2005, 105, 247–258. [Google Scholar] [CrossRef]
- Fisher, P.; Hedeler, C.; Wolstencroft, K.; Hulme, H.; Noyes, H.; Kemp, S.; Stevens, R.; Brass, A. A systematic strategy for large-scale analysis of genotype–phenotype correlations: Identification of candidate genes involved in African trypanosomiasis. Nucleic Acids Res. 2007, 35, 5625–5633. [Google Scholar] [CrossRef]
- O’Gorman, G.M.; Park, S.D.; Hill, E.W.; Meade, K.G.; Coussens, P.M.; Agaba, M.; Naessens, J.; Kemp, S.J.; MacHugh, D.E. Transcriptional profiling of cattle infected with Trypanosoma congolense highlights gene expression signatures underlying trypanotolerance and trypanosusceptibility. BMC Genom. 2009, 10, 207. [Google Scholar] [CrossRef] [Green Version]
- Noyes, H.; Brass, A.; Obara, I.; Anderson, S.; Archibald, A.L.; Bradley, D.G.; Fisher, P.; Freeman, A.; Gibson, J.; Gicheru, M.; et al. Genetic and expression analysis of cattle identifies candidate genes in pathways responding to Trypanosoma congolense infection. Proc. Natl. Acad. Sci. USA 2011, 108, 9304–9309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gautier, M.; Flori, L.; Riebler, A.; Jaffrézic, F.; Laloé, D.; Gut, I.; Moazami-Goudarzi, K.; Foulley, J.L. A whole genome Bayesian scan for adaptive genetic divergence in West African cattle. BMC Genom. 2009, 10, 550. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yaro, M.; Munyard, K.; Stear, M.; Groth, D. Combatting African animal trypanosomiasis (AAT) in livestock: The potential role of trypanotolerance. Vet. Parasitol. 2016, 225, 43–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rajavel, A.; Heinrich, F.; Schmitt, A.O.; Gültas, M. Identifying Cattle Breed-Specific Partner Choice of Transcription Factors during the African Trypanosomiasis Disease Progression Using Bioinformatics Analysis. Vaccines 2020, 8, 246. [Google Scholar] [CrossRef] [PubMed]
- Rajavel, A.; Schmitt, A.O.; Gültas, M. Computational Identification of Master Regulators Influencing Trypanotolerance in Cattle. Int. J. Mol. Sci. 2021, 22, 562. [Google Scholar] [CrossRef] [PubMed]
- Shook, G. Major advances in determining appropriate selection goals. J. Dairy Sci. 2006, 89, 1349–1361. [Google Scholar] [CrossRef] [Green Version]
- Ron, M.; Israeli, G.; Seroussi, E.; Weller, J.I.; Gregg, J.P.; Shani, M.; Medrano, J.F. Combining mouse mammary gland gene expression and comparative mapping for the identification of candidate genes for QTL of milk production traits in cattle. BMC Genom. 2007, 8, 183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogorevc, J.; Kunej, T.; Razpet, A.; Dovc, P. Database of cattle candidate genes and genetic markers for milk production and mastitis. Anim. Genet. 2009, 40, 832–851. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cannistraci, C.V.; Ogorevc, J.; Zorc, M.; Ravasi, T.; Dovc, P.; Kunej, T. Pivotal role of the muscle-contraction pathway in cryptorchidism and evidence for genomic connections with cardiomyopathy pathways in RASopathies. BMC Med. Genom. 2013, 6, 5. [Google Scholar] [CrossRef]
- Hayes, B.J.; MacLeod, I.M.; Daetwyler, H.D.; Bowman, P.J.; Chamberlian, A.; Vander Jagt, C.; Capitan, A.; Pausch, H.; Stothard, P.; Liao, X.; et al. Genomic prediction from whole genome sequence in livestock: The 1000 bull genomes project. In Proceedings of the World Congress of Genetics Applied to Livestock Production, Vancouver, BC, Canada, 17–22 August 2014; ASAS: Champaign, IL, USA, 2014. [Google Scholar]
- Klees, S.; Lange, T.M.; Bertram, H.; Rajavel, A.; Schlüter, J.S.; Lu, K.; Schmitt, A.O.; Gültas, M. In Silico Identification of the Complex Interplay between Regulatory SNPs, Transcription Factors, and Their Related Genes in Brassica napus L. Using Multi-Omics Data. Int. J. Mol. Sci. 2021, 22, 789. [Google Scholar] [CrossRef]
- Klees, S.; Heinrich, F.; Schmitt, A.O.; Gültas, M. agReg-SNPdb: A Database of Regulatory SNPs for Agricultural Animal Species. Biology 2021, 10, 790. [Google Scholar] [CrossRef] [PubMed]
- Heinrich, F.; Wutke, M.; Das, P.P.; Kamp, M.; Gültas, M.; Link, W.; Schmitt, A.O. Identification of regulatory SNPs associated with vicine and convicine content of Vicia faba based on genotyping by sequencing data using deep learning. Genes 2020, 11, 614. [Google Scholar] [CrossRef] [PubMed]
- Buckland, P.R. The importance and identification of regulatory polymorphisms and their mechanisms of action. Biochim. Biophys. Acta-Mol. Basis Dis. 2006, 1762, 17–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, H.J.; Jung, S.; DebRoy, A.R.; Davuluri, R.V. Identification and validation of regulatory SNPs that modulate transcription factor chromatin binding and gene expression in prostate cancer. Oncotarget 2016, 7, 54616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramzan, F.; Klees, S.; Schmitt, A.O.; Cavero, D.; Gültas, M. Identification of Age-Specific and Common Key Regulatory Mechanisms Governing Eggshell Strength in Chicken Using Random Forests. Genes 2020, 11, 464. [Google Scholar] [CrossRef]
- Chang, C.C.; Chow, C.C.; Tellier, L.C.; Vattikuti, S.; Purcell, S.M.; Lee, J.J. Second-generation PLINK: Rising to the challenge of larger and richer datasets. Gigascience 2015, 4, 7. [Google Scholar] [CrossRef]
- Wingender, E.; Chen, X.; Fricke, E.; Geffers, R.; Hehl, R.; Liebich, I.; Krull, M.; Matys, V.; Michael, H.; Ohnhaeuser, R.; et al. Match-a tool for searching transcription factor binding sites in DNA sequences. Nucl. Acids Res. 2001, 29, 281–283. [Google Scholar] [CrossRef] [Green Version]
- Stegmaier, P.; Kel, A.; Wingender, E. geneXplainR: An R interface for the geneXplain platform. J. Open Source Softw. 2017, 2, 412. [Google Scholar] [CrossRef] [Green Version]
- Krull, M.; Pistor, S.; Voss, N.; Kel, A.; Reuter, I.; Kronenberg, D.; Michael, H.; Schwarzer, K.; Potapov, A.; Choi, C.; et al. TRANSPATH®: An information resource for storing and visualizing signaling pathways and their pathological aberrations. Nucleic Acids Res. 2006, 34, D546–D551. [Google Scholar] [CrossRef] [Green Version]
- Smith, C. Genomics: SNPs and human disease. Nature 2005, 435, 993. [Google Scholar] [CrossRef]
- Singh, M.; Singh, P.; Juneja, P.K.; Singh, S.; Kaur, T. SNP–SNP interactions within APOE gene influence plasma lipids in postmenopausal osteoporosis. Rheumatol. Int. 2011, 31, 421–423. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, Y.; Mackey, J.R.; Lai, R.; Franco-Villalobos, C.; Lupichuk, S.; Robson, P.J.; Kopciuk, K.; Cass, C.E.; Yasui, Y.; Damaraju, S. Assessing SNP-SNP interactions among DNA repair, modification and metabolism related pathway genes in breast cancer susceptibility. PLoS ONE 2013, 8, e64896. [Google Scholar] [CrossRef] [PubMed]
- Onay, V.Ü.; Briollais, L.; Knight, J.A.; Shi, E.; Wang, Y.; Wells, S.; Li, H.; Rajendram, I.; Andrulis, I.L.; Ozcelik, H. SNP-SNP interactions in breast cancer susceptibility. BMC Cancer 2006, 6, 114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moszyńska, A.; Gebert, M.; Collawn, J.F.; Bartoszewski, R. SNPs in microRNA target sites and their potential role in human disease. Open Biol. 2017, 7, 170019. [Google Scholar] [CrossRef]
- Soares-Silva, M.; Diniz, F.F.; Gomes, G.N.; Bahia, D. The mitogen-activated protein kinase (MAPK) pathway: Role in immune evasion by trypanosomatids. Front. Microbiol. 2016, 7, 183. [Google Scholar] [CrossRef]
- Parihar, S.; Ozturk, M.; Marakalala, M.; Loots, D.; Hurdayal, R.; Maasdorp, D.B.; Van Reenen, M.; Zak, D.; Darboe, F.; Penn-Nicholson, A.; et al. Protein kinase C-delta (PKCδ), a marker of inflammation and tuberculosis disease progression in humans, is important for optimal macrophage killing effector functions and survival in mice. Mucosal Immunol. 2018, 11, 496–511. [Google Scholar] [CrossRef] [Green Version]
- Berg, L.J.; Finkelstein, L.D.; Lucas, J.A.; Schwartzberg, P.L. Tec family kinases in T lymphocyte development and function. Annu. Rev. Immunol. 2005, 23, 549–600. [Google Scholar] [CrossRef]
- Felices, M.; Falk, M.; Kosaka, Y.; Berg, L.J. Tec kinases in T cell and mast cell signaling. Adv. Immunol. 2007, 93, 145–184. [Google Scholar]
- Readinger, J.A.; Mueller, K.L.; Venegas, A.M.; Horai, R.; Schwartzberg, P.L. Tec kinases regulate T-lymphocyte development and function: New insights into the roles of Itk and Rlk/Txk. Immunol. Rev. 2009, 228, 93–114. [Google Scholar] [CrossRef]
- Fan, K.; Jia, Y.; Wang, S.; Li, H.; Wu, D.; Wang, G.; Chen, J.L. Role of Itk signalling in the interaction between influenza A virus and T-cells. J. Gen. Virol. 2012, 93, 987–997. [Google Scholar] [CrossRef]
- Clements, J.L.; Ross-Barta, S.E.; Tygrett, L.T.; Waldschmidt, T.J.; Koretzky, G.A. SLP-76 expression is restricted to hemopoietic cells of monocyte, granulocyte, and T lymphocyte lineage and is regulated during T cell maturation and activation. J. Immunol. 1998, 161, 3880–3889. [Google Scholar] [PubMed]
- Koretzky, G.A.; Abtahian, F.; Silverman, M.A. SLP76 and SLP65: Complex regulation of signalling in lymphocytes and beyond. Nat. Rev. Immunol. 2006, 6, 67–78. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Littman, D.R. A kinase-independent function of Lck in potentiating antigen-specific T cell activation. Cell 1993, 74, 633–643. [Google Scholar] [CrossRef]
- Lattanzio, R.; Iezzi, M.; Sala, G.; Tinari, N.; Falasca, M.; Alberti, S.; Buglioni, S.; Mottolese, M.; Perracchio, L.; Natali, P.G.; et al. PLC-gamma-1 phosphorylation status is prognostic of metastatic risk in patients with early-stage Luminal-A and-B breast cancer subtypes. BMC Cancer 2019, 19, 747. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.R.; Choi, J.H.; Chang, J.S.; Kwon, H.M.; Jang, H.J.; Ryu, S.H.; Suh, P.G. Diverse Cellular and Physiological Roles of Phospholipase C-γ1; Elsevier BV: Amsterdam, The Netherlands, 2012; Volume 52, pp. 138–151. [Google Scholar]
- Herman, J.A.; Miller, M.P.; Biggins, S. chTOG is a conserved mitotic error correction factor. Elife 2020, 9, e61773. [Google Scholar] [CrossRef] [PubMed]
- Jang, C.W.; Shibata, Y.; Starmer, J.; Yee, D.; Magnuson, T. Histone H3. 3 maintains genome integrity during mammalian development. Genes Dev. 2015, 29, 1377–1392. [Google Scholar] [CrossRef] [Green Version]
- Weaver, D.C.; Shpakovski, G.V.; Caputo, E.; Levin, H.L.; Bocke, J. Sequence analysis of closely related retrotransposon families from fission yeast. Gene 1993, 131, 135–139. [Google Scholar] [CrossRef]
- Mellor, P.; Furber, L.A.; Nyarko, J.N.; Anderson, D.H. Multiple roles for the p85α isoform in the regulation and function of PI3K signalling and receptor trafficking. Biochem. J. 2012, 441, 23–37. [Google Scholar] [CrossRef] [Green Version]
- Chagpar, R.B.; Links, P.H.; Pastor, M.C.; Furber, L.A.; Hawrysh, A.D.; Chamberlain, M.D.; Anderson, D.H. Direct positive regulation of PTEN by the p85 subunit of phosphatidylinositol 3-kinase. Proc. Natl. Acad. Sci. USA 2010, 107, 5471–5476. [Google Scholar] [CrossRef] [Green Version]
- Morchikh, M.; Cribier, A.; Raffel, R.; Amraoui, S.; Cau, J.; Severac, D.; Dubois, E.; Schwartz, O.; Bennasser, Y.; Benkirane, M. HEXIM1 and NEAT1 long non-coding RNA form a multi-subunit complex that regulates DNA-mediated innate immune response. Mol. Cell 2017, 67, 387–399. [Google Scholar] [CrossRef]
- Mascareno, E.; Gupta, R.; Martello, L.A.; Dhar-Mascareno, M.; Salciccioli, L.; Beckles, D.; Walsh, M.G.; Machado, F.S.; Tanowitz, H.B.; Haseeb, M. Rapidly progressive course of Trypanosoma cruzi infection in mice heterozygous for hexamethylene bis-acetamide inducible 1 (Hexim1) gene. Microbes Infect. 2018, 20, 25–36. [Google Scholar] [CrossRef] [PubMed]
- Vousden, K.H.; Prives, C. Blinded by the light: The growing complexity of p53. Cell 2009, 137, 413–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Efeyan, A.; Serrano, M. p53: Guardian of the genome and policeman of the oncogenes. Cell Cycle 2007, 6, 1006–1010. [Google Scholar] [CrossRef] [PubMed]
- Komarova, E.A.; Krivokrysenko, V.; Wang, K.; Neznanov, N.; Chernov, M.V.; Komarov, P.G.; Brennan, M.L.; Golovkina, T.V.; Rokhlin, O.; Kuprash, D.V.; et al. p53 is a suppressor of inflammatory response in mice. FASEB J. 2005, 19, 1030–1032. [Google Scholar] [CrossRef] [PubMed]
- Madenspacher, J.H.; Azzam, K.M.; Gowdy, K.M.; Malcolm, K.C.; Nick, J.A.; Dixon, D.; Aloor, J.J.; Draper, D.W.; Guardiola, J.J.; Shatz, M.; et al. p53 Integrates host defense and cell fate during bacterial pneumonia. J. Exp. Med. 2013, 210, 891–904. [Google Scholar] [CrossRef]
- Chung, J.H. The role of DNA-PK in aging and energy metabolism. FEBS J. 2018, 285, 1959–1972. [Google Scholar] [CrossRef] [Green Version]
- Ferguson, B.J.; Mansur, D.S.; Peters, N.E.; Ren, H.; Smith, G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife 2012, 1, e00047. [Google Scholar] [CrossRef]
- Black, J.A.; Crouch, K.; Lemgruber, L.; Lapsley, C.; Dickens, N.; Tosi, L.R.; Mottram, J.C.; McCulloch, R. Trypanosoma brucei ATR links DNA damage signaling during antigenic variation with regulation of RNA polymerase I-transcribed surface antigens. Cell Rep. 2020, 30, 836–851. [Google Scholar] [CrossRef] [Green Version]
- Baker, N.; Catta-Preta, C.M.; Neish, R.; Sadlova, J.; Powell, B.; Alves-Ferreira, E.V.; Geoghegan, V.; Carnielli, J.B.; Newling, K.; Hughes, C.; et al. Systematic functional analysis of Leishmania protein kinases identifies regulators of differentiation or survival. Nat. Commun. 2021, 12, 1244. [Google Scholar] [CrossRef]
- Xu, J.; Cai, R.; Lu, L.; Duan, C.; Tao, X.; Chen, D.; Liu, Y.; Wang, X.; Cao, M.; Chen, Y. Genetic regulatory network analysis reveals that low density lipoprotein receptor-related protein 11 is involved in stress responses in mice. Psychiatry Res. 2014, 220, 1131–1137. [Google Scholar] [CrossRef]
- Seguel, M.; Perez-Venegas, D.; Gutierrez, J.; Crocker, D.E.; DeRango, E.J. Parasitism elicits a stress response that allocates resources for immune function in South American fur seals (Arctocephalus australis). Physiol. Biochem. Zool. 2019, 92, 326–338. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A. On a remarkable bacteriolytic element found in tissues and secretions. Proc. R. Soc. London Ser. Contain. Pap. Biol. Character 1922, 93, 306–317. [Google Scholar]
- Ng, C.J.; Wadleigh, D.J.; Gangopadhyay, A.; Hama, S.; Grijalva, V.R.; Navab, M.; Fogelman, A.M.; Reddy, S.T. Paraoxonase-2 is a ubiquitously expressed protein with antioxidant properties and is capable of preventing cell-mediated oxidative modification of low density lipoprotein. J. Biol. Chem. 2001, 276, 44444–44449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourquard, N.; Ng, C.J.; Reddy, S.T. Impaired hepatic insulin signalling in PON2-deficient mice: A novel role for the PON2/apoE axis on the macrophage inflammatory response. Biochem. J. 2011, 436, 91–100. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Devarajan, A.; Bourquard, N.; Grijalva, V.R.; Gao, F.; Ganapathy, E.; Verma, J.; Reddy, S.T. Role of PON2 in innate immune response in an acute infection model. Mol. Genet. Metab. 2013, 110, 362–370. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sprecher, C.A.; Grant, F.J.; Baumgartner, J.W.; Presnell, S.R.; Schrader, S.K.; Yamagiwa, T.; Whitmore, T.E.; O’Hara, P.J.; Foster, D.F. Cloning and characterization of a novel class I cytokine receptor. Biochem. Biophys. Res. Commun. 1998, 246, 82–90. [Google Scholar] [CrossRef]
- Pflanz, S.; Hibbert, L.; Mattson, J.; Rosales, R.; Vaisberg, E.; Bazan, J.F.; Phillips, J.H.; McClanahan, T.K.; de Waal Malefyt, R.; Kastelein, R.A. WSX-1 and glycoprotein 130 constitute a signal-transducing receptor for IL-27. J. Immunol. 2004, 172, 2225–2231. [Google Scholar] [CrossRef]
- Villarino, A.; Hibbert, L.; Lieberman, L.; Wilson, E.; Mak, T.; Yoshida, H.; Kastelein, R.A.; Saris, C.; Hunter, C.A. The IL-27R (WSX-1) is required to suppress T cell hyperactivity during infection. Immunity 2003, 19, 645–655. [Google Scholar] [CrossRef] [Green Version]
- Reczek, D.; Schwake, M.; Schröder, J.; Hughes, H.; Blanz, J.; Jin, X.; Brondyk, W.; Van Patten, S.; Edmunds, T.; Saftig, P. LIMP-2 is a receptor for lysosomal mannose-6-phosphate-independent targeting of β-glucocerebrosidase. Cell 2007, 131, 770–783. [Google Scholar] [CrossRef] [Green Version]
- Neculai, D.; Schwake, M.; Ravichandran, M.; Zunke, F.; Collins, R.F.; Peters, J.; Neculai, M.; Plumb, J.; Loppnau, P.; Pizarro, J.C.; et al. Structure of LIMP-2 provides functional insights with implications for SR-BI and CD36. Nature 2013, 504, 172–176. [Google Scholar] [CrossRef]
- Heybrock, S.; Kanerva, K.; Meng, Y.; Ing, C.; Liang, A.; Xiong, Z.J.; Weng, X.; Kim, Y.A.; Collins, R.; Trimble, W.; et al. Lysosomal integral membrane protein-2 (LIMP-2/SCARB2) is involved in lysosomal cholesterol export. Nat. Commun. 2019, 10, 3521. [Google Scholar] [CrossRef] [PubMed]
- Mori, H.; Sakakibara, S.i.; Imai, T.; Nakamura, Y.; Iijima, T.; Suzuki, A.; Yuasa, Y.; Takeda, M.; Okano, H. Expression of mouse igf2 mRNA-binding protein 3 and its implications for the developing central nervous system. J. Neurosci. Res. 2001, 64, 132–143. [Google Scholar] [CrossRef] [PubMed]
- Natkunam, Y.; Vainer, G.; Chen, J.; Zhao, S.; Marinelli, R.J.; Hammer, A.S.; Hamilton-Dutoit, S.; Pikarsky, E.; Amir, G.; Levy, R.; et al. Expression of the RNA-binding protein VICKZ in normal hematopoietic tissues and neoplasms. Haematologica 2007, 92, 176–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patel, S.B.; Bellini, M. The assembly of a spliceosomal small nuclear ribonucleoprotein particle. Nucleic Acids Res. 2008, 36, 6482–6493. [Google Scholar] [CrossRef] [Green Version]
- Luo, H.R.; Moreau, G.A.; Levin, N.; Moore, M.J. The human Prp8 protein is a component of both U2-and U12-dependent spliceosomes. RNA 1999, 5, 893–908. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.Y.; Park, E.J.; Kim, S.; Lee, C.W. Ssu72 Phosphatase Is Involved in Immunometabolism. J. Immunol. 2018, 200, 108. [Google Scholar]
- Silva, J.S.; Twardzik, D.R.; Reed, S.G. Regulation of Trypanosoma cruzi infections in vitro and in vivo by transforming growth factor beta (TGF-beta). J. Exp. Med. 1991, 174, 539–545. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Li, Q.; Ni, S.; Huang, Y.; Wei, J.; Liu, J.; Yu, Y.; Wang, S.; Qin, Q. The roles of grouper clathrin light chains in regulating the infection of a novel marine DNA virus, Singapore grouper iridovirus. Sci. Rep. 2019, 9, 15647. [Google Scholar] [CrossRef]
- Ni, H.; Wang, X.S.; Diener, K.; Yao, Z. MAPKAPK5, a novel mitogen-activated protein kinase (MAPK)-activated protein kinase, is a substrate of the extracellular-regulated kinase (ERK) and p38 kinase. Biochem. Biophys. Res. Commun. 1998, 243, 492–496. [Google Scholar] [CrossRef]
- Kress, T.R.; Cannell, I.G.; Brenkman, A.B.; Samans, B.; Gaestel, M.; Roepman, P.; Burgering, B.M.; Bushell, M.; Rosenwald, A.; Eilers, M. The MK5/PRAK kinase and Myc form a negative feedback loop that is disrupted during colorectal tumorigenesis. Mol. Cell 2011, 41, 445–457. [Google Scholar] [CrossRef]
- Casacuberta-Serra, S.; Soucek, L. Myc and Ras, the Bonnie and Clyde of immune evasion. Transl. Cancer Res. 2018, 7, S457. [Google Scholar] [CrossRef] [PubMed]
- Okada, M. Regulation of the SRC family kinases by Csk. Int. J. Biol. Sci. 2012, 8, 1385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niki, M.; Di Cristofano, A.; Zhao, M.; Honda, H.; Hirai, H.; Van Aelst, L.; Cordon-Cardo, C.; Pandolfi, P.P. Role of Dok-1 and Dok-2 in leukemia suppression. J. Exp. Med. 2004, 200, 1689–1695. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Kim, S.; Bae, D.J.; Park, S.Y.; Lee, G.Y.; Park, G.M.; Kim, I.S. Coordinated balance of Rac1 and RhoA plays key roles in determining phagocytic appetite. PLoS ONE 2017, 12, e0174603. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Beier, U.H.; Han, R.; Bhatti, T.R.; Akimova, T.; Hancock, W.W. Foxp3+ T-regulatory cells require DNA methyltransferase 1 expression to prevent development of lethal autoimmunity. Blood J. Am. Soc. Hematol. 2013, 121, 3631–3639. [Google Scholar] [CrossRef] [Green Version]
| Boran | N’Dama | |||
|---|---|---|---|---|
| Ascending | Descending | Ascending | Descending | |
| Liver | 741 | 308 | 757 | 124 |
| Spleen | 669 | 126 | 13 | 139 |
| Lymph node | 87 | 5 | 119 | 114 |
| Boran | N’Dama | |||
|---|---|---|---|---|
| Gain of TFBS | Loss of TFBS | Gain of TFBS | Loss of TFBS | |
| Liver | 365 | 403 | 342 | 385 |
| Spleen | 152 | 154 | 3 | 8 |
| Lymph node | 10 | 12 | 3 | 12 |
| Boran | N’Dama | |
|---|---|---|
| Liver | 194 | 102 |
| Spleen | 157 | 9 |
| Lymph node | 13 | 5 |
| Cattle Breed | Tissue | Effectors |
|---|---|---|
| Boran | Liver | Itk:Lck:PLCgamma1:SLP-76 |
| Boran | Liver | PKCdelta |
| Boran | Liver | SRF |
| Boran | Spleen | histone H3:DNA-PKcs |
| Boran | Spleen | p53:HEXIM1 |
| Boran | Spleen | HEXIM1:p53 |
| Boran | Lymph node | LIMPII:Prpf8 |
| Boran | Lymph node | VICKZ3:Prpf8 |
| Boran | Lymph node | SNRPGP15:Prpf8 |
| N’Dama | Liver | CHTOG:h3f3a |
| N’Dama | Liver | p85alpha |
| N’Dama | Liver | TFII-I |
| N’Dama | Spleen | LYZL2-isoform2:LRP11 |
| N’Dama | Spleen | PON 2-isoform1:LRP11 |
| N’Dama | Spleen | WSX-1:LRP11 |
| N’Dama | Lymph node | Ssu72 |
| N’Dama | Lymph node | MTMR4 |
| N’Dama | Lymph node | Clathrin LCb |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rajavel, A.; Klees, S.; Hui, Y.; Schmitt, A.O.; Gültas, M. Deciphering the Molecular Mechanism Underlying African Animal Trypanosomiasis by Means of the 1000 Bull Genomes Project Genomic Dataset. Biology 2022, 11, 742. https://doi.org/10.3390/biology11050742
Rajavel A, Klees S, Hui Y, Schmitt AO, Gültas M. Deciphering the Molecular Mechanism Underlying African Animal Trypanosomiasis by Means of the 1000 Bull Genomes Project Genomic Dataset. Biology. 2022; 11(5):742. https://doi.org/10.3390/biology11050742
Chicago/Turabian StyleRajavel, Abirami, Selina Klees, Yuehan Hui, Armin Otto Schmitt, and Mehmet Gültas. 2022. "Deciphering the Molecular Mechanism Underlying African Animal Trypanosomiasis by Means of the 1000 Bull Genomes Project Genomic Dataset" Biology 11, no. 5: 742. https://doi.org/10.3390/biology11050742
APA StyleRajavel, A., Klees, S., Hui, Y., Schmitt, A. O., & Gültas, M. (2022). Deciphering the Molecular Mechanism Underlying African Animal Trypanosomiasis by Means of the 1000 Bull Genomes Project Genomic Dataset. Biology, 11(5), 742. https://doi.org/10.3390/biology11050742






