Optimizing the Growth Conditions of the Selected Plant-Growth-Promoting Rhizobacteria Paenibacillus sp. MVY-024 for Industrial Scale Production
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sample Collection
2.2. Isolation of N2-Fixer Rhizobacteria
2.3. Nitrogen Fixation Capabilities with N-Free Media
2.4. Endospore Formation Test
2.5. Molecular Identification of Isolated Microorganisms
2.6. Vegetative Growth Promotion in Wheat Plants by Paenibacillus spp.
2.7. Soil and Plant Analysis
2.8. Optimization of Carbon and Nitrogen Sources for Paenibacillus sp. MVY-024 Cultivation
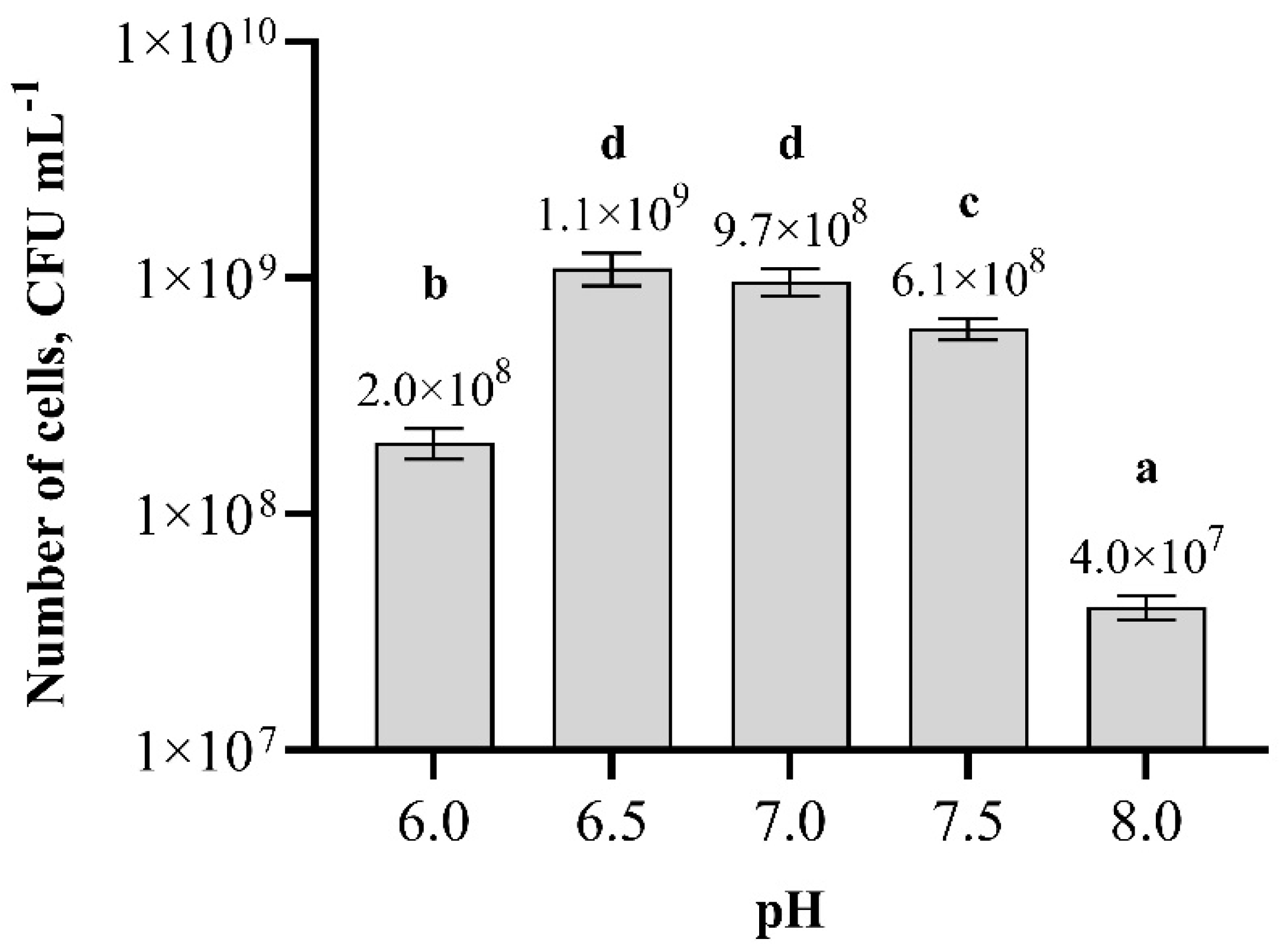
2.9. pH and Temperature Optimization
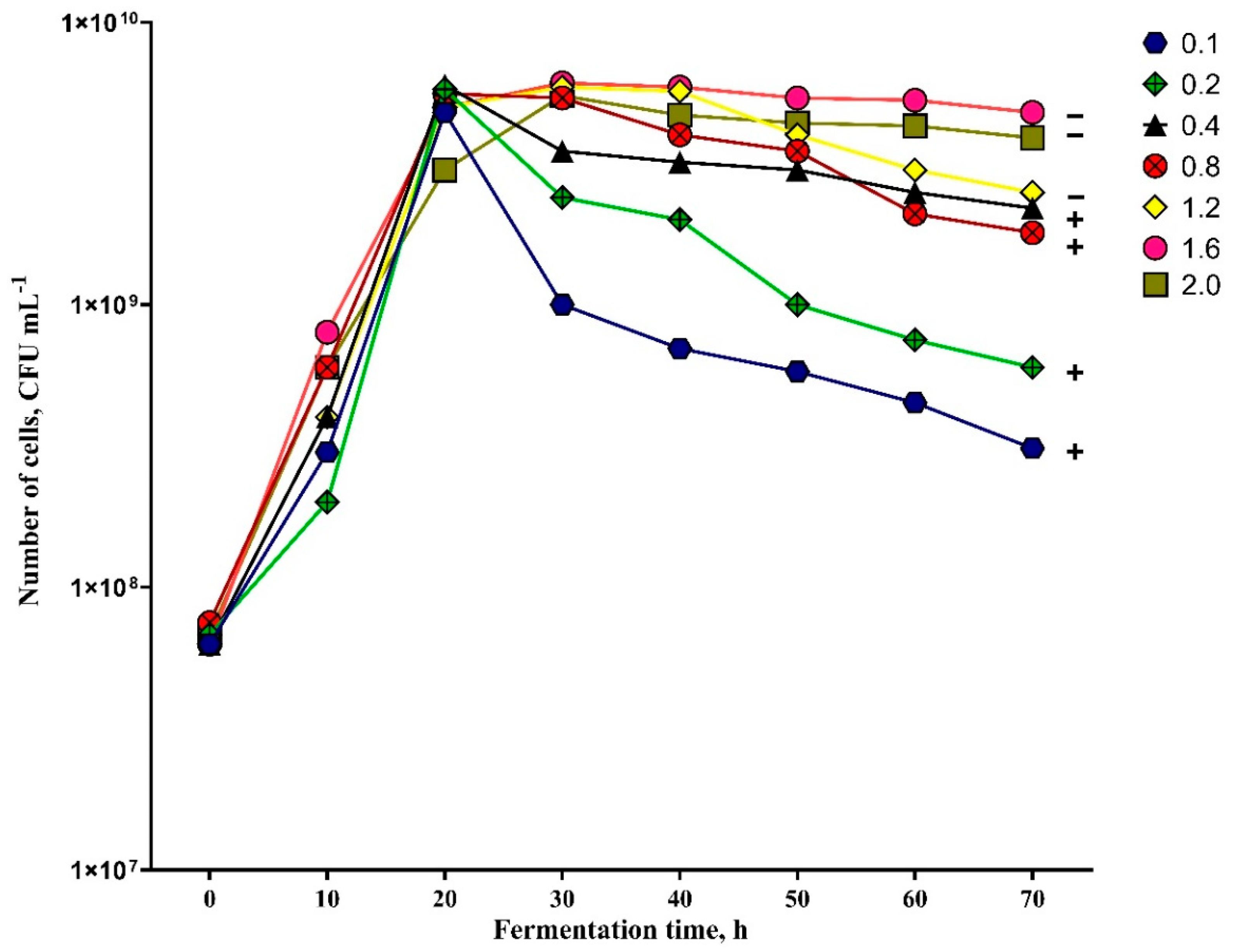
2.10. Optimization of Air Flow
2.11. Statistical Analysis
3. Results
3.1. Isolation and Description of the Physiological Properties of the Isolates
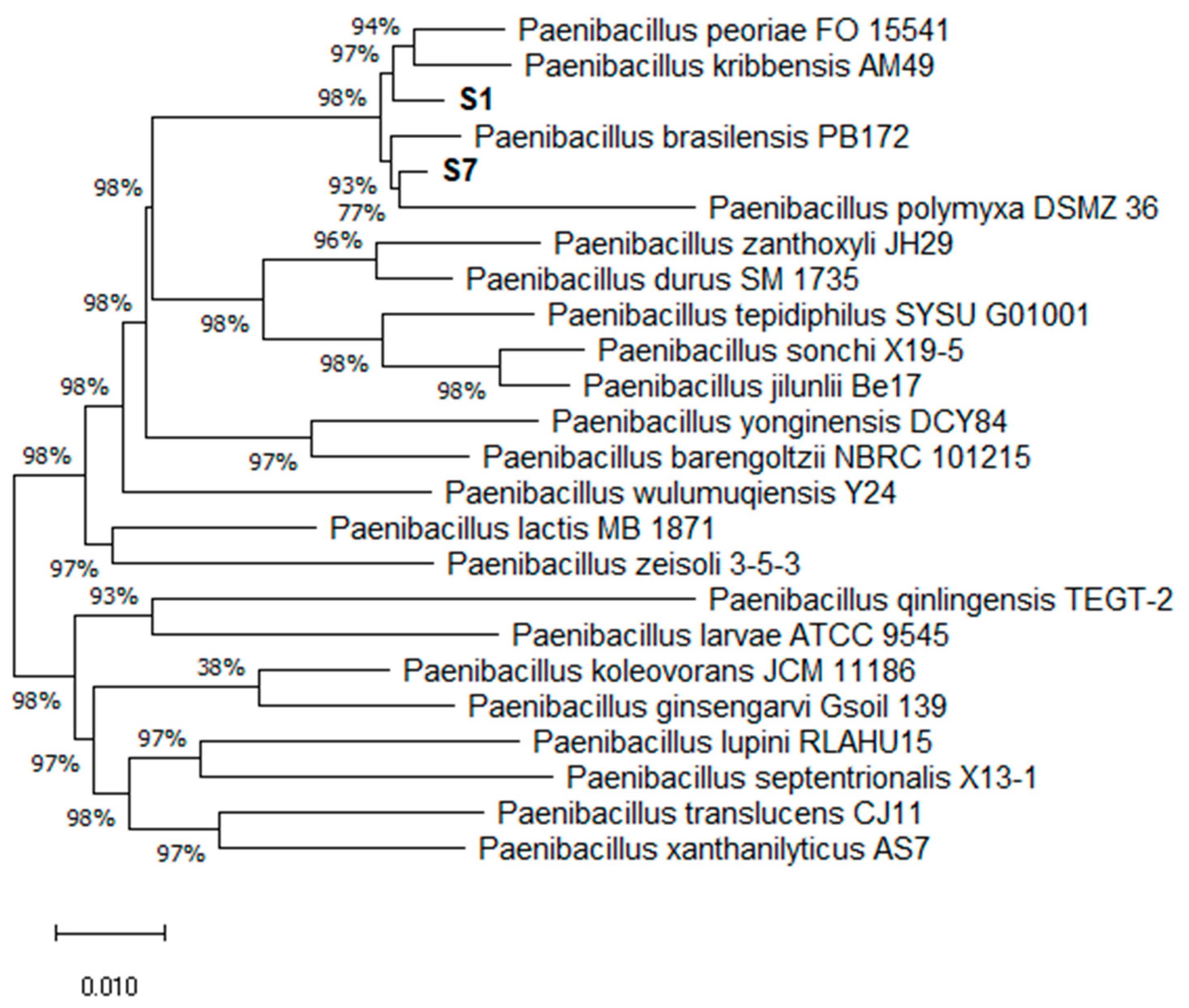
3.2. Phylogenetic Analysis
3.3. Soil Nitrogen Changes as Affected by Paenibacillus sp. S1 and Paenibacillus sp. S7
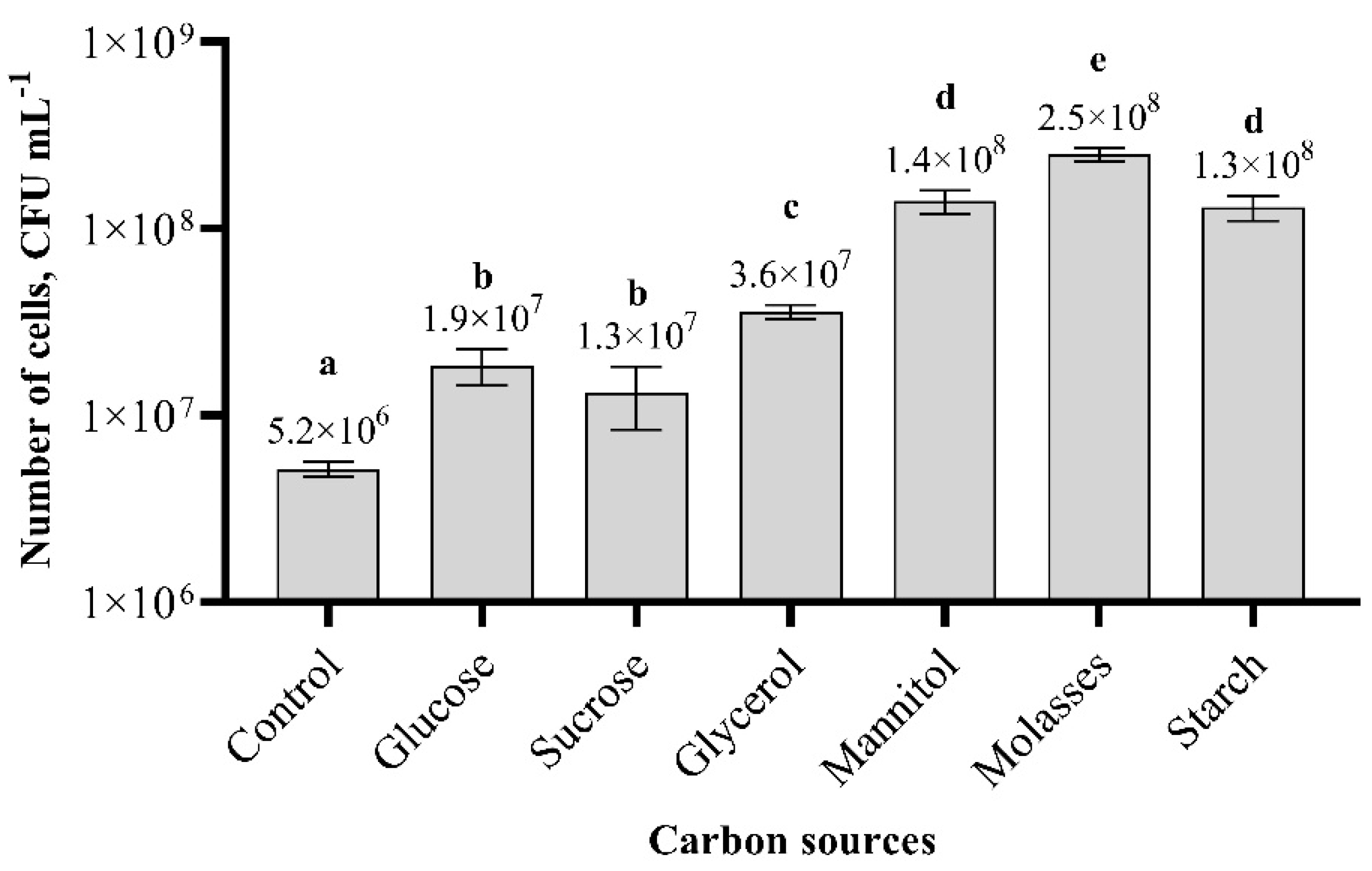
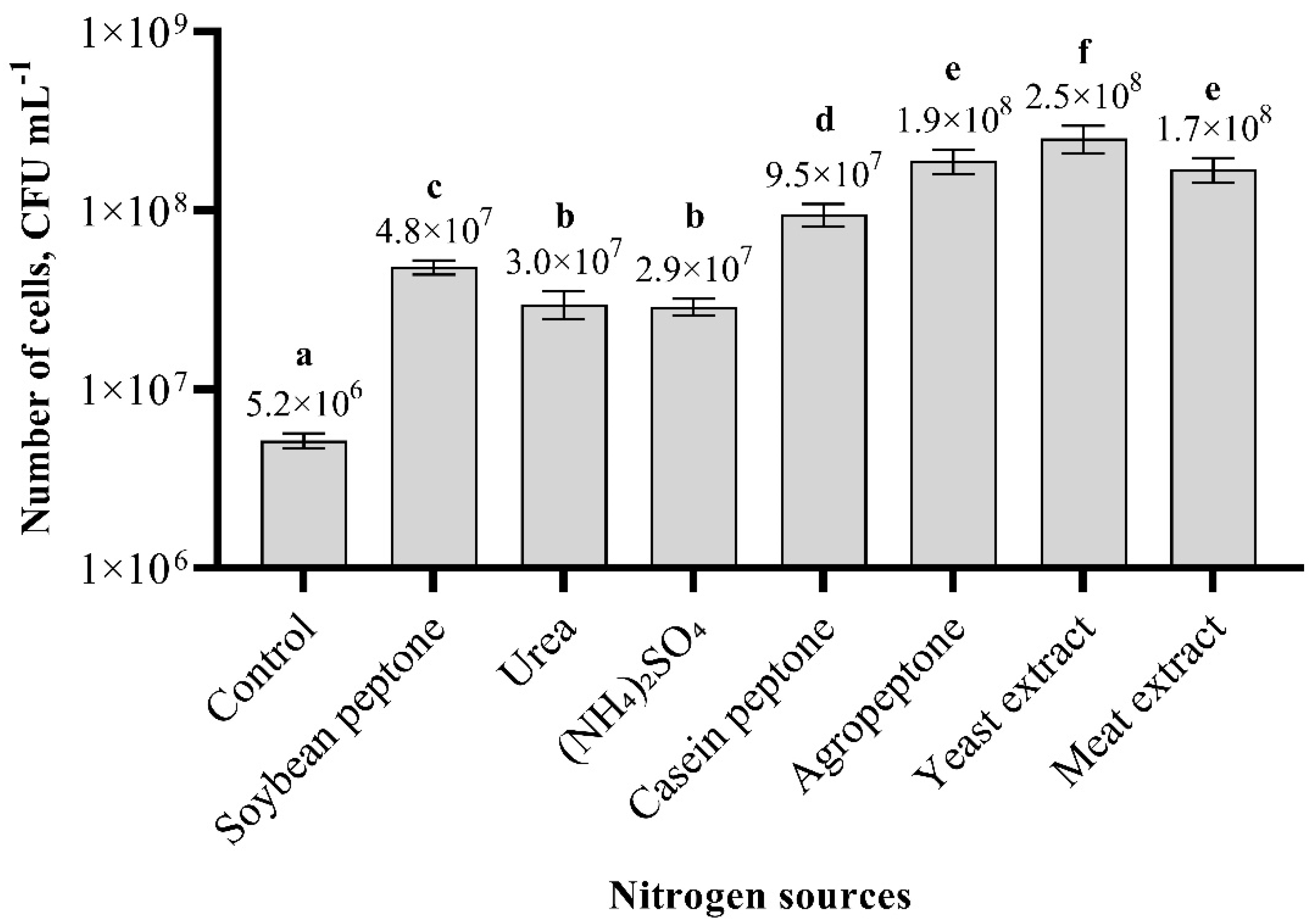
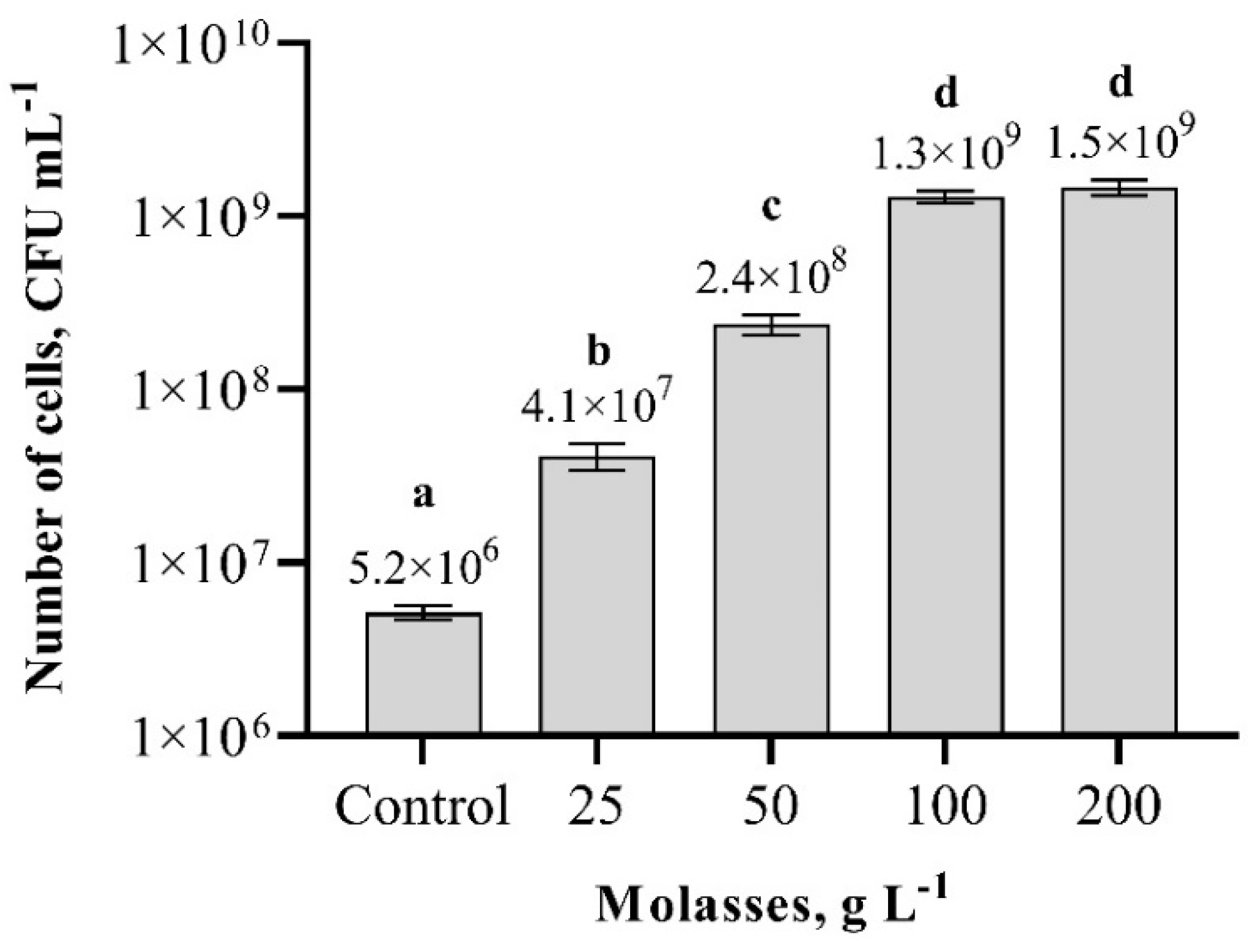
3.4. Effect of Carbon and Nitrogen Sources on Paenibacillus sp. MVY-024 Biomass Production
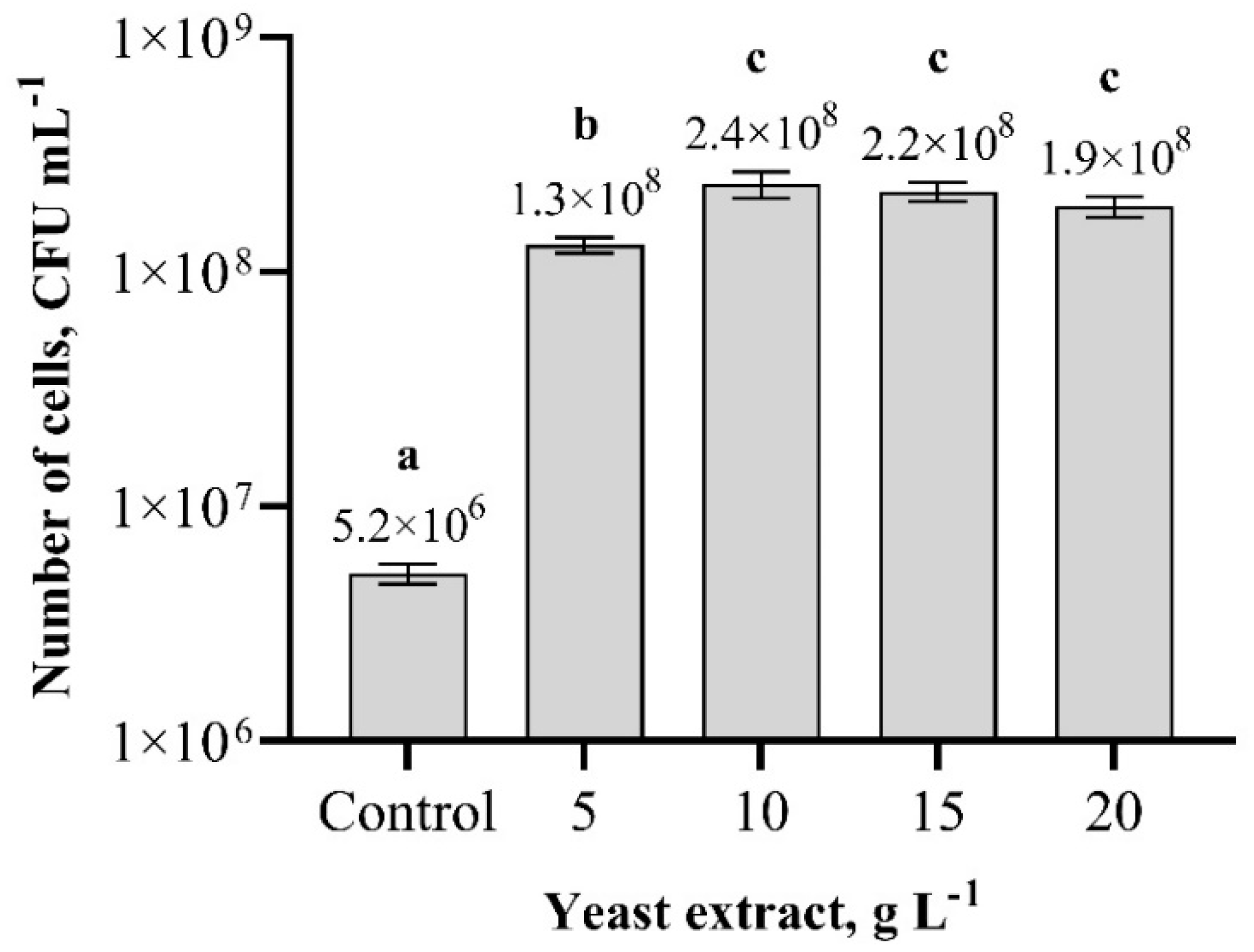
3.5. Optimization of Nutrition Broth pH and Temperature for Paenibacillus sp. MVY-024 Biomass Production
3.6. Optimization of Air Flow for Paenibacillus sp. MVY-024 Biomass Production
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Weselowski, B.; Nathoo, N.; Eastman, A.W.; MacDonald, J.; Yuan, Z.C. Isolation, identification and characterization of Paenibacillus polymyxa CR1 with potentials for biopesticide, biofertilization, biomass degradation and biofuel production. BMC Microbiol. 2016, 16, 244. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Langendries, S.; Goormachtig, S. Paenibacillus polymyxa, a Jack of all trades. Environ. Microbiol. 2021, 23, 5659–5669. [Google Scholar] [CrossRef] [PubMed]
- Puri, A.; Padda, K.P.; Chanway, C.P. Seedling growth promotion and nitrogen fixation by a bacterial endophyte Paenibacillus polymyxa P2b-2R and its GFP derivative in corn in a long-term trial. Symbiosis 2016, 69, 123–129. [Google Scholar] [CrossRef]
- Cherchali, A.; Boukhelata, N.; Kaci, Y.; Abrous-Belbachir, O.; Djebbar, R. Isolation and identification of a phosphate-solubilizing Paenibacillus polymyxa strain GOL 0202 from durum wheat (Triticum durum Desf.) rhizosphere and its effect on some seedlings morphophysiological parameters. Biocatal. Agric. Biotechnol. 2019, 19, 101087. [Google Scholar] [CrossRef]
- Xie, J.; Shi, H.; Du, Z.; Wang, T.; Liu, X.; Chen, S. Comparative genomic and functional analysis reveal conservation of plant growth promoting traits in Paenibacillus polymyxa and its closely related species. Sci. Rep. 2016, 6, 21329. [Google Scholar] [CrossRef]
- Abdallah, Y.; Yang, M.; Zhang, M.; Masum, M.; Ogunyemi, S.; Hossain, A.; An, Q.; Yan, C.; Li, B. Plant growth promotion and suppression of bacterial leaf blight in rice by Paenibacillus polymyxa Sx3. Lett. Appl. Microbiol. 2019, 68, 423–429. [Google Scholar] [CrossRef]
- Khan, M.S.; Gao, J.; Chen, X.; Zhang, M.; Yang, F.; Du, Y.; Moe, T.S.; Munir, I.; Xue, J.; Zhang, X. Isolation and Characterization of Plant Growth-Promoting Endophytic Bacteria Paenibacillus polymyxa SK1 from Lilium lancifolium. BioMed Res. Int. 2020, 2020, 8650957. [Google Scholar] [CrossRef] [Green Version]
- He, Z.; Kisla, D.; Zhang, L.; Yuan, C.; Green-Church, K.B.; Yousef, A.E. Isolation and Identification of a Paenibacillus polymyxa Strain That Coproduces a Novel Lantibiotic and Polymyxin. Appl. Environ. Microbiol. 2007, 73, 168–178. [Google Scholar] [CrossRef] [Green Version]
- Choi, S.-K.; Park, S.-Y.; Kim, R.; Lee, C.-H.; Kim, J.F.; Park, S.-H. Identification and functional analysis of the fusaricidin biosynthetic gene of Paenibacillus polymyxa E681. Biochem. Biophys. Res. Commun. 2008, 365, 89–95. [Google Scholar] [CrossRef]
- Shaheen, M.; Li, J.; Ross, A.C.; Vederas, J.; Jensen, S.E. Paenibacillus polymyxa PKB1 Produces Variants of Polymyxin B-Type Antibiotics. Chem. Biol. 2011, 18, 1640–1648. [Google Scholar] [CrossRef] [Green Version]
- Timmusk, S.; Grantcharova, N.; Wagner, E.G.H. Paenibacillus polymyxa Invades Plant Roots and Forms Biofilms. Appl. Environ. Microbiol. 2005, 71, 7292–7300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timmusk, S.; Copolovici, D.; Copolovici, L.; Teder, T.; Nevo, E.; Behers, L. Paenibacillus polymyxa biofilm polysaccharides antagonise Fusarium graminearum. Sci. Rep. 2019, 9, 662. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Li, X.-L.; Zhu, S.-J.; Ojaghian, M.R.; Zhang, J.-Z. Biocontrol potential of Paenibacillus polymyxa against Verticillium dahliae infecting cotton plants. Biol. Control 2018, 127, 70–77. [Google Scholar] [CrossRef]
- Hou, X.; Yu, X.; Du, B.; Liu, K.; Yao, L.; Zhang, S.; Selin, C.; Fernando, W.; Wang, C.; Ding, Y. A single amino acid mutation in Spo0A results in sporulation deficiency of Paenibacillus polymyxa SC2. Res. Microbiol. 2016, 167, 472–479. [Google Scholar] [CrossRef] [Green Version]
- Savci, S. Investigation of Effect of Chemical Fertilizers on Environment. APCBEE Procedia 2012, 1, 287–292. [Google Scholar] [CrossRef] [Green Version]
- Jimoh, A.A.; Lin, J. Enhancement of Paenibacillus sp. D9 Lipopeptide Biosurfactant Production Through the Optimization of Medium Composition and Its Application for Biodegradation of Hydrophobic Pollutants. Appl. Biochem. Biotechnol. 2018, 187, 724–743. [Google Scholar] [CrossRef]
- Yu, Z.; Sun, Z.; Yin, J.; Qiu, J. Enhanced Production of Polymyxin E in Paenibacillus polymyxa by Replacement of Glucose by Starch. BioMed Res. Int. 2018, 2018, 1934309. [Google Scholar] [CrossRef] [Green Version]
- Doty, S.L.; Oakley, B.; Xin, G.; Kang, J.W.; Singleton, G.; Khan, Z.; Vajzovic, A.; Staley, J.T. Diazotrophic endophytes of native black cottonwood and willow. Symbiosis 2009, 47, 23–33. [Google Scholar] [CrossRef]
- Hara, S.; Hashidoko, Y.; Desyatkin, R.V.; Morishita, T.; Hatano, R. Clear increases in acetylene reduction by soil bacteria from an East Siberian Taiga forest bed under conditions mimicking the natural soil environment. Soil Sci. Plant Nutr. 2010, 56, 716–724. [Google Scholar] [CrossRef]
- Kifle, M.H.; Laing, M.D. Isolation and Screening of Bacteria for Their Diazotrophic Potential and Their Influence on Growth Promotion of Maize Seedlings in Greenhouses. Front. Plant Sci. 2016, 6, 2. [Google Scholar] [CrossRef] [Green Version]
- Shukla, A.K.; Vishwakarma, P.; Singh, R.; Upadhyay, S.; Dubey, S.K. Bio-filtration of trichloroethylene using diazotrophic bacterial community. Bioresour. Technol. 2010, 101, 2126–2133. [Google Scholar] [CrossRef] [PubMed]
- Duangkhet, M.; Chikoti, Y.; Thepsukhon, A.; Thapanapongworakul, P.; Chungopast, S.; Tajima, S.; Nomura, M. Isolation and characterization of rhizobia from nodules of Clitoria ternatea in Thailand. Plant Biotechnol. 2018, 35, 123–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vidic, J.; Chaix, C.; Manzano, M.; Heyndrickx, M. Food Sensing: Detection of Bacillus cereus Spores in Dairy Products. Biosensors 2020, 10, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galkiewicz, J.P.; Kellogg, C.A. Cross-Kingdom Amplification Using Bacteria-Specific Primers: Complications for Studies of Coral Microbial Ecology. Appl. Environ. Microbiol. 2008, 74, 7828–7831. [Google Scholar] [CrossRef] [Green Version]
- IUSS Working Group WRB. International Soil Classification System for Naming Soils and Creating Legends for Soil Maps. In World Reference Base for Soil Resources 2014; World Soil Resources Reports No. 106; Food and Agriculture Organization of the United Nations: Rome, Italy, 2014. [Google Scholar] [CrossRef]
- Sáez-Plaza, P.; Navas, M.J.; Wybraniec, S.; Michałowski, T.; Asuero, A.G. An Overview of the Kjeldahl Method of Nitrogen Determination. Part II. Sample Preparation, Working Scale, Instrumental Finish, and Quality Control. Crit. Rev. Anal. Chem. 2013, 43, 224–272. [Google Scholar] [CrossRef]
- McKenney, P.T.; Driks, A.; Eichenberger, P. The Bacillus subtilis endospore: Assembly and functions of the multilayered coat. Nat. Rev. Genet. 2012, 11, 33–44. [Google Scholar] [CrossRef]
- Faulds-Pain, A.; Martin, M.J.; Wren, B.W. Clostridium difficile—A Pathogen on the Move. In Molecular Medical Microbiology, 2nd ed.; Academic Press: Cambridge, MA, USA, 2014. [Google Scholar] [CrossRef]
- Yegorenkova, I.V.; Tregubova, K.V.; Krasov, A.I.; Evseeva, N.V.; Matora, L.Y. Effect of exopolysaccharides of Paenibacillus polymyxa rhizobacteria on physiological and morphological variables of wheat seedlings. J. Microbiol. 2021, 59, 729–735. [Google Scholar] [CrossRef]
- Karnwal, A. Screening, isolation and characterization of culturable stress-tolerant bacterial endophytes associated with Salicornia brachiata and their effect on wheat (Triticum aestivum L.) and maize (Zea mays) growth. J. Plant Prot. Res. 2019, 59, 293–303. [Google Scholar] [CrossRef]
- Padda, K.P.; Puri, A.; Chanway, C.P. Effect of GFP tagging of Paenibacillus polymyxa P2b-2R on its ability to promote growth of canola and tomato seedlings. Biol. Fertil. Soils 2015, 52, 377–387. [Google Scholar] [CrossRef]
- Gong, X.-Y.; Luan, Z.-K.; Pei, Y.-S.; Wang, S.-G. Culture Conditions for Flocculant Production by Paenibacillus polymyxa BY-28. J. Environ. Sci. Health Part A Toxic/Hazard. Subst. Environ. Eng. 2003, 38, 657–669. [Google Scholar] [CrossRef]
- Raza, W.; Makeen, K.; Wang, Y.; Xu, Y.; Qirong, S. Optimization, purification, characterization and antioxidant activity of an extracellular polysaccharide produced by Paenibacillus polymyxa SQR-21. Bioresour. Technol. 2011, 102, 6095–6103. [Google Scholar] [CrossRef] [PubMed]
- Okonkwo, C.C.; Ujor, V.C.; Mishra, P.K.; Ezeji, T.C. Process Development for Enhanced 2,3-Butanediol Production by Paenibacillus polymyxa DSM 365. Fermentation 2017, 3, 18. [Google Scholar] [CrossRef] [Green Version]
- Tinôco, D.; de Castro, A.M.; Seldin, L.; Freire, D.M. Production of (2R,3R)-butanediol by Paenibacillus polymyxa PM 3605 from crude glycerol supplemented with sugarcane molasses. Process Biochem. 2021, 106, 88–95. [Google Scholar] [CrossRef]
- De Mas, C.; Jansen, N.B.; Tsao, G.T. Production of optically active 2,3-butanediol by Bacillus polymyxa. Biotechnol. Bioeng. 1988, 31, 366–377. [Google Scholar] [CrossRef]
- Nakashimada, Y.; Kanai, K.; Nishio, N. Optimization of dilution rate, pH and oxygen supply on optical purity of 2, 3-butanediol produced by Paenibacillus polymyxa in chemostat culture. Biotechnol. Lett. 1998, 20, 1133–1138. [Google Scholar] [CrossRef]
- Häßler, T.; Schieder, D.; Pfaller, R.; Faulstich, M.; Sieber, V. Enhanced fed-batch fermentation of 2,3-butanediol by Paenibacillus polymyxa DSM 365. Bioresour. Technol. 2012, 124, 237–244. [Google Scholar] [CrossRef]
- Barros-Rodríguez, A.; Rangseekaew, P.; Lasudee, K.; Pathom-Aree, W.; Manzanera, M. Regulatory risks associated with bacteria as biostimulants and biofertilizers in the frame of the European Regulation (EU) 2019/1009. Sci. Total Environ. 2020, 740, 140239. [Google Scholar] [CrossRef]
- Regulation (EU) 2019/1009 of the European Parliament and of the Council. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/?uri=CELEX%3A32019R1009 (accessed on 5 May 2022).

| Carbon Source Optimization | Nitrogen Source Optimization | ||||||
|---|---|---|---|---|---|---|---|
| Carbon Source, g L−1 | Nitrogen Source, g L−1 | Nitrogen Source, g L−1 | Carbon Source, g L−1 | ||||
| Glucose | 40 | Yeast extract | 10 | Soybean peptone | 10 | Molasses | 57 |
| Sucrose | 40 | Yeast extract | 10 | Carbamide | 10 | Molasses | 57 |
| Glycerol | 40 | Yeast extract | 10 | Ammonium suphate | 10 | Molasses | 57 |
| Mannitol | 40 | Yeast extract | 10 | Casein peptone | 10 | Molasses | 57 |
| Molasses | 57 | Yeast extract | 10 | Agropeptone | 10 | Molasses | 57 |
| Starch | 40 | Yeast extract | 10 | Yeast extract | 10 | Molasses | 57 |
| Meat extract | 10 | Molasses | 57 | ||||
| Molasses concentration optimization | Yeast extract concentration optimization | ||||||
| Molasses | 25 | Yeast extract | 10 | Yeast extract | 5 | Molasses | 57 |
| Molasses | 50 | Yeast extract | 10 | Yeast extract | 10 | Molasses | 57 |
| Molasses | 100 | Yeast extract | 10 | Yeast extract | 15 | Molasses | 57 |
| Molasses | 200 | Yeast extract | 10 | Yeast extract | 20 | Molasses | 57 |
| Isolates | Ashby’s | Winogradsky’s | NF | NFB | Gram Reaction | Endospore Formation |
|---|---|---|---|---|---|---|
| Isolates from soil | ||||||
| Isolate S1 | +++ | + | +++ | + | + | + |
| Isolate S2 | ++ | ++ | + | + | − | − |
| Isolate S3 | ++ | ++ | +++ | + | − | − |
| Isolate S4 | − | − | + | − | + | + |
| Isolate S5 | ++ | +++ | ++ | − | + | + |
| Isolate S6 | + | ++ | + | − | + | + |
| Isolate S7 | ++ | ++ | +++ | + | + | + |
| Isolate S8 | + | + | + | − | + | + |
| Isolate S9 | + | ++ | ++ | − | + | + |
| Isolates from pea root nodules | ||||||
| Isolate R1 | + | + | ++ | − | + | + |
| Isolate R2 | − | − | ++ | − | + | + |
| Isolate R3 | +++ | + | ++ | + | − | − |
| Isolate R4 | +++ | − | +++ | + | − | − |
| Isolate | Hit Taxon | Similarity, % | Sequence Length, bp | Accession |
|---|---|---|---|---|
| S1 | Paenibacillus peoriae DSM 8320(T) | 99.28 | 1390 | AJ320494 |
| S2 | Ensifer meliloti LMG 6133(T) | 100.00 | 1320 | X67222 |
| S3 | Ensifer medicae WSM419(T) | 99.70 | 1320 | CP000738 |
| S4 | Bacillus siamensis KCTC 13613(T) | 99.86 | 1390 | AJVF01000043 |
| S5 | Bacillus mojavensis RO-H-1(T) | 99.93 | 1390 | JH600280 |
| S6 | Bacillus paramycoides NH24A2(T) | 99.93 | 1400 | MAOI01000012 |
| S7 | Paenibacillus polymyxa ATCC 842(T) | 99.71 | 1380 | AFOX01000032 |
| S8 | Bacillus paralicheniformis KJ-16(T) | 100.00 | 1390 | KY694465 |
| S9 | Bacillus siamensis KCTC 13613(T) | 99.93 | 1390 | AJVF01000043 |
| R1 | Bacillus zanthoxyli 1433(T) | 99.51 | 1421 | KX865140 |
| R2 | Bacillus velezensis CR-502(T) | 99.85 | 1399 | AY603658 |
| R3 | Lelliottia amnigena NBRC 105700(T) | 99.42 | 1379 | BCNN01000001 |
| R4 | Rhizobium leguminosarum USDA 2370(T) | 100.00 | 1334 | MRDL01000029 |
| No. | Treatment | Soil N-NH4, after 4 Days | Soil N-NH4, after 2 Weeks | Soil N-NH4, after 2 Months | Soil Nmin, after 2 Months |
|---|---|---|---|---|---|
| 1. | Control | 36.5 ab | 36.5 a | 1.12 a | 3.13 ab |
| 2. | Paenibacillus sp. S7 | 42.3 b | 47.1 b | 2.87 b | 4.48 b |
| 3. | Paenibacillus sp. S1 | 41.6 ab | 48.1 c | 0.29 a | 1.48 a |
| Probability Pr>F | 0.1415 | 0.001 | 0.026 | 0.042 | |
| No. | Treatment | N Yield in Grain, g−1 per Plant | Leaf Chlorophyll, SPAD | Grain Yield per Plant, g | TKW, g | Protein in Grain, % | Kernels per Spike, Unit |
|---|---|---|---|---|---|---|---|
| 1. | Control | 0.0355 ab | 45.8 a | 1.42 a | 40.9 a | 11.5 a | 34.7 a |
| 2. | Paenibacillus sp. S7 | 0.0361 b | 46.8 b | 1.54 b | 43.1 b | 12.8 b | 36.0 b |
| 3. | Paenibacillus sp. S1 | 0.0297 a | 45.1 a | 1.46 ab | 40.4 a | 11.1 a | 36.2 b |
| Probability Pr > F | 0.041 | 0.025 | 0.361 | 0.196 | 0.118 | 0.831 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kaziūnienė, J.; Mažylytė, R.; Krasauskas, A.; Toleikienė, M.; Gegeckas, A. Optimizing the Growth Conditions of the Selected Plant-Growth-Promoting Rhizobacteria Paenibacillus sp. MVY-024 for Industrial Scale Production. Biology 2022, 11, 745. https://doi.org/10.3390/biology11050745
Kaziūnienė J, Mažylytė R, Krasauskas A, Toleikienė M, Gegeckas A. Optimizing the Growth Conditions of the Selected Plant-Growth-Promoting Rhizobacteria Paenibacillus sp. MVY-024 for Industrial Scale Production. Biology. 2022; 11(5):745. https://doi.org/10.3390/biology11050745
Chicago/Turabian StyleKaziūnienė, Justina, Raimonda Mažylytė, Aurimas Krasauskas, Monika Toleikienė, and Audrius Gegeckas. 2022. "Optimizing the Growth Conditions of the Selected Plant-Growth-Promoting Rhizobacteria Paenibacillus sp. MVY-024 for Industrial Scale Production" Biology 11, no. 5: 745. https://doi.org/10.3390/biology11050745
APA StyleKaziūnienė, J., Mažylytė, R., Krasauskas, A., Toleikienė, M., & Gegeckas, A. (2022). Optimizing the Growth Conditions of the Selected Plant-Growth-Promoting Rhizobacteria Paenibacillus sp. MVY-024 for Industrial Scale Production. Biology, 11(5), 745. https://doi.org/10.3390/biology11050745







