Isolation of Listeria ivanovii from Bulk-Tank Milk of Sheep and Goat Farms—From Clinical Work to Bioinformatics Studies: Prevalence, Association with Milk Quality, Antibiotic Susceptibility, Predictors, Whole Genome Sequence and Phylogenetic Relationships
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
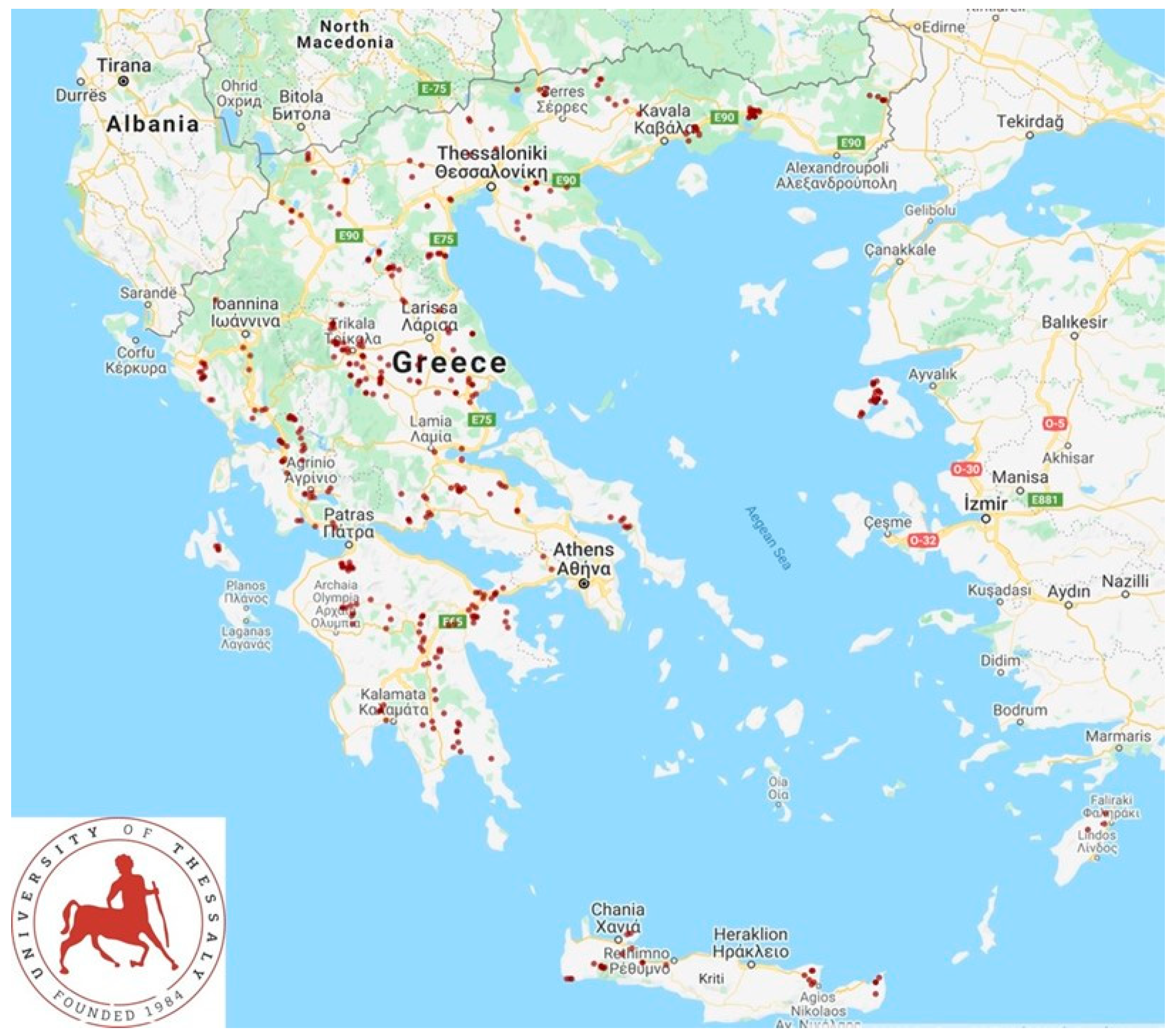
2.1. Farms and Sampling
2.2. Overview of Laboratory Examinations
2.3. Procedures for Listeria spp. Isolation and Identification
2.4. Testing for Susceptibility to Antibiotics
2.5. Data Management and Analysis
2.6. Whole Genome Analysis of L. ivanovii Isolates
2.7. Phylogenetic Relationships of L. ivanovii Isolates
3. Results
3.1. Isolation of Listeria monocytogenes and Listeria ivanovii from Bulk-Tank Raw Milk
3.2. Lack of an Association of Isolation of L. monocytogenes or L. ivanovii with Milk Quality
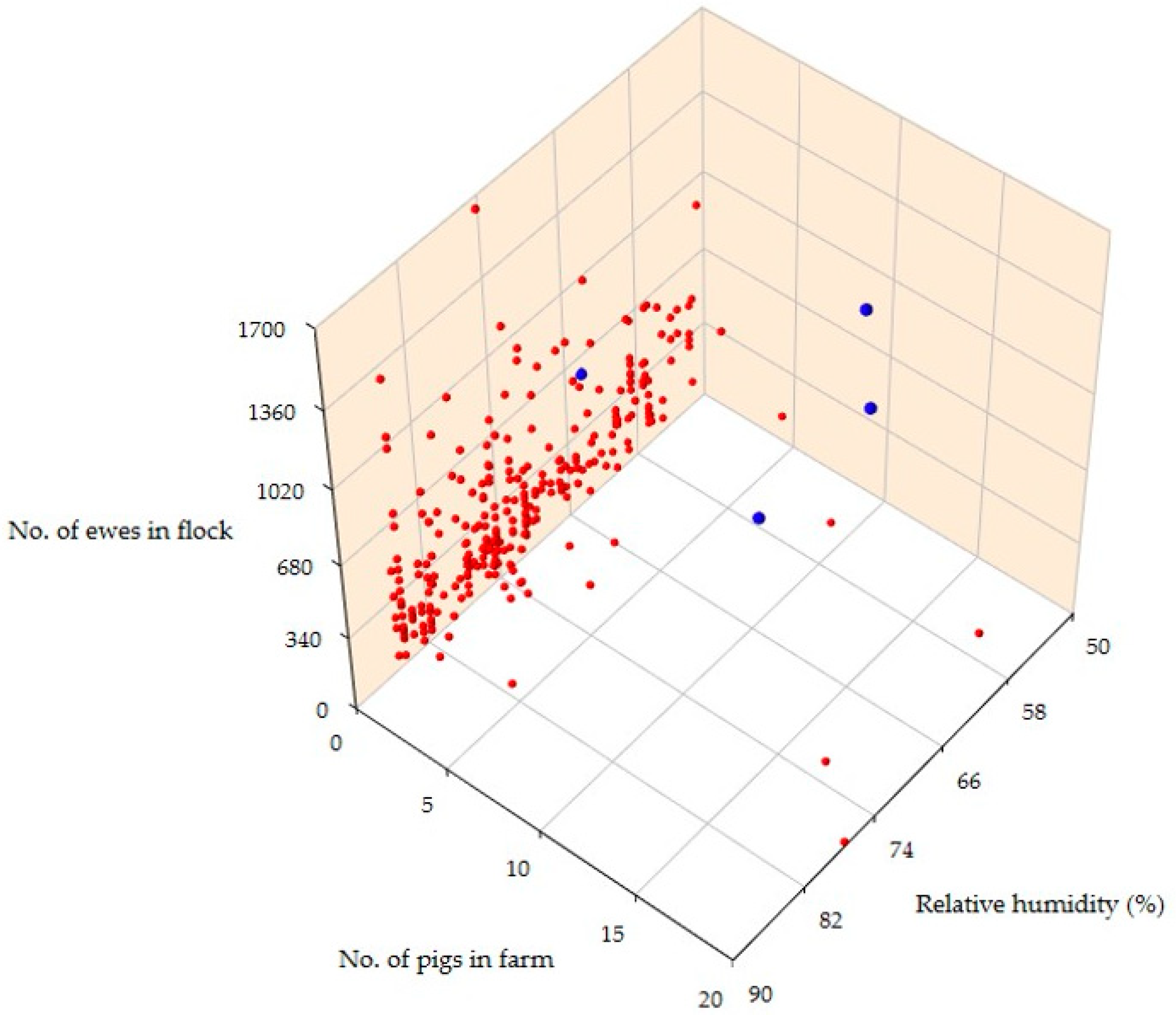
3.3. Predictors of the Isolation of L. monocytogenes or L. ivanovii from Bulk-Tank Raw Milk
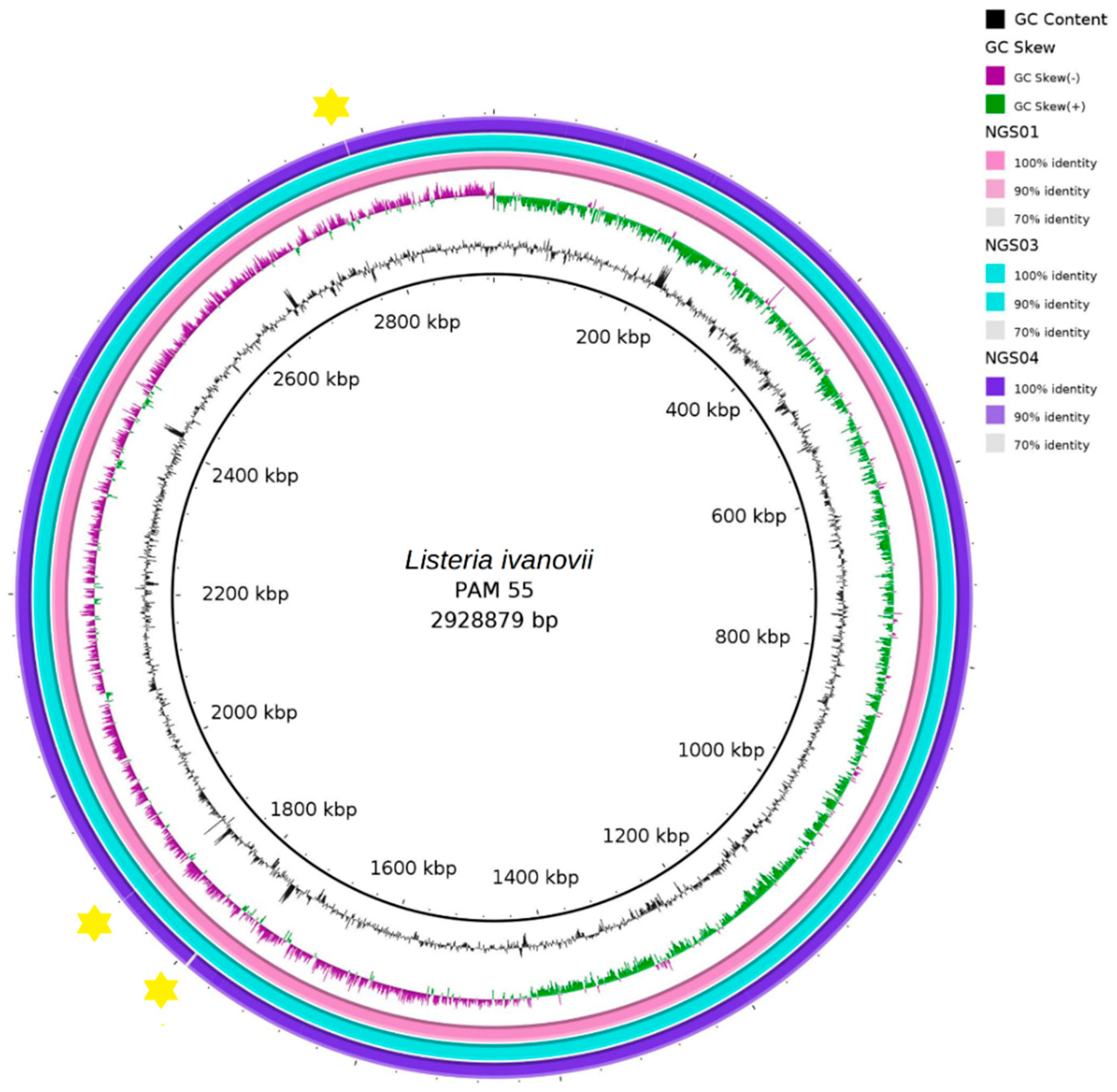
3.4. Whole Genome Analysis of L. ivanovii Isolates
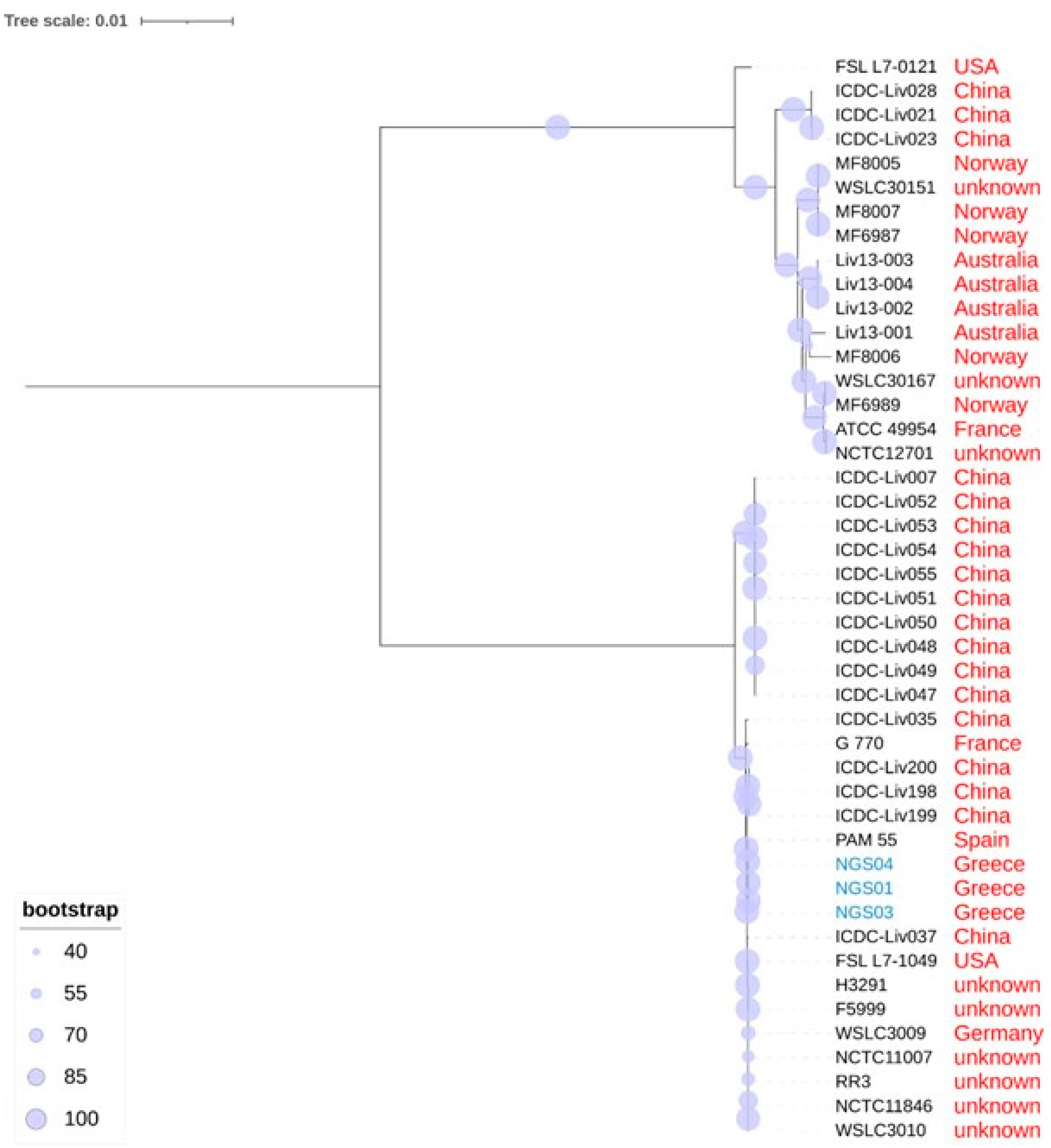
3.5. Phylogenetic relationships of L. ivanovii Isolates
4. Discussion
4.1. Presence and Identification of L. monocytogenes or L. ivanovii in Bulk-Tank Milk
4.2. Predictors of the Ιsolation of L. monocytogenes or L. ivanovii in Βulk-Τank Μilk
4.3. Bioinformatics Assessment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seeliger, H.P.R.; Rocourt, J.; Schretten-Brunner, A.; Grimont, P.A.D.; Jones, D. Listeria ivanovii sp. nov. Int. J. Syst. Bacteriol. 1984, 34, 336–337. [Google Scholar] [CrossRef]
- Parte, A.C.; Carbasse, J.S.; Meier-Kolthoff, J.P.; Reimer, L.C.; Goeker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef] [PubMed]
- Chiara, M.; Caruso, M.; D’Erchia, A.M.; Manzari, C.; Fraccalvieri, R.; Goffredo, E.; Latorre, L.; Miccolupo, A.; Padalino, I.; Santagada, G.; et al. Comparative genomics of Listeria sensu lato: Genus-wide differences in evolutionary dynamics and the progressive gain of complex, potentially pathogenicity-related traits through lateral gene transfer. Genome Biol. Evol. 2015, 7, 2154–2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buchrieser, C.; Rusniok, C.; Garrido, P.; Hain, T.; Scortti, M.; Lampidis, R.; Karst, U.; Chakraborty, T.; Cossart, P.; Kreft, J.; et al. Complete genome sequence of the animal pathogen Listeria ivanovii, which provides insights into host specificities and evolution of the genus Listeria. J. Bacteriol. 2011, 193, 6787–6788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sarangi, L.N.; Panda, H.K. Isolation, characterization and antibiotic sensitivity test of pathogenic Listeria species in livestock, poultry and farm environment of Odisha. Ind. J. Anim. Res. 2011, 46, 242–247. [Google Scholar]
- Dunnett, E.; Florea, L.; Thurston, L.; Floyd, T.; Collins, R.; Otter, A. Deaths of weaned lambs with visceral Listeria ivanovii infections. Vet. Rec. Case Rep. 2020, 8, e001254. [Google Scholar] [CrossRef]
- Rodriguez-Lazaro, D.; Lopez-Enriquez, L.; Hernandez, M. smcL as a novel diagnostic marker for quantitative detection of Listeria ivanovii in biological samples. J. Appl. Microbiol. 2010, 109, 863–872. [Google Scholar] [CrossRef]
- Osman, K.M.; Zolnikov, T.R.; Samir, A.; Orabi, A. Prevalence, pathogenic capability, virulence genes, biofilm formation, and antibiotic resistance of Listeria in goat and sheep milk confirms need of hygienic milking conditions. Pathog. Glob. Health 2014, 108, 21–29. [Google Scholar] [CrossRef] [Green Version]
- Soncini, G.; Valnegri, L. Analysis of bulk goats’ milk and milk-filters from Valtellina and Valchiavenna (Lombardy Prealps) for the presence of Listeria species. Small Rumin. Res. 2005, 58, 143–147. [Google Scholar] [CrossRef]
- Elezebeth, G.; Malik, S.V.S.; Chaudhari, S.P.; Barbuddhe, S.B. The occurrence of Listeria species and antibodies against listeriolysin-O in naturally infected goats. Small Rumin. Res. 2007, 67, 173–178. [Google Scholar] [CrossRef]
- Arimi, S.M.; Ryser, E.T.; Pritchard, T.J.; Donnelly, C.W. Diversity of Listeria ribotypes recovered from dairy cattle, silage, and dairy processing environments. J. Food Prot. 1997, 60, 811–816. [Google Scholar] [CrossRef] [PubMed]
- Terentjeva, M.; Steingolde, Z.; Meistere, I.; Elferts, D.; Avsejenko, J.; Streikisa, M.; Gradovska, S.; Alksne, L.; Kibilds, J.; Berzins, A. Prevalence, genetic diversity and factors associated with distribution of Listeria monocytogenes and other Listeria spp. in cattle farms in Latvia. Pathogens 2021, 10, 851. [Google Scholar] [CrossRef] [PubMed]
- Rawool, D.B.; Malik, S.V.S.; Shakuntala, I.; Sahare, A.M.; Barbuddhe, S.B. Detection of multiple virulence-associated genes in Listeria monocytogenes, isolated from bovine mastitis cases. Int. J. Food Microbiol. 2007, 113, 201–207. [Google Scholar] [CrossRef] [PubMed]
- Ramage, C.P.; Low, J.C.; McLauchlin, J.; Donachie, W. Characterisation of Listeria ivanovii isolates from the U.K. using pulsed-field gel electrophoresis. FEBS Microbiol. Lett. 1999, 170, 349–353. [Google Scholar] [CrossRef] [PubMed]
- Snapir, Y.M.; Vaisbein, E.; Nassar, F. Low virulence but potentially fatal outcome—Listeria ivanovii. Eur. J. Intern. Med. 2006, 17, 286–287. [Google Scholar] [CrossRef]
- Guillet, C.; Join-Lambert, O.; Le Monnier, A.; Leclercq, A.; Mechai, F.; Mamzer-Bruneel, M.F.; Bielecka, M.K.; Scortti, M.; Disson, O.; Berche, P.; et al. Human listeriosis caused by Listeria ivanovii. Emerg. Infect Dis. 2010, 16, 136–138. [Google Scholar] [CrossRef]
- Giacone, V.; Bertoja, G. Listeria and listeriosis in the milk production’s chain: An up-to-date. Large Anim. Rev. 2013, 19, 280–286. [Google Scholar]
- Hellenic Statistical Authority. Farm Structure Surveys. Available online: https://www.statistics.gr (accessed on 15 February 2022).
- Pulina, G.; Milan, M.J.; Lavin, M.P.; Theodoridis, A.; Morin, E.; Capote, J.; Thomas, D.L.; Francesconi, A.H.D.; Caja, G. Invited review: Current production trends, farm structures, and economics of the dairy sheep and goat sector. J. Dairy Sci. 2018, 101, 6715–6729. [Google Scholar] [CrossRef] [Green Version]
- Hellenic Agricultural Organisation—Demeter. Deliveries of Ovine and Caprine Milk by Region and Regional Authority and Average Milk Price—Calendar Year 2019. Available online: https://www.elgo.gr/images/ELOGAK_files/Statistics/2021/Milk21/6.%CE%95%CE%9B%CE%93%CE%9F_STATS_%CE%A0%CE%91%CE%A1%CE%91%CE%94_%CE%A0%CE%9F%CE%A3_%CE%A0%CE%A1%CE%9F%CE%92%CE%95%CE%99%CE%9F%CE%A5_%CE%93%CE%99%CE%94%CE%99%CE%9D%CE%9F%CE%A5_%CE%91%CE%9D%CE%91_%CE%A0%CE%95%CE%A1%CE%99%CE%A6_2019.pdf (accessed on 12 January 2022).
- Lianou, D.T.; Michael, C.K.; Vasileiou, N.G.C.; Petinaki, E.; Cripps, P.J.; Tsilipounidaki, K.; Katsafadou, A.I.; Politis, A.P.; Kordalis, N.G.; Ioannidi, K.S.; et al. Extensive countrywide field investigation of somatic cell counts and total bacterial counts in bulk-tank raw milk in sheep flocks in Greece. Foods 2021, 10, 268. [Google Scholar] [CrossRef]
- Lianou, D.T.; Michael, C.K.; Vasileiou, N.G.C.; Petinaki, E.; Cripps, P.J.; Tsilipounidaki, K.; Katsafadou, A.I.; Politis, A.P.; Kordalis, N.G.; Ioannidi, K.S.; et al. Extensive countrywide field investigation of somatic cell counts and total bacterial counts in bulk-tank raw milk in goat herds in Greece. J. Dairy Res. 2021, 88, 307–313. [Google Scholar] [CrossRef]
- Lianou, D.T.; Chatziprodromidou, I.P.; Vasileiou, N.G.C.; Michael, C.K.; Mavrogianni, V.S.; Politis, A.P.; Kordalis, N.G.; Billinis, C.; Giannakopoulos, A.; Papadopoulos, E.; et al. A detailed questionnaire for the evaluation of health management in dairy sheep and goats. Animals 2020, 10, 1489. [Google Scholar] [CrossRef] [PubMed]
- Laird, D.T.; Gambrel-Lenarz, S.A.; Scher, F.M.; Graham, T.E.; Reddy, R. Microbiological count methods. In Standard Methods for the Examination of Dairy Products, 17th ed.; Wehr, H.M., Frank, J.F., Eds.; APHA Press: Washington, DC, USA, 2004; pp. 153–186. [Google Scholar]
- ISO 11290-1:2017; Microbiology of the Food Chain—Horizontal Method for the Detection and Enumeration of Listeria Monocytogenes and of Listeria spp.—Part 1: Detection Method. 2nd ed. International Organization for Standardization: Geneva, Switzerland, 2017; 36p.
- Ojima-Kato, T.; Yamamoto, N.; Takahashi, H.; Tamura, H. Matrix-assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry (MALDI-TOF MS) can precisely discriminate the lineages of Listeria monocytogenes and species of Listeria. PLoS ONE 2016, 11, e0159730. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thouvenot, P.; Vales, G.; Bracq-Dieye, H.; Tessaud-Rita, N.; Maury, M.M.; Moura, A.; Lecuit, M.; Leclercq, A. MALDI-TOF mass spectrometry-based identification of Listeria species in surveillance: A prospective study. J. Microbiol. Methods 2018, 144, 29–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrews, S. FastQC: A quality Control Tool for High Throughput Sequence Data, 2015. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/ (accessed on 15 January 2022).
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Bortolaia, V.; Kaas, R.F.; Ruppe, E.; Roberts, M.C.; Schwarz, S.; Cattoir, V.; Philippon, A.; Allesoe, R.L.; Rebelo, A.R.; Florensa, A.R.; et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 2020, 75, 3491–3500. [Google Scholar] [CrossRef]
- Zankari, E.; Allesøe, R.; Joensen, K.G.; Cavaco, L.M.; Lund, O.; Aarestrup, F.M. PointFinder: A novel web tool for WGS-based detection of antimicrobial resistance associated with chromosomal point mutations in bacterial pathogens. J. Antimicrob. Chemother. 2020, 72, 2764–2768. [Google Scholar] [CrossRef] [Green Version]
- Joensen, K.G.; Scheutz, F.; Lund, O.; Hasman, H.; Kaas, R.S.; Nielsen, E.M.; Aarestrup, F.M. Real-time whole-genome sequencing for routine typing, surveillance, and outbreak detection of verotoxigenic Escherichia coli. J. Clin. Microbiol. 2014, 52, 1501–1510. [Google Scholar] [CrossRef] [Green Version]
- Malberg Tetzschner, A.M.; Johnson, J.R.; Johnston, B.D.; Lund, O.; Scheutz, F. In Silico genotyping of Escherichia coli isolates for extraintestinal virulence genes by uUse of whole-genome sequencing data. J. Clin. Microbiol. 2020, 58, e01269-20. [Google Scholar] [CrossRef] [PubMed]
- Johansson, M.H.K.; Bortolaia, V.; Tansirichaiya, S.; Aarestrup, F.M.; Roberts, A.P.; Petersen, T.N. Detection of mobile genetic elements associated with antibiotic resistance in Salmonella enterica using a newly developed web tool: MobileElementFinder. J. Antimicrob. Chemother. 2021, 76, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, Y.; Lynch, K.H.; Dennis, J.J.; Wishart, D.S. PHAST: A fast phage search tool. Nucleic Acids Res. 2011, 39, W347–W352. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carattoli, A.; Zankari, E.; Garcia-Fernandez, A.; Voldby Larsen, M.; Lund, O.; Villa, L.; Aarestrup, F.M.; Hasman, H. PlasmidFinder and pMLST: In silico detection and typing of plasmids. Antimicrob. Agents Chemother. 2014, 58, 3895–3903. [Google Scholar] [CrossRef] [Green Version]
- Kurtz, S.; Schleiermacher, C. REPuter: Fast computation of maximal repeats in complete genomes. Bioinformatics 1999, 15, 426–427. [Google Scholar] [CrossRef] [Green Version]
- Abouelhoda, M.I.; Kurtz, S.; Ohlebusch, E. Replacing suffix trees with enhanced suffix arrays. J. Discret. Algorithms 2004, 2, 53–86. [Google Scholar] [CrossRef] [Green Version]
- Hyatt, D.; Chen, G.-L.; Locascio, P.F.; Land, M.L.; Larimer, F.W.; Hauser, L.J. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinform. 2010, 11, 119. [Google Scholar] [CrossRef] [Green Version]
- Abby, S.S.; Neron, B.; Menager, H.; Touchon, M.; Rocha, E.P. MacSyFinder: A program to mine genomes for molecular systems with an application to CRISPR-Cas systems. PLoS ONE 2014, 9, e110726. [Google Scholar] [CrossRef]
- Biswas, A.; Fineran, P.C.; Brown, C.M. Accurate computational prediction of the transcribed strand of CRISPR non-coding R.N.As. Bioinformatics 2014, 30, 1805–1813. [Google Scholar] [CrossRef]
- Clark, K.; Karsch-Mizrachi, I.; Lipman, D.J.; Ostell, J.; Sayers, E.W. GenBank. Nucleic Acids Res. 2016, 44, D67–D72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tettelin, H.; Riley, D.; Cattuto, C.; Medini, D. Comparative genomics: The bacterial pan-genome. Curr. Opin. Microbiol. 2008, 11, 472–477. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Niu, B.; Zhu, Z.; Wu, S.; Li, W. CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 2012, 28, 3150–3152. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.G.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Sievers, F.; Higgins, D.G. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018, 27, 135–145. [Google Scholar] [CrossRef] [Green Version]
- Sievers, F.; Barton, G.J.; Higgins, D.G. Multiple Sequence Alignment. In Bioinformatics, 4th ed.; Baxevanis, A.D., Bader, G.D., Wishart, D.S., Eds.; Wiley: Hoboken, NJ, USA, 2020; pp. 227–250. [Google Scholar]
- Stamatakis, A. RAxML version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014, 30, 1312. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics 2006, 23, 127–128. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life v2: Online annotation and display of phylogenetic trees made easy. Nucleic Acids Res. 2011, 39, W475–W478. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v3: An online tool for the display and annotation of phylogenetic and othertrees. Nucleic Acids Res. 2016, 44, W242–W245. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Letunic, I.; Bork, P. Interactive Tree of Life (iTOL) v5: An online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Boc, A.; Diallo, A.B.; Makarenkov, V. T-REX: A web server for inferring, validating and visualizing phylogenetic trees and networks. Nucleic Acids Res. 2012, 40, W573–W579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gonzales-Barron, U.; Goncalves-Tenorio, A.; Rodrigues, V.; Cadavez, V. Foodborne pathogens in raw milk and cheese of sheep and goat origin: A meta-analysis approach. Curr. Opin. Food Sci. 2017, 18, 7–13. [Google Scholar] [CrossRef]
- Amagliani, G.; Petruzzelli, A.; Carloni, E.; Tonucci, F.; Foglini, M.; Micci, E.; Ricci, M.; Di Lullo, S.; Rotundo, L.; Brandi, G. Presence of Escherichia coli O157, Salmonella spp., and Listeria monocytogenes in raw ovine milk destined for cheese production and evaluation of the equivalence between the analytical methods applied. Foodborne Pathog. Dis. 2016, 13, 626–632. [Google Scholar] [CrossRef]
- Condoleo, R.; Giangolini, G.; Chiaverini, A.; Patriarca, D.; Scaramozzino, P.; Mezher, Z. Occurrence of Listeria monocytogenes and Escherichia coli in raw sheep’s milk from farm bulk tanks in Central Italy. J. Food Prot. 2020, 83, 1929–1933. [Google Scholar] [CrossRef]
- Bogdanovicova, K.; Vyletelova-Klimesova, M.; Babak, V.; Kalhotka, L.; Kolackova, I.; Karpiskova, R. Microbiological quality of raw milk in the Czech Republic. Czech J. Food Sci. 2016, 34, 189–196. [Google Scholar]
- Artursson, K.; Schelin, J.; Lambertz, S.T.; Hansson, I.; Engvall, E.O. Foodborne pathogens in unpasteurized milk in Sweden. Int. J. Food Microbiol. 2018, 284, 120–127. [Google Scholar] [CrossRef]
- Al-Tahiri, R.; Omar, S.; Rewashdeh, A. A study of the occurrence of Listeria species in raw sheep milk. Int. J. Dairy Technol. 2008, 61, 347–351. [Google Scholar] [CrossRef]
- Atil, E.; Ertas, H.B.; Ozbey, G. Isolation and molecular characterization of Listeria spp. from animals, food and environmental samples. Vet. Med. 2011, 56, 386–394. [Google Scholar] [CrossRef] [Green Version]
- Tzora, A.; Nelli, A.; Voidarou, C.; Fthenakis, G.; Rozos, G.; Theodorides, G.; Bonos, E.; Skoufos, I. Microbiota “fingerprint” of Greek feta cheese through ripening. Appl. Sci. 2021, 11, 5631. [Google Scholar] [CrossRef]
- Pappa, E.C.; Bontinis, T.G.; Samelis, J.; Sotirakoglou, K. Assessment of the microbiological quality and biochemical parameters of traditional hard xinotyri cheese made from raw or pasteurized goat milk. Fermentation 2022, 8, 20. [Google Scholar] [CrossRef]
- Rhoades, J.; Anastasiou, I.; Michailidou, S.; Koinidis, A.; Doulgerakis, C.; Alexa, E.A.; Alvarez-Ordonez, A.; Argiriou, A.; Likotrafiti, E. Microbiological analysis of Greek Protected Designation of Origin cheeses and characterisation of the isolated lactic acid bacteria. Int. Dairy J. 2021, 123, 105183. [Google Scholar] [CrossRef]
- Vazquez-Villanueva, J.; Orgaz, B.; Ortiz, S.; Lopez, V.; Martinez-Suarez, J.V.; San Jose, C. Predominance and persistence of a single clone of Listeria ivanovii in a manchego cheese factory over 6 months. Zoonoses Public Health 2010, 57, 402–410. [Google Scholar] [CrossRef]
- Spanu, C.; Scarano, C.; Ibba, M.; Spanu, V.; De Santis, E.P.L. Occurrence and traceability of Listeria monocytogenes strains isolated from sheep’s milk cheese-making plants environment. Food Control 2015, 47, 318–325. [Google Scholar] [CrossRef]
- Hellenic Center for Disease Control and Prevention. Epidemiological Data for Listeriosis in Greece 2004–2017. Available online: https://eody.gov.gr/wp-content/uploads/2019/01/Epidemiological_data_for_listeriosis_Greece_2004_2017.pdf (accessed on 18 February 2022).
- Farzan, A.; Friendship, R.M.; Cook, A.; Pollari, F. Occurrence of Salmonella, Campylobacter, Yersinia enterocolitica, Escherichia coli O157 and Listeria monocytogenes in swine. Zoonoses Public Health 2010, 57, 388–396. [Google Scholar] [CrossRef]
- Gomez-Laguna, J.; Cardoso-Toset, F.; Meza-Torres, J.; Pizarro-Cerda, J.; Quereda, J.J. Virulence potential of Listeria monocytogenes strains recovered from pigs in Spain. Vet. Rec. 2020, 187, 105945. [Google Scholar] [CrossRef]
- Ikeh, M.A.C.; Obi, S.K.C.; Ezeasor, D.N.; Ezeonu, I.M.; Moneke, A.N. Incidence and pathogenicity profile of Listeria spp. isolated from food and environmental samples in Nsukka, Nigeria. Afr. J. Biotechnol. 2010, 9, 4776–4782. [Google Scholar]
- Sarno, E.; Stephan, R.; Zweifel, C. Occurrence of Erysipelothrix spp., Salmonella spp., and Listeria spp. in tonsils of healthy Swiss pigs at slaughter. Arch. Lebensmittelhyg. 2012, 63, 11–15. [Google Scholar]
- Gray, M.L.; Killinger, A.H. Listeria monocytogenes and listeric infections. Bacteriol. Rev. 1966, 30, 309–382. [Google Scholar] [CrossRef] [PubMed]
- Moerner, T. Monitoring disease in wildlife—A review of diseases in the orders lagomorpha and rodentia in Sweden. Verhber. Erkg. Zootiere. 1999, 39, 255–262. [Google Scholar]
- Gyles, C.L.; Prescott, J.F.; Songer, G.; Thoen, C.O. Pathogenesis of Bacterial Infections in Animals, 4th ed.; Wiley-Blackwell: Ames, IA, USA, 2020. [Google Scholar]
- Wacheck, S.; Giezendanner, N.; König, M.; Fredriksson-Ahomaa, M.; Stephan, R. Phenotypical and genotypical traits of Listeria monocytogens strains isolated from tonsils of wild boars hunted in Switzerland. Int. J. Infect. Dis. 2010, 14, e162. [Google Scholar] [CrossRef] [Green Version]
- Sannö, A.; Rosendal, T.; Aspán, A.; Backhans, A.; Jacobson, M. Distribution of enteropathogenic Yersinia spp. and Salmonella spp. in the Swedish wild boar population, and assessment of risk factors that may affect their prevalence. Acta Vet. Scand. 2018, 60, 40. [Google Scholar] [CrossRef] [Green Version]
- Skovgaard, N.; Nørrung, B. The incidence of Listeria spp. in faeces of Danish pigs and in minced pork meat. Int. J. Food Microbiol. 1989, 8, 59–63. [Google Scholar] [CrossRef]
- Felon, D.R.; Wilson, J.; Donachie, W. The incidence and level of Listeria monocytogenes contamination of food sources at primary production and initial processing. J. Appl. Microbiol. 1996, 81, 641–650. [Google Scholar] [CrossRef]
- Zoz, F.; Iaconelli, C.; Lang, E.; Iddir, H.; Guyot, S.; Grandvalet, C.; Gervais, P.; Beney, L. Control of relative air humidity as a potential means to improve hygiene on surfaces: A preliminary approach with Listeria monocytogenes. PLoS ONE 2016, 11, e0148418. [Google Scholar] [CrossRef]
- Zoz, F.; Guyot, S.; Grandvalet, C.; Ragon, M.; Lesniewska, E.; Dupont, S.; Firmesse, O.; Carpentier, B.; Beney, L. Management of Listeria monocytogenes on surfaces via relative air humidity: Key role of cell envelope. Foods 2021, 10, 2002. [Google Scholar] [CrossRef]
- Redfern, J.; Verran, J. Effect of humidity and temperature on the survival of Listeria monocytogenes on surfaces. Appl. Microbiol. 2017, 64, 276–282. [Google Scholar] [CrossRef]
- Tettelin, H.; Masignani, V.; Cieslewicz, M.J.; Donati, C.; Medini, D.; Ward, N.L.; Angiuoli, S.V.; Crabtree, J.; Jones, A.L.; Scott Durkin, A.; et al. Genome analysis of multiple pathogenic isolates of Streptococcus agalactiae: Implications for the microbial “pan-genome”. Proc. Natl. Acad. Sci. USA 2005, 102, 13950–13955. [Google Scholar] [CrossRef] [Green Version]
- Gan, L.; Mao, P.; Jiang, H.; Zhang, L.; Liu, D.; Cao, X.; Wang, Y.; Wang, Y.; Sun, H.; Huang, Y.; et al. Two prevalent Listeria ivanovii subsp. ivanovii clonal strains with different virulence exist in wild rodents and pikas of China. Front. Veter Sci. 2020, 7, 88. [Google Scholar]
- De Sousa, J.A.M.; Buffet, A.; Haudiquet, M.; Rocha, E.P.C.; Rendueles, O. Modular prophage interactions driven by capsule serotype select for capsule loss under phage predation. ISME J. 2020, 14, 2980. [Google Scholar] [CrossRef] [PubMed]
- Hupfeld, M.; Fouts, D.E.; Loessner, M.J.; Klumpp, J. Genome Sequences of the Listeria ivanovii subsp. ivanovii type strain and two Listeria ivanovii subsp. londoniensis strains. Genome Announc. 2015, 3, e01440-14. [Google Scholar] [PubMed] [Green Version]
- Knudsen, G.M.; Olsen, J.E.; Dons, L. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 2004, 15, 171–179. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.; Bhunia, A.K. Secreted Listeria adhesion protein (Lap) influences Lap-mediated Listeria monocytogenes paracellular translocation through epithelial barrier. Gut Pathog. 2013, 5, 16. [Google Scholar] [CrossRef] [Green Version]
- Rajabian, T.; Gavicherla, B.; Heisig, M.; Müller-Altrock, S.; Goebel, W.; Gray-Owen, S.D.; Ireton, K. The bacterial virulence factor InlC perturbs apical cell junctions and promotes cell-to-cell spread of Listeria. Nat. Cell Biol. 2009, 11, 1212–1218. [Google Scholar] [CrossRef] [Green Version]
- Jasnin, M.; Asanoa, S.; Gouin, E.; Hegerla, R.; Plitzkoa, J.M.; Villa, E.; Cossart, P.; Baumeister, W. Three-dimensional architecture of actin filaments in Listeria monocytogenes comet tails. Proc. Natl Acad. Sci. USA 2013, 110, 20521–20526. [Google Scholar] [CrossRef] [Green Version]
- Plastino, J.; Sykes, C. The actin slingshot. Curr. Opin. Cell Biol. 2005, 17, 62–66. [Google Scholar] [CrossRef]
- Beye, M.; Gouriet, F.; Michelle, C.; Casalta, J.P.; Habib, G.; Raoult, D.; Fournier, P.E. Genome analysis of Listeria ivanovii strain G770 that caused a deadly aortic prosthesis infection. New Microbes New Infect. 2016, 10, 87–92. [Google Scholar] [CrossRef] [Green Version]
| Variables (n = 3) | Odds Ratios 1 (95% CI) | p |
|---|---|---|
| Isolation of L. monocytogenes | ||
| Presence of pigs on the farm | <0.0001 | |
| Yes (1/36 = 2.8%) | 24.46 (0.98–612.06) | 0.05 |
| No (0/289 = 0.0%) | reference | - |
| Average relative humidity at 2 m | 0.006 | |
| Per unit decrease | 6.09 (0.00–13.19) | 0.006 |
| Number of female animals on the farm | ||
| ≤165 (0/88 = 0.0%) | 1.36 (0.03–69.29) | 0.88 |
| 166–330 (0/120 = 0.0%) | reference | - |
| 331–500 (0/66 = 0.0%) | 1.81 (0.04–92.38) | 0.77 |
| >500 (1/51 = 2.0%) | 7.16 (0.2 9–178.71) | 0.23 |
| Isolation of L. ivanovii | ||
| Presence of pigs on the farm | <0.0001 | |
| Yes (2/36 = 5.6%) | 17.00 (1.50–192.46) | 0.022 |
| No (1/289 = 0.3%) | reference | - |
| Average relative humidity at 2 m | 0.006 | |
| Per unit decrease | 6.09 (0.00–13.19) | 0.006 |
| Number of female animals on the farm | 0.049 | |
| ≤165 (0/88 = 0.0%) | 1.36 (0.03–69.29) | 0.88 |
| 166–330 (0/120 = 0.0%) | reference | - |
| 331– 500 (1/66 = 1.5%) | 5.52 (0.22–137.41) | 0.30 |
| >500 (2/51 = 3.9%) | 12.17 (0.57–258.14) | 0.11 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lianou, D.T.; Skoulakis, A.; Michael, C.K.; Katsarou, E.I.; Chatzopoulos, D.C.; Solomakos, N.; Tsilipounidaki, K.; Florou, Z.; Cripps, P.J.; Katsafadou, A.I.; et al. Isolation of Listeria ivanovii from Bulk-Tank Milk of Sheep and Goat Farms—From Clinical Work to Bioinformatics Studies: Prevalence, Association with Milk Quality, Antibiotic Susceptibility, Predictors, Whole Genome Sequence and Phylogenetic Relationships. Biology 2022, 11, 871. https://doi.org/10.3390/biology11060871
Lianou DT, Skoulakis A, Michael CK, Katsarou EI, Chatzopoulos DC, Solomakos N, Tsilipounidaki K, Florou Z, Cripps PJ, Katsafadou AI, et al. Isolation of Listeria ivanovii from Bulk-Tank Milk of Sheep and Goat Farms—From Clinical Work to Bioinformatics Studies: Prevalence, Association with Milk Quality, Antibiotic Susceptibility, Predictors, Whole Genome Sequence and Phylogenetic Relationships. Biology. 2022; 11(6):871. https://doi.org/10.3390/biology11060871
Chicago/Turabian StyleLianou, Daphne T., Anargyros Skoulakis, Charalambia K. Michael, Eleni I. Katsarou, Dimitris C. Chatzopoulos, Nikolaos Solomakos, Katerina Tsilipounidaki, Zoe Florou, Peter J. Cripps, Angeliki I. Katsafadou, and et al. 2022. "Isolation of Listeria ivanovii from Bulk-Tank Milk of Sheep and Goat Farms—From Clinical Work to Bioinformatics Studies: Prevalence, Association with Milk Quality, Antibiotic Susceptibility, Predictors, Whole Genome Sequence and Phylogenetic Relationships" Biology 11, no. 6: 871. https://doi.org/10.3390/biology11060871
APA StyleLianou, D. T., Skoulakis, A., Michael, C. K., Katsarou, E. I., Chatzopoulos, D. C., Solomakos, N., Tsilipounidaki, K., Florou, Z., Cripps, P. J., Katsafadou, A. I., Vasileiou, N. G. C., Dimoveli, K. S., Bourganou, M. V., Liagka, D. V., Papatsiros, V. G., Kontou, P. I., Mavrogianni, V. S., Caroprese, M., Petinaki, E., & Fthenakis, G. C. (2022). Isolation of Listeria ivanovii from Bulk-Tank Milk of Sheep and Goat Farms—From Clinical Work to Bioinformatics Studies: Prevalence, Association with Milk Quality, Antibiotic Susceptibility, Predictors, Whole Genome Sequence and Phylogenetic Relationships. Biology, 11(6), 871. https://doi.org/10.3390/biology11060871














