OsbHLHq11, the Basic Helix-Loop-Helix Transcription Factor, Involved in Regulation of Chlorophyll Content in Rice
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Field Design
2.2. Measurement of Leaf Color, Chlorophyll Content, and Fv/Fm
2.3. Response Surface Methodology (RSM) Analysis
2.4. Construction of Genetic Map and QTL Analysis
2.5. Gene Information Analysis
2.6. RNA Extraction
2.7. Analysis of Relative Expression Level
2.8. Statistical Analysis
3. Results
3.1. Measurement of Leaf Color, Chlorophyll Content, and Fv/Fm after Heading
3.2. Analysis of QTLs Associated with Leaf Color, Chlorophyll Content, and Fv/Fm
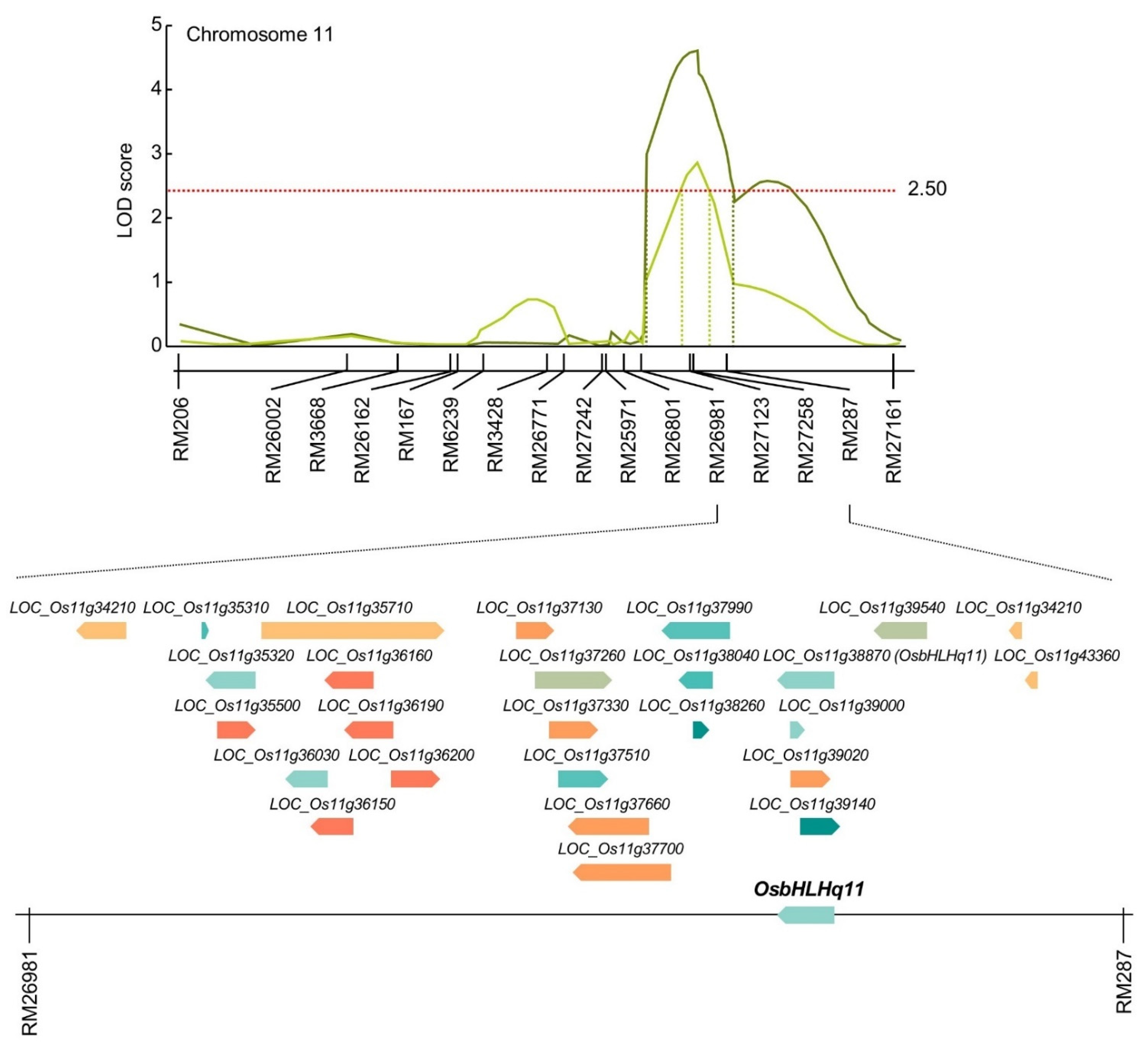
3.3. Search for Chlorophyll-Related Gene Based on QTL Mapping
3.4. Relative Expression Levels of Chlorophyll Content Related Genes
3.5. Analysis of Homology Sequence and Protein Interaction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Muthayya, S.; Sugimoto, J.D.; Montgomery, S.; Maberly, G.F. An overview of global rice production, supply, trade, and consumption. Ann. N. Y. Acad. Sci. 2014, 1324, 7–14. [Google Scholar] [CrossRef] [PubMed]
- Van Nguyen, N.; Ferrero, A. Meeting the challenges of global rice production. Paddy Water Environ. 2006, 4, 1–9. [Google Scholar] [CrossRef]
- Baslam, M.; Mitsui, T.; Hodges, M.; Priesack, E.; Herritt, M.T.; Aranjuelo, I.; Sanz-Sáez, Á. Photosynthesis in a Changing Global Climate: Scaling Up and Scaling Down in Crops. Front. Plant Sci. 2020, 11, 882. [Google Scholar] [CrossRef] [PubMed]
- Zhu, D.; Cheng, S.; Zhang, Y.; Lin, X.; Chen, H. Analysis of status and constraints of rice production in the world. Sci. Agric. Sin. 2010, 43, 474–479. [Google Scholar]
- Furbank, R.T.; Quick, W.P.; Sirault, X.R.R. Improving photosynthesis and yield potential in cereal crops by targeted genetic manipulation: Prospects, progress and challenges. Field Crops Res. 2015, 182, 19–29. [Google Scholar] [CrossRef] [Green Version]
- Sheehy, J.E.; Mitchell, P.L. Developments in rice research: Visions and pragmatism. World Agric. 2011, 2, 1–19. [Google Scholar]
- Simkin, A.J. Genetic engineering for global food security: Photosynthesis and biofortification. Plants 2019, 8, 586. [Google Scholar] [CrossRef] [Green Version]
- Gu, J.; Zhou, Z.; Li, Z.; Chen, Y.; Wang, Z.; Zhang, H. Rice (Oryza sativa L.) with reduced chlorophyll content exhibit higher photosynthetic rate and efficiency, improved canopy light distribution, and greater yields than normally pigmented plants. Field Crops Res. 2017, 200, 58–70. [Google Scholar] [CrossRef]
- Uphoff, N.; Dazzo, F.B. Making rice production more environmentally-friendly. Environments 2016, 3, 12. [Google Scholar] [CrossRef] [Green Version]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.-G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef] [Green Version]
- Long, S.P. Photosynthesis engineered to increase rice yield. Nat. Food 2020, 1, 105. [Google Scholar] [CrossRef] [Green Version]
- Makino, A. Photosynthesis, grain yield, and nitrogen utilization in rice and wheat. Plant Physiol. 2011, 155, 125–129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, R.D.; Black, R.A. Growth, photosynthesis, and resource investment for vegetative and reproductive modules of Artemisia tridentata. Ecology 1993, 74, 1516–1528. [Google Scholar] [CrossRef]
- Austin, R.B.; Morgan, C.L.; Ford, M.A.; Bhagwat, S.G. Flag leaf photosynthesis of Triticum aestivum and related diploid and tetraploid species. Ann. Bot. 1982, 49, 177–189. [Google Scholar] [CrossRef]
- Qu, M.; Zheng, G.; Hamdani, S.; Essemine, J.; Song, Q.; Wang, H.; Chu, C.; Sirault, X.; Zhu, X.-G. Leaf photosynthetic parameters related to biomass accumulation in a global rice diversity survey. Plant Physiol. 2017, 175, 248–258. [Google Scholar] [CrossRef] [Green Version]
- Teng, S.; Qian, Q.; Zeng, D.; Kunihiro, Y.; Fujimoto, K.; Huang, D.; Zhu, L. QTL analysis of leaf photosynthetic rate and related physiological traits in rice (Oryza sativa L.). Euphytica 2004, 135, 1–7. [Google Scholar] [CrossRef]
- Sun, J.; Yang, L.; Wang, Y.; Ort, D.R. FACE-ing the global change: Opportunities for improvement in photosynthetic radiation use efficiency and crop yield. Plant Sci. 2009, 177, 511–522. [Google Scholar] [CrossRef]
- Wang, Q.; Xie, W.; Xing, H.; Yan, J.; Meng, X.; Li, X.; Fu, X.; Xu, J.; Lian, X.; Yu, S. Genetic architecture of natural variation in rice chlorophyll content revealed by a genome-wide association study. Mol. Plant 2015, 8, 946–957. [Google Scholar] [CrossRef] [Green Version]
- Kurniawan, N.S.H.; Kirana, I.A.P.; Abidin, A.S.; Jupri, A.; Widyastuti, S.; Hernawan, A.; Nikmatullah, A.; Sunarpi, H.; Prasedya, E.S. Analysis of leaf chlorophyll content of paddy plants during vegetative stage grown in soil media containing macroalgae organic fertilizer. IOP Conf. Ser. 2021, 913, 012025. [Google Scholar] [CrossRef]
- Peng, Y.; Gitelson, A.A.; Keydan, G.; Rundquist, D.C.; Moses, W. Remote estimation of gross primary production in maize and support for a new paradigm based on total crop chlorophyll content. Remote Sens. Environ. 2011, 115, 978–989. [Google Scholar] [CrossRef]
- Miglani, G.S.; Kaur, R.; Sharma, P.; Gupta, N. Leveraging photosynthetic efficiency toward improving crop yields. J. Crop Improv. 2021, 35, 361–402. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Wang, R.; Mo, G.; Luo, S.; Luo, X.; He, L.; Gonsamo, A.; Arabian, J.; Zhang, Y. The global distribution of leaf chlorophyll content. Remote Sens. Environ. 2020, 236, 111479. [Google Scholar] [CrossRef]
- Evans, J.R. Photosynthesis and nitrogen relationships in leaves of C3 plants. Oecologia 1989, 78, 9–19. [Google Scholar] [CrossRef] [PubMed]
- Kura-Hotta, M.; Satoh, K.; Katoh, S. Relationship between photosynthesis and chlorophyll content during leaf senescence of rice seedlings. Plant Cell Physiol. 1987, 28, 1321–1329. [Google Scholar]
- Peng, S.; Khush, G.S.; Virk, P.; Tang, Q.; Zou, Y. Progress in ideotype breeding to increase rice yield potential. Field Crops Res. 2008, 108, 32–38. [Google Scholar] [CrossRef]
- Yano, M.; Sasaki, T. Genetic and molecular dissection of quantitative traits in rice. Oryza: Mol. Plant 1997, 145–153. [Google Scholar]
- Ishimaru, K.; Yano, M.; Aoki, N.; Ono, K.; Hirose, T.; Lin, S.Y.; Monna, L.; Sasaki, T.; Ohsugi, R. Toward the mapping of physiological and agronomic characters on a rice function map: QTL analysis and comparison between QTLs and expressed sequence tags. Theor. Appl. Genet. 2001, 102, 793–800. [Google Scholar] [CrossRef]
- Wang, F.; Wang, G.; Li, X.; Huang, J.; Zheng, J. Heredity, physiology and mapping of a chlorophyll content gene of rice (Oryza sativa L.). J. Plant Physiol. 2008, 165, 324–330. [Google Scholar] [CrossRef]
- Takai, T.; Kondo, M.; Yano, M.; Yamamoto, T. A Quantitative Trait Locus for Chlorophyll Content and its Association with Leaf Photosynthesis in Rice. Rice 2010, 3, 172–180. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wandell, B.A. A spatial extension of CIELAB for digital color-image reproduction. J. Soc. Inf. Disp. 1997, 5, 61–63. [Google Scholar] [CrossRef]
- Bible, B.B.; Singha, S. Canopy Position Influences CIELAB Coordinates of Peach Color. HortSci. 1993, 28, 992–993. [Google Scholar] [CrossRef] [Green Version]
- Strasser, R.J.; Tsimilli-Michael, M.; Srivastava, A. Analysis of the Chlorophyll a Fluorescence Transient. In Chlorophyll a Fluorescence: A Signature of Photosynthesis; Papageorgiou, G.C., Govindjee, Eds.; Springer: Dordrecht, The Netherlands, 2004; pp. 321–362. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- McCouch, S.R. Gene nomenclature system for rice. Rice 2008, 1, 72–84. [Google Scholar] [CrossRef] [Green Version]
- Kubo, M.; Purevdorj, M. The future of rice production and consumption. J. Food Distrib. Res. 2004, 35, 128–142. [Google Scholar]
- Prasad, R.; Shivay, Y.S.; Kumar, D. Current status, challenges, and opportunities in rice production. In Rice Production Worldwide; Springer: Dordrecht, The Netherlands, 2017; pp. 1–32. [Google Scholar]
- Cao, S.; Zhai, H.; Yang, T.; Zhang, R.; Kuang, T. Studies on photosynthetic rate and function duration of rice germplasm resources. Zhongguo Shuidao Kexue 2001, 15, 29–34. [Google Scholar]
- Huang, M.; Shan, S.; Zhou, X.; Chen, J.; Cao, F.; Jiang, L.; Zou, Y. Leaf photosynthetic performance related to higher radiation use efficiency and grain yield in hybrid rice. Field Crops Res. 2016, 193, 87–93. [Google Scholar] [CrossRef]
- Buttery, B.R.; Buzzell, R.I. The Relationship Between Chlorophyll Content and Rate of Photosynthesis in Soybeans. Can. J. Plant Sci. 1977, 57, 1–5. [Google Scholar] [CrossRef]
- Eggink, L.L.; Park, H.; Hoober, J.K. The role of chlorophyll b in photosynthesis: Hypothesis. BMC Plant Biol. 2001, 1, 2. [Google Scholar] [CrossRef]
- Horton, P.; Ruban, A.V.; Walters, R.G. Regulation of Light Harvesting in Green Plants (Indication by Nonphotochemical Quenching of Chlorophyll Fluorescence). Plant Physiol. 1994, 106, 415–420. [Google Scholar] [CrossRef] [Green Version]
- Masuda, T.; Fujita, Y. Regulation and evolution of chlorophyll metabolism. Photochem. Photobiol. Sci. 2008, 7, 1131–1149. [Google Scholar] [CrossRef]
- Shibghatallah, M.A.H.; Khotimah, S.N.; Suhandono, S.; Viridi, S.; Kesuma, T. Measuring leaf chlorophyll concentration from its color: A way in monitoring environment change to plantations. AIP Conf. Proc. 2013, 1554, 210–213. [Google Scholar]
- Zhang, H.; Ge, Y.; Xie, X.; Atefi, A.; Wijewardane, N.K.; Thapa, S. High throughput analysis of leaf chlorophyll content in sorghum using RGB, hyperspectral, and fluorescence imaging and sensor fusion. Plant Methods 2022, 18, 60. [Google Scholar] [CrossRef] [PubMed]
- Hu, H.; Zhang, J.; Sun, X.; Zhang, X. Estimation of leaf chlorophyll content of rice using image color analysis. Can. J. Remote Sens. 2013, 39, 185–190. [Google Scholar] [CrossRef]
- Madeira, A.C.; Ferreira, A.; de Varennes, A.; Vieira, M.I. SPAD Meter Versus Tristimulus Colorimeter to Estimate Chlorophyll Content and Leaf Color in Sweet Pepper. Commun. Soil Sci. Plant Anal. 2003, 34, 2461–2470. [Google Scholar] [CrossRef]
- Borges, C.S.; Vega, R.R.A.; Chakraborty, S.; Weindorf, D.C.; Lopes, G.; Guilherme, L.R.G.; Curi, N.; Li, B.; Ribeiro, B.T. Pocket-sized sensor for controlled, quantitative and instantaneous color acquisition of plant leaves. J. Plant Physiol. 2022, 272, 153686. [Google Scholar] [CrossRef] [PubMed]
- Friedman, J.M.; Hunt, E.R., Jr.; Mutters, R.G. Assessment of Leaf Color Chart Observations for Estimating Maize Chlorophyll Content by Analysis of Digital Photographs. J. Agron. 2016, 108, 822–829. [Google Scholar] [CrossRef] [Green Version]
- Amandeep, S.; Maninder Lal, S. Performance evaluation of various classifiers for color prediction of rice paddy plant leaf. J. Electron. Imaging 2016, 25, 1–10. [Google Scholar]
- Kumagai, E.; Araki, A.; Kubota, F. Correlation of Chlorophyll Meter Readings with Gas exchange and Chlorophyll Fluorescence in Flag Leaves of Rice (Oryza sativa L.) Plants. Plant Prod. Sci. 2009, 12, 50–53. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.; Wan, L.; Igathinathane, C.; Zhang, Z.; Guo, Y.; Sun, D.; Cen, H. Spatiotemporal Heterogeneity of Chlorophyll Content and Fluorescence Response Within Rice (Oryza sativa L.) Canopies Under Different Nitrogen Treatments. Front. Plant Sci. 2021, 12, 645977. [Google Scholar] [CrossRef]
- Wang, Y.W.; Xu, C.; Lv, C.F.; Wu, M.; Cai, X.J.; Liu, Z.T.; Song, X.M.; Chen, G.X.; Lv, C.G. Chlorophyll a fluorescence analysis of high-yield rice (Oryza sativa L.) LYPJ during leaf senescence. Photosynthetica 2016, 54, 422–429. [Google Scholar] [CrossRef]
- Baruffo, L.; Tretiach, M. Seasonal variations of Fo, Fm, and Fv/Fm in an epiphytic population of the lichen Punctelia subrudecta (Nyl.) Krog. Lichenologist 2007, 39, 555–565. [Google Scholar] [CrossRef]
- Tsai, Y.-C.; Chen, K.-C.; Cheng, T.-S.; Lee, C.; Lin, S.-H.; Tung, C.-W. Chlorophyll fluorescence analysis in diverse rice varieties reveals the positive correlation between the seedlings salt tolerance and photosynthetic efficiency. BMC Plant Biol. 2019, 19, 403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takai, T.; Adachi, S.; Taguchi-Shiobara, F.; Sanoh-Arai, Y.; Iwasawa, N.; Yoshinaga, S.; Hirose, S.; Taniguchi, Y.; Yamanouchi, U.; Wu, J.; et al. A natural variant of NAL1, selected in high-yield rice breeding programs, pleiotropically increases photosynthesis rate. Sci. Rep. 2013, 3, 2149. [Google Scholar] [CrossRef]
- San-oh, Y.; Sugiyama, T.; Yoshita, D.; Ookawa, T.; Hirasawa, T. The effect of planting pattern on the rate of photosynthesis and related processes during ripening in rice plants. Field Crops Res. 2006, 96, 113–124. [Google Scholar] [CrossRef]
- Murchie, E.H.; Yang, J.; Hubbart, S.; Horton, P.; Peng, S. Are there associations between grain-filling rate and photosynthesis in the flag leaves of field-grown rice? J. Exp. Bot. 2002, 53, 2217–2224. [Google Scholar] [CrossRef]
- Yue, B.; Xue, W.-Y.; Luo, L.-J.; Xing, Y.-Z. QTL Analysis for Flag Leaf Characteristics and Their Relationships with Yield and Yield Traits in Rice. Acta Anat. Sin. 2006, 33, 824–832. [Google Scholar] [CrossRef]
- Zhu, M.; Yu, M.; Zhao, S. Understanding quantitative genetics in the systems biology era. Int. J. Biol. Sci. 2009, 5, 161–170. [Google Scholar] [CrossRef] [Green Version]
- Asíns, M.J. Present and future of quantitative trait locus analysis in plant breeding. Plant Breed. 2002, 121, 281–291. [Google Scholar] [CrossRef]
- Sharma, D.K.; Andersen, S.B.; Ottosen, C.-O.; Rosenqvist, E. Wheat cultivars selected for high Fv/Fm under heat stress maintain high photosynthesis, total chlorophyll, stomatal conductance, transpiration and dry matter. Physiol. Plant. 2015, 153, 284–298. [Google Scholar] [CrossRef]
- Mazumdar, D.; Saha, S.P.; Ghosh, S. RSM based optimization of plant growth promoting rhizobacteria and nitrogen dosage for enhanced growth and yield of mustard (Brassica campestris L.). J. Plant Nutr. 2021, 44, 2228–2244. [Google Scholar] [CrossRef]
- Kumari, M.; Gupta, S.K. Response surface methodological (RSM) approach for optimizing the removal of trihalomethanes (THMs) and its precursor’s by surfactant modified magnetic nanoadsorbents (sMNP)—An endeavor to diminish probable cancer risk. Sci. Rep. 2019, 9, 18339. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Pan, Y.; Zhao, H.; Ji, A.; Shi, J.; Guo, P. Response surface optimization of cultivation conditions for yield of tomato (Lycopersicon esculentum Mill.) in greenhouses. J. Plant Nutr. 2018, 41, 210–220. [Google Scholar]
- Abdelkhalik, A.F.; Shishido, R.; Nomura, K.; Ikehashi, H. QTL-based analysis of leaf senescence in an indica/japonica hybrid in rice (Oryza sativa L.). Theor. Appl. Genet. 2005, 110, 1226–1235. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Zeng, J.; He, Y. Analysis of quantitative trait loci affecting chlorophyll content of rice leaves in a double haploid population and two backcross populations. Gene 2014, 536, 287–295. [Google Scholar] [CrossRef]
- Lin, P.-C.; Tsai, Y.-C.; Hsu, S.-K.; Ou, J.-H.; Liao, C.-T.; Tung, C.-W. Identification of natural variants affecting chlorophyll content dynamics during rice seedling development. Plant Breed. 2018, 137, 355–363. [Google Scholar] [CrossRef]
- Walker, M.B.; Roy, L.M.; Coleman, E.; Voelker, R.; Barkan, A. The Maize tha4 Gene Functions in Sec-Independent Protein Transport in Chloroplasts and Is Related to hcf106, tatA, and tatB. J. Biol. Chem. 1999, 147, 267–276. [Google Scholar] [CrossRef] [Green Version]
- Wunnakup, T.; Vimolmangkang, S.; De-Eknamkul, W. Transient expression of the homogentisate phytyltransferase gene from Clitoria ternatea causes metabolic enhancement of α-tocopherol biosynthesis and chlorophyll degradation in tomato leaves. J. Plant Biochem. Biotechnol. 2018, 27, 55–67. [Google Scholar] [CrossRef]
- Martinoia, E.; Klein, M.; Geisler, M.; Bovet, L.; Forestier, C.; Kolukisaoglu, Ü.; Müller-Röber, B.; Schulz, B. Multifunctionality of plant ABC transporters—More than just detoxifiers. Planta 2002, 214, 345–355. [Google Scholar] [CrossRef]
- Motose, H.; Iwamoto, K.; Endo, S.; Demura, T.; Sakagami, Y.; Matsubayashi, Y.; Moore, K.L.; Fukuda, H. Involvement of Phytosulfokine in the Attenuation of Stress Response during the Transdifferentiation of Zinnia Mesophyll Cells into Tracheary Elements. Plant Physiol. 2009, 150, 437–447. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Li, X.; Khatab, A.A.; Xie, G. Phylogeny, structural diversity and genome-wide expression analysis of fibrillin family genes in rice. Phytochemistry 2020, 175, 112377. [Google Scholar] [CrossRef]
- Shen, L.; Zhang, Q.; Wang, Z.; Wen, H.; Hu, G.; Ren, D.; Hu, J.; Zhu, L.; Gao, Z.; Zhang, G.; et al. OsCAF2 contains two CRM domains and is necessary for chloroplast development in rice. BMC Plant Biol. 2020, 20, 381. [Google Scholar] [CrossRef] [PubMed]
- Trifa, Y.; Privat, I.; Gagnon, J.; Baeza, L.; Lerbs-Mache, S. The Nuclear RPL4 Gene Encodes a Chloroplast Protein That Co-purifies with the T7-like Transcription Complex as Well as Plastid Ribosomes. J. Biol. Chem. 1998, 273, 3980–3985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Zheng, T.; Yu, J.; Wu, T.; Wang, X.; Chen, G.; Tian, Y.; Zhang, H.; Wang, Y.; Terzaghi, W.; et al. TSV, a putative plastidic oxidoreductase, protects rice chloroplasts from cold stress during development by interacting with plastidic thioredoxin Z. New Phytol. 2017, 215, 240–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wong, G.R.; Mazumdar, P.; Lau, S.-E. Harikrishna, J.A.; Ectopic expression of a Musa acuminata root hair defective 3 (MaRHD3) in Arabidopsis enhances drought tolerance. J. Plant Physiol. 2018, 231, 219–233. [Google Scholar] [CrossRef] [PubMed]
- Yan, J.; He, C.; Wang, J.; Mao, Z.; Holaday, S.A.; Allen, R.D.; Zhang, H. Overexpression of the Arabidopsis 14-3-3 Protein GF14λ in Cotton Leads to a “Stay-Green” Phenotype and Improves Stress Tolerance under Moderate Drought Conditions. Plant Cell Physiol. 2004, 45, 1007–1014. [Google Scholar] [CrossRef]
- Bonardi, V.; Pesaresi, P.; Becker, T.; Schleiff, E.; Wagner, R.; Pfannschmidt, T.; Jahns, P.; Leister, D. Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 2005, 437, 1179–1182. [Google Scholar] [CrossRef]
- Shin, N.-H.; Trang, D.T.; Hong, W.-J.; Kang, K.; Chuluuntsetseg, J.; Moon, J.-K.; Yoo, Y.-H.; Jung, K.-H.; Yoo, S.-C. Rice Senescence-Induced Receptor-Like Kinase (OsSRLK) Is Involved in Phytohormone-Mediated Chlorophyll Degradation. Int. J. Mol. Sci. 2020, 21, 260. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Niu, X.; Huang, L.; Gross, R.; Lu, H.; Hawkins, M.; Yuan, Y.; Miao, M.; Liu, Y.; Xiao, F. A novel BSD domain-containing transcription factor controls vegetative growth, leaf senescence, and fruit quality in tomato. J. Exp. Bot. 2020, 71, 6945–6957. [Google Scholar] [CrossRef]
- Martínez-García, J.F.; Quail, P.H. The HMG-I/Y protein PF1 stimulates binding of the transcriptional activator GT-2 to the PHYA gene promoter. Plant J. 1999, 18, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Xia, Y.; Li, Z.; Wang, J.; Li, Y.; Ren, Y.; Du, J.; Song, Q.; Ma, S.; Song, Y.; Zhao, H.; et al. Isolation and Identification of a TaTDR-Like Wheat Gene Encoding a bHLH Domain Protein, Which Negatively Regulates Chlorophyll Biosynthesis in Arabidopsis. Int. J. Mol. Sci. 2020, 21, 629. [Google Scholar] [CrossRef] [Green Version]
- Moser, J.; Schubert, W.-D.; Beier, V.; Bringemeier, I.; Jahn, D.; Heinz, D.W. V-shaped structure of glutamyl-tRNA reductase, the first enzyme of tRNA-dependent tetrapyrrole biosynthesis. EMBO J. 2001, 20, 6583–6590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Singh, B.; Sharma, R.A. Plant terpenes: Defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 2015, 5, 129–151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, Y.; Morita, R.; Katsuma, S.; Nishimura, M.; Tanaka, A.; Kusaba, M. Two short-chain dehydrogenase/reductases, NON-YELLOW COLORING 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J. 2009, 57, 120–131. [Google Scholar] [CrossRef]
- Wang, J.; Hu, Z.; Zhao, T.; Yang, Y.; Chen, T.; Yang, M.; Yu, W.; Zhang, B. Genome-wide analysis of bHLH transcription factor and involvement in the infection by yellow leaf curl virus in tomato (Solanum lycopersicum). BMC Genom. 2015, 16, 39. [Google Scholar] [CrossRef] [Green Version]
- Atchley, W.R.; Terhalle, W.; Dress, A. Positional Dependence, Cliques, and Predictive Motifs in the bHLH Protein Domain. J. Mol. Evol. 1999, 48, 501–516. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Zong, X.; Ren, P.; Qian, Y.; Fu, A. Basic Helix-Loop-Helix (bHLH) Transcription Factors Regulate a Wide Range of Functions in Arabidopsis. Int. J. Mol. Sci. 2021, 22, 7152. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.; Nie, X.; Liu, Y.; Zheng, L.; Zhao, H.; Zhang, B.; Huo, L.; Wang, Y. A bHLH gene from Tamarix hispida improves abiotic stress tolerance by enhancing osmotic potential and decreasing reactive oxygen species accumulation. Tree Physiol. 2016, 36, 193–207. [Google Scholar]
- Qian, Y.; Zhang, T.; Yu, Y.; Gou, L.; Yang, J.; Xu, J.; Pi, E. Regulatory mechanisms of bHLH transcription factors in plant adaptive responses to various abiotic stresses. Front. Plant Sci. 2021, 12, 1143. [Google Scholar] [CrossRef]
- Verma, D.; Jalmi, S.K.; Bhagat, P.K.; Verma, N.; Sinha, A.K. A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 2020, 287, 2560–2576. [Google Scholar] [CrossRef]
- Nozoye, T. The Nicotianamine Synthase Gene Is a Useful Candidate for Improving the Nutritional Qualities and Fe-Deficiency Tolerance of Various Crops. Front. Plant Sci. 2018, 9, 340. [Google Scholar] [CrossRef]
- Ogo, Y.; Itai, R.N.; Nakanishi, H.; Inoue, H.; Kobayashi, T.; Suzuki, M.; Takahashi, M.; Mori, S.; Nishizawa, N.K. Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 2006, 57, 2867–2878. [Google Scholar] [CrossRef]
- Kobayashi, T.; Nagasaka, S.; Senoura, T.; Itai, R.N.; Nakanishi, H.; Nishizawa, N.K. Iron-binding haemerythrin RING ubiquitin ligases regulate plant iron responses and accumulation. Nat. Commun. 2013, 4, 2792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Danisman, S. TCP Transcription Factors at the Interface between Environmental Challenges and the Plant’s Growth Responses. Front. Plant Sci. 2016, 7, 1930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Terry, N.; Low, G. Leaf chlorophyll content and its relation to the intracellular localization of iron. J. Plant Nutr. 1982, 5, 301–310. [Google Scholar] [CrossRef]
- Xiao, J.; Li, J.; Yuan, L.; Tanksley, S.D. Identification of QTLs affecting traits of agronomic importance in a recombinant inbred population derived from a subspecific rice cross. Theor. Appl. Genet. 1996, 92, 230–244. [Google Scholar] [CrossRef] [PubMed]
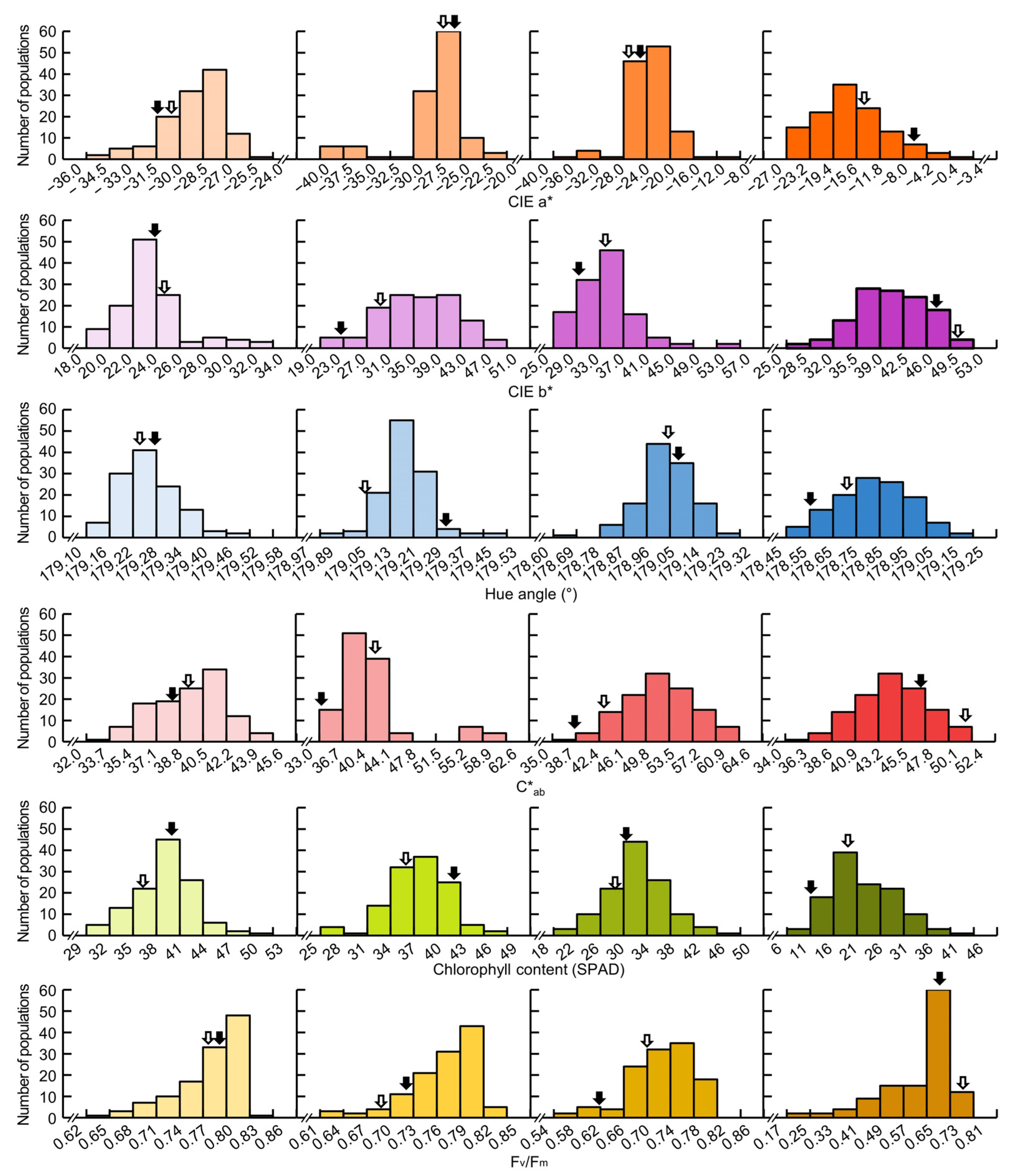
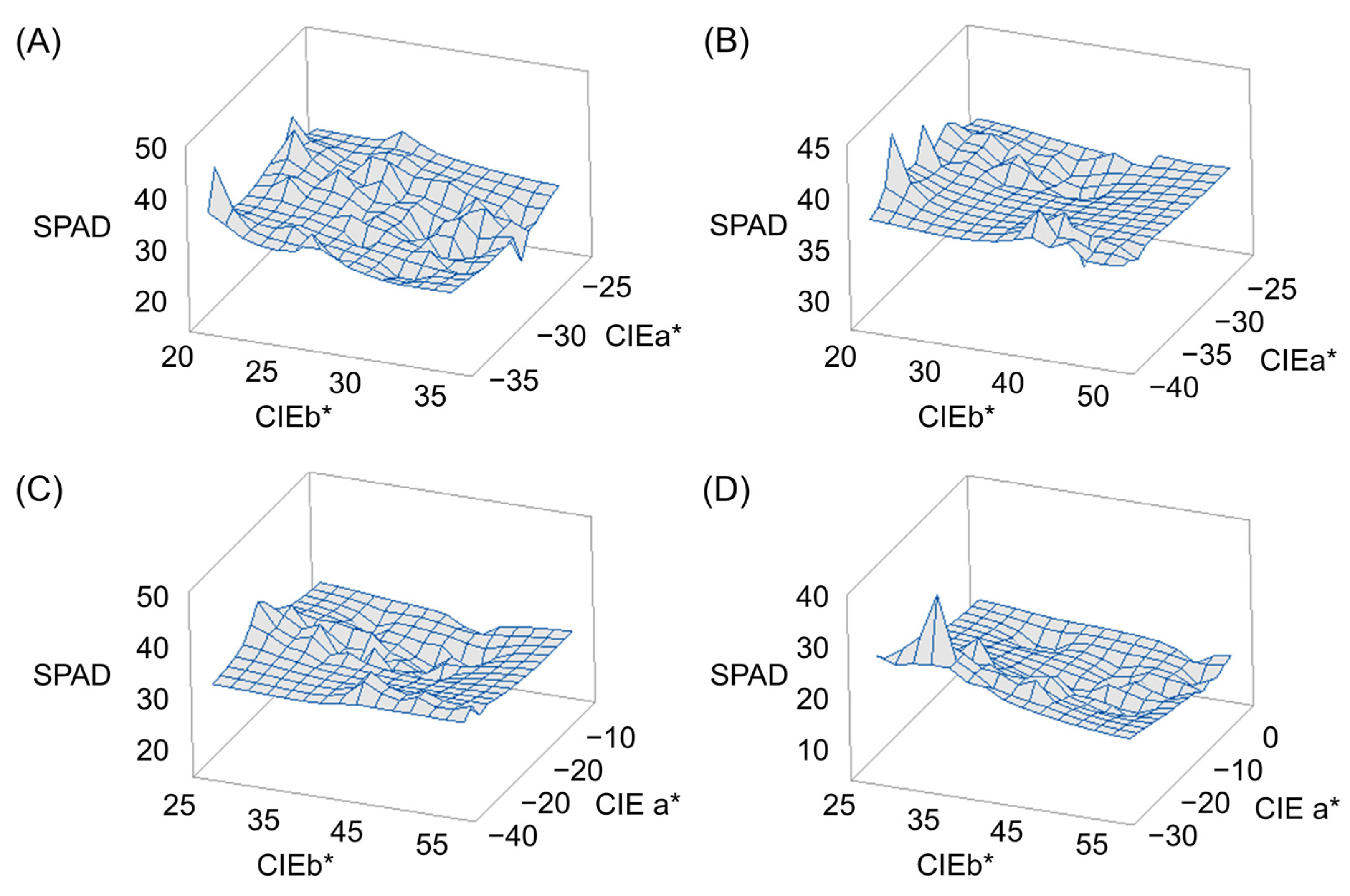
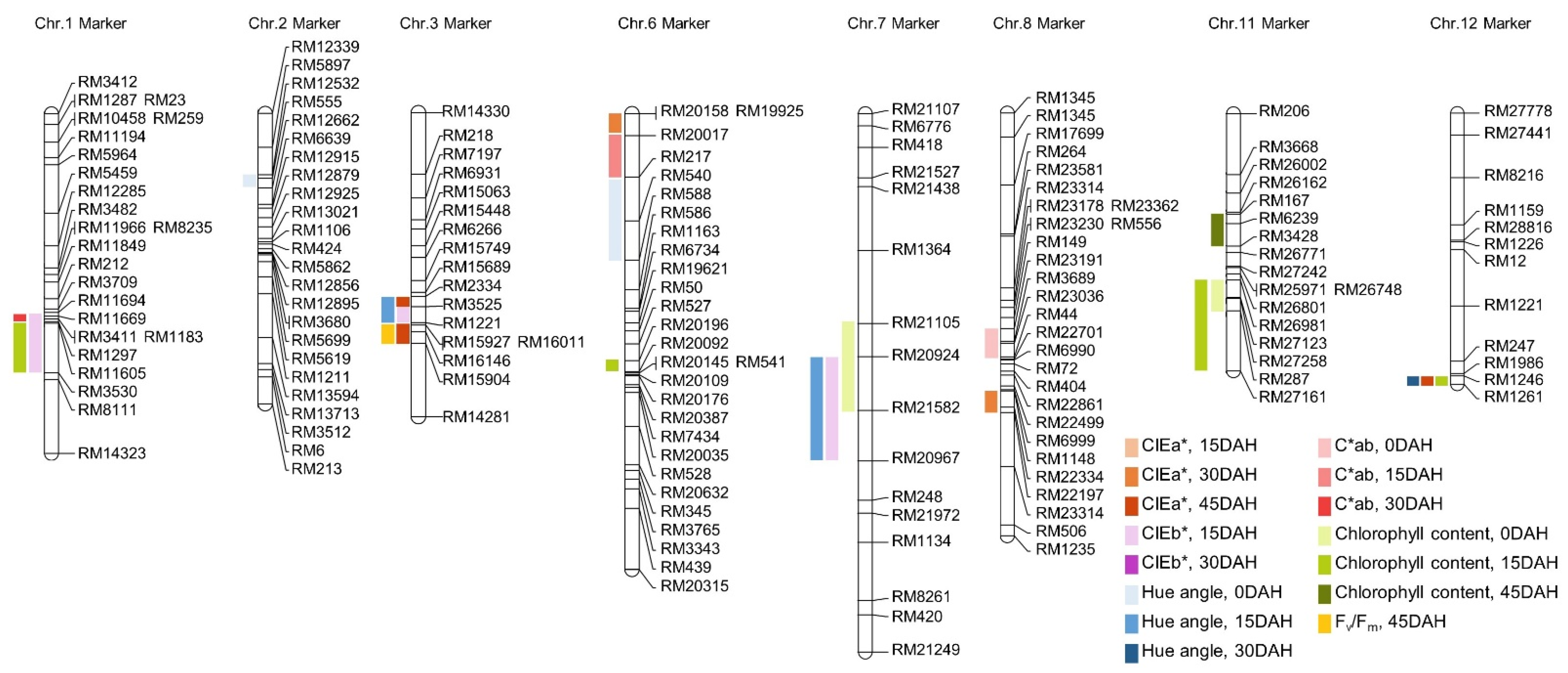
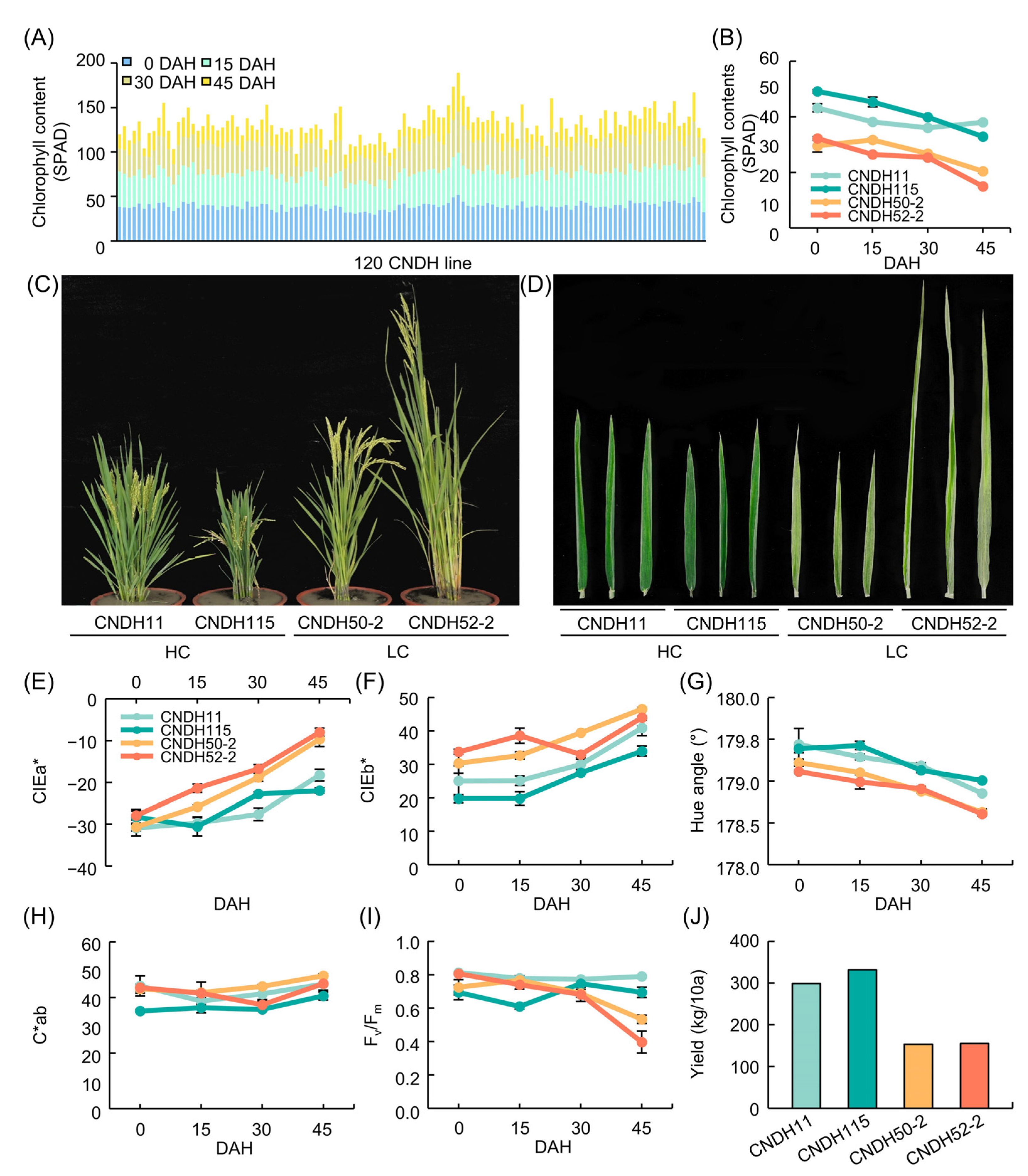
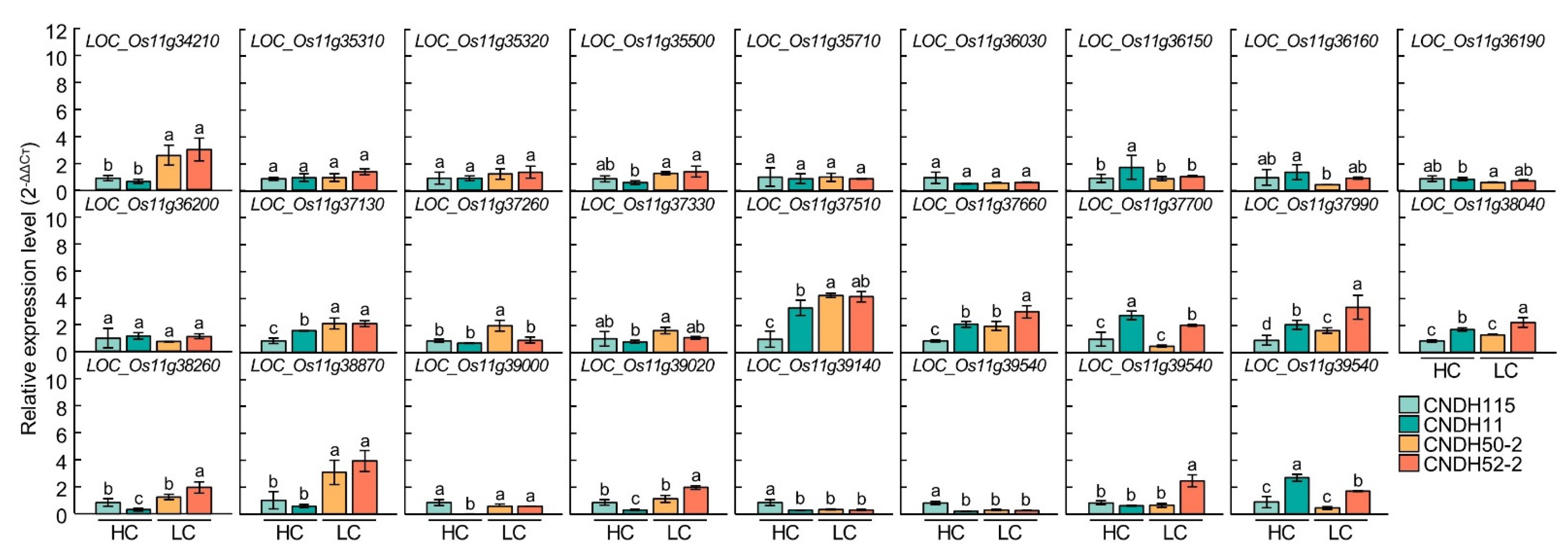

| Plant Traits | DAH z | Parents | DH Line | |
|---|---|---|---|---|
| Cheongcheong | Nagdong | |||
| CIEL* | 0 | 51.36 ± 2.32 y | 50.42 ± 0.99 | 49.32 ± 3.41 |
| 15 | 46.34 ± 0.66 | 51.65 ± 4.32 | 49.69 ± 3.47 | |
| 30 | 53.86 ± 0.70 | 53.90 ± 0.99 | 52.96 ± 4.06 | |
| 45 | 65.66 ± 1.60 | 64.97 ± 1.10 | 57.88 ± 7.31 | |
| CIEa* | 0 | −29.90 ± 0.49 | −30.01 ± 1.20 | −28.90 ± 3.05 |
| 15 | −25.85 ± 1.41 | −26.16 ± 1.12 | −27.84 ± 3.56 | |
| 30 | −23.79 ± 2.04 | −23.79 ± 1.74 | −23.78 ± 4.28 | |
| 45 | −5.53 ± 2.06 | −5.53 ± 2.13 | −15.73 ± 6.65 | |
| CIEb* | 0 | 25.48 ± 2.63 | 27.54 ± 2.11 | 26.42 ± 4.02 |
| 15 | 21.73 ± 2.30 | 32.71 ± 3.61 | 29.69 ± 6.22 | |
| 30 | 31.36 ± 2.06 | 34.77 ± 1.61 | 34.44 ± 5.85 | |
| 45 | 47.61 ± 2.57 | 49.16 ± 2.71 | 40.54 ± 6.64 | |
| Hue angle (°) | 0 | 179.28 ± 0.01 | 179.25 ± 0.04 | 179.26 ± 0.07 |
| 15 | 179.31 ± 0.06 | 179.09 ± 0.03 | 179.19 ± 0.09 | |
| 30 | 179.08 ± 0.07 | 179.05 ± 0.03 | 179.04 ± 0.10 | |
| 45 | 178.54 ± 0.03 | 178.73 ± 0.02 | 178.83 ± 0.16 | |
| C*ab | 0 | 39.52 ± 0.47 | 40.99 ± 0.72 | 39.54 ± 2.67 |
| 15 | 33.80 ± 1.39 | 42.06 ± 1.84 | 41.32 ± 5.98 | |
| 30 | 39.43 ± 0.92 | 42.49 ± 0.45 | 41.99 ± 4.84 | |
| 45 | 47.69 ± 1.35 | 51.94 ± 1.00 | 44.49 ± 3.45 | |
| Chlorophyll contents (SPAD) | 0 | 40.05 ± 0.66 | 36.55 ± 1.01 | 39.10 ± 0.92 |
| 15 | 42.05 ± 1.57 | 36.38 ± 0.62 | 37.62 ± 3.91 | |
| 30 | 30.45 ± 0.34 | 28.10 ± 0.50 | 32.32 ± 5.08 | |
| 45 | 12.73 ± 0.37 | 20.15 ± 0.79 | 22.16 ± 6.86 | |
| Fv/Fm | 0 | 0.80 ± 0.01 | 0.80 ± 0.00 | 0.80 ± 0.04 |
| 15 | 0.73 ± 0.01 | 0.78 ± 0.04 | 0.77 ± 0.04 | |
| 30 | 0.62 ± 0.07 | 0.71 ± 0.02 | 0.73 ± 0.05 | |
| 45 | 0.69 ± 0.01 | 0.74 ± 0.02 | 0.62 ± 0.12 | |
| DAH z | Plant Traits | Yield | CIEa* | CIEb* | Hue Angle | C*ab | Chlorophyll Content (SPAD) | Fv/Fm |
|---|---|---|---|---|---|---|---|---|
| 0 | Yield | 1.000 | ||||||
| CIEa* | −0.025 | 1.000 | ||||||
| CIEb* | −0.091 | 0.042 | 1.000 | |||||
| Hue angle (°) | 0.059 | −0.491 ** | −0.861 ** | 1.000 | ||||
| C*ab | −0.076 | −0.540 ** | 0.784 ** | −0.381 ** | 1.000 | |||
| Chlorophyll content (SPAD) | 0.246 ** | 0.041 | −0.596 ** | 0.489 ** | −0.499 ** | 1.000 | ||
| Fv/Fm | −0.126 | 0.005 | 0.112 | −0.096 | 0.092 | −0.016 | 1.000 | |
| 15 | Yield | 1.000 | ||||||
| CIEa* | −0.114 | 1.000 | ||||||
| CIEb* | 0.037 | −0.485 ** | 1.000 | |||||
| Hue angle (°) | 0.040 | −0.213 * | −0.742 ** | 1.000 | ||||
| C*ab | 0.074 | −0.776 ** | 0.925 ** | −0.437 ** | 1.000 | |||
| Chlorophyll content (SPAD) | 0.303 ** | −0.243 ** | −0.270 ** | 0.496 ** | −0.078 | 1.000 | ||
| Fv/Fm | 0.092 | −0.080 | 0.323 ** | −0.335 ** | 0.262 ** | 0.017 | 1.000 | |
| 30 | Yield | 1.000 | ||||||
| CIEa* | 0.135 | 1.000 | ||||||
| CIEb* | −0.177 | −0.077 | 1.000 | |||||
| Hue angle (°) | 0.021 | −0.693 ** | −0.656 ** | 1.000 | ||||
| C*ab | −0.214 * | −0.472 ** | 0.906 ** | −0.288 ** | 1.000 | |||
| Chlorophyll content (SPAD) | 0.273 ** | −0.379 ** | −0.409 ** | 0.573 ** | −0.211 ** | 1.000 | ||
| Fv/Fm | −0.027 | −0.326 ** | −0.290 ** | 0.448 ** | −0.136 | 0.479 ** | 1.000 | |
| 45 | Yield | 1.000 | ||||||
| CIEa* | −0.005 | 1.000 | ||||||
| CIEb* | −0.011 | 0.725 ** | 1.000 | |||||
| Hue angle (°) | −0.051 | −0.816 ** | −0.756 ** | 1.000 | ||||
| C*ab | −0.020 | 0.431 ** | 0.917 ** | −0.506 ** | 1.000 | |||
| Chlorophyll content (SPAD) | 0.032 | −0.674 ** | −0.668 ** | 0.644 ** | −0.481 ** | 1.000 | ||
| Fv/Fm | −0.057 | −0.589 ** | −0.300 ** | 0.443 ** | −0.078 | 0.474 ** | 1.000 |
| Plant Traits z | DAH z | QTL z | Chromosome z | Interval Markers y | LOD z | Additive Effect x | R2 w | Increasing Effects v |
|---|---|---|---|---|---|---|---|---|
| CIEa* | 30 | qCa6 | 6 | RM20158-RM20017 | 3.45 | −01.31 | 0.30 | Cheongcheong |
| qCa8 | 8 | RM1148-RM22197 | 4.41 | −01.52 | 0.26 | Cheongcheong | ||
| 45 | qCa3 | 3 | RM2334-RM3525 | 2.63 | 0–2.89 | 0.32 | Nagdong | |
| qCa3-1 | 3 | RM15927-RM16146 | 3.82 | −03.08 | 0.26 | Cheongcheong | ||
| qCa12 | 12 | RM1246-RM1261 | 3.51 | −01.96 | 0.27 | Cheongcheong | ||
| CIEb* | 15 | qCb3 | 3 | RM3525-RM1221 | 2.50 | −01.93 | 0.20 | Cheongcheong |
| qCb7 | 7 | RM20924-RM20967 | 3.70 | −02.57 | 0.27 | Cheongcheong | ||
| qCb1 | 1 | RM11694-RM3530 | 2.92 | −01.63 | 0.20 | Cheongcheong | ||
| Hue angle | 0 | qHa2 | 2 | RM12532-RM12662 | 3.63 | −00.03 | 0.26 | Cheongcheong |
| qHa6 | 6 | RM217-RM588 | 3.71 | 0–0.03 | 0.29 | Nagdong | ||
| 15 | qHa3 | 3 | RM2334-RM1221 | 4.38 | −00.04 | 0.26 | Cheongcheong | |
| qHa7 | 7 | RM20924-RM20967 | 2.75 | 0–0.03 | 0.23 | Nagdong | ||
| 30 | qHa12 | 12 | RM1246-RM1261 | 2.80 | 0–0.03 | 0.24 | Nagdong | |
| C*ab | 0 | qCab8 | 8 | RM23191-RM44 | 3.19 | 0–0.93 | 0.24 | Nagdong |
| 15 | qCab6 | 6 | RM20017-RM217 | 2.63 | 0–2.45 | 0.28 | Nagdong | |
| 30 | qCab1 | 1 | RM11694-RM1297 | 3.10 | −01.59 | 0.21 | Cheongcheong | |
| Chlorophyll contents | 0 | qCc7 | 7 | RM21105-RM21582 | 3.84 | 0–1.69 | 0.36 | Nagdong |
| qCc11 | 11 | RM26981-RM287 | 2.95 | −01.28 | 0.31 | Cheongcheong | ||
| 15 | qCc1 | 1 | RM11605-RM3530 | 2.50 | −01.35 | 0.36 | Cheongcheong | |
| qCc6 | 6 | RM20196-RM20092 | 2.78 | 0–1.21 | 0.32 | Nagdong | ||
| qCc11-1 | 11 | RM26981-RM287 | 4.78 | −01.66 | 0.33 | Cheongcheong | ||
| qCc11-2 | 11 | RM287-RM27161 | 2.65 | −01.68 | 0.33 | Cheongcheong | ||
| qCc12 | 12 | RM1246-RM1261 | 2.93 | 0–1.20 | 0.32 | Nagdong | ||
| 45 | qCc11-3 | 11 | RM167-RM3428 | 3.12 | 0–2.36 | 0.26 | Nagdong | |
| Fv/Fm | 45 | qPqy3 | 3 | RM15927-RM15904 | 3.45 | 0–0.06 | 0.25 | Nagdong |
| Yield | - | qYd6 | 6 | RM20176-RM20387 | 2.74 | −46.19 | 0.35 | Cheongcheong |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jang, Y.-H.; Park, J.-R.; Kim, E.-G.; Kim, K.-M. OsbHLHq11, the Basic Helix-Loop-Helix Transcription Factor, Involved in Regulation of Chlorophyll Content in Rice. Biology 2022, 11, 1000. https://doi.org/10.3390/biology11071000
Jang Y-H, Park J-R, Kim E-G, Kim K-M. OsbHLHq11, the Basic Helix-Loop-Helix Transcription Factor, Involved in Regulation of Chlorophyll Content in Rice. Biology. 2022; 11(7):1000. https://doi.org/10.3390/biology11071000
Chicago/Turabian StyleJang, Yoon-Hee, Jae-Ryoung Park, Eun-Gyeong Kim, and Kyung-Min Kim. 2022. "OsbHLHq11, the Basic Helix-Loop-Helix Transcription Factor, Involved in Regulation of Chlorophyll Content in Rice" Biology 11, no. 7: 1000. https://doi.org/10.3390/biology11071000
APA StyleJang, Y.-H., Park, J.-R., Kim, E.-G., & Kim, K.-M. (2022). OsbHLHq11, the Basic Helix-Loop-Helix Transcription Factor, Involved in Regulation of Chlorophyll Content in Rice. Biology, 11(7), 1000. https://doi.org/10.3390/biology11071000







