Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis
Abstract
Simple Summary
Abstract
1. Introduction
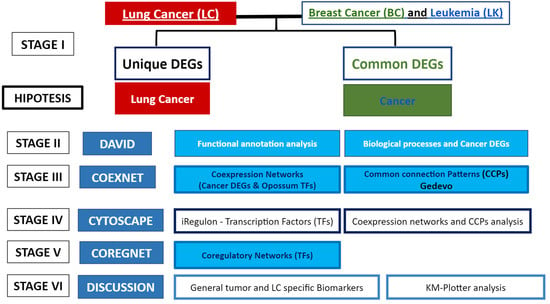
2. Materials and Methods
3. Results
3.1. Differentially Expressed Genes (DEGs) Common between the 3 Types of Cancer, and Unique Lung Cancer DEGs
3.2. Functional Annotation and Enrichment Analysis
3.3. Coexpression Network Analysis
3.4. Coregulatory Network Analysis
3.5. Survival Analysis of Top Winning Transcription Factors in Lung Cancer
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Siegel, R.L.; Miller, K.D.; Fuchs, H.E.; Jemal, A. Cancer Statistics, 2022. CA. Cancer J. Clin. 2022, 72, 7–33. [Google Scholar] [CrossRef] [PubMed]
- Dela Cruz, C.S.; Tanoue, L.T.; Matthay, R.A. Lung Cancer: Epidemiology, Etiology, and Prevention. Clin. Chest Med. 2011, 32, 605–644. [Google Scholar] [CrossRef] [PubMed]
- Hassanpour, S.H.; Dehghani, M. Review of Cancer from Perspective of Molecular. J. Cancer Res. Pract. 2017, 4, 127–129. [Google Scholar] [CrossRef]
- Gridelli, C.; Rossi, A.; Carbone, D.P.; Guarize, J.; Karachaliou, N.; Mok, T.; Petrella, F.; Spaggiari, L.; Rosell, R. Non-Small-Cell Lung Cancer. Nat. Rev. Dis. Primer 2015, 1, 15009. [Google Scholar] [CrossRef]
- Vajpeyi, R. WHO Classification of Tumours: Pathology and Genetics of Tumours of the Breast and Female Genital Organs. J. Clin. Pathol. 2005, 58, 671–672. [Google Scholar]
- Eliyatkın, N.; Yalçın, E.; Zengel, B.; Aktaş, S.; Vardar, E. Molecular Classification of Breast Carcinoma: From Traditional, Old-Fashioned Way to A New Age, and A New Way. J. Breast Health 2015, 11, 59–66. [Google Scholar] [CrossRef]
- Chennamadhavuni, A.; Lyengar, V.; Shimanovsky, A. Leukemia. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: https://www.ncbi.nlm.nih.gov/books/NBK560490/ (accessed on 26 May 2022).
- Kamel, H.F.M.; Al-Amodi, H.S.A.B. Exploitation of Gene Expression and Cancer Biomarkers in Paving the Path to Era of Personalized Medicine. Genom. Proteom. Bioinform. 2017, 15, 220–235. [Google Scholar] [CrossRef]
- Dong, Z.-Y.; Wu, Y.-L. What Is the Significance of TP53 and KRAS Mutation for Immunotherapy in Non-Small Cell Lung Cancer? Transl. Cancer Res. 2017, 6, S1115–S1117. [Google Scholar] [CrossRef]
- Fathi, Z.; Mousavi, S.A.J.; Roudi, R.; Ghazi, F. Distribution of KRAS, DDR2, and TP53 Gene Mutations in Lung Cancer: An Analysis of Iranian Patients. PLoS ONE 2018, 13, e0200633. [Google Scholar] [CrossRef]
- Braun, M.M.; Caporaso, N.E.; Page, W.F.; Hoover, R.N. Genetic Component of Lung Cancer: Cohort Study of Twins. Lancet Lond. Engl. 1994, 344, 440–443. [Google Scholar] [CrossRef]
- Blanchon, F.; Grivaux, M.; Asselain, B.; Lebas, F.-X.; Orlando, J.-P.; Piquet, J.; Zureik, M. 4-Year Mortality in Patients with Non-Small-Cell Lung Cancer: Development and Validation of a Prognostic Index. Lancet Oncol. 2006, 7, 829–836. [Google Scholar] [CrossRef]
- Hutter, C.M.; Mechanic, L.E.; Chatterjee, N.; Kraft, P.; Gillanders, E.M. Gene-Environment Interactions in Cancer Epidemiology: A National Cancer Institute Think Tank Report. Genet. Epidemiol. 2013, 37, 643–657. [Google Scholar] [CrossRef] [PubMed]
- Bahrami, S.; Drabløs, F. Gene Regulation in the Immediate-Early Response Process. Adv. Biol. Regul. 2016, 62, 37–49. [Google Scholar] [CrossRef] [PubMed]
- Palstra, R.-J. Transcription Factor Binding at Enhancers: Shaping a Genomic Regulatory Landscape in Flux. Front. Genet. 2012, 3, 195. [Google Scholar] [CrossRef] [PubMed]
- Lambert, S.A.; Jolma, A.; Campitelli, L.F.; Das, P.K.; Yin, Y.; Albu, M.; Chen, X.; Taipale, J.; Hughes, T.R.; Weirauch, M.T. The Human Transcription Factors. Cell 2018, 172, 650–665. [Google Scholar] [CrossRef] [PubMed]
- Huilgol, D.; Venkataramani, P.; Nandi, S.; Bhattacharjee, S. Transcription Factors That Govern Development and Disease: An Achilles Heel in Cancer. Genes 2019, 10, 794. [Google Scholar] [CrossRef]
- Bhagwat, A.S.; Vakoc, C.R. Targeting Transcription Factors in Cancer. Trends Cancer 2015, 1, 53. [Google Scholar] [CrossRef] [PubMed]
- Henao, J.D. Coexnet: An R Package to Build CO-EXpression NETworks from Microarray Data. 2018. [Software Version 0.1]. Available online: https://bioconductor.org/packages/coexnet/ (accessed on 26 May 2022).
- Otálora-Otálora, B.A.; Florez, M.; López-Kleine, L.; Canas Arboleda, A.; Grajales Urrego, D.M.; Rojas, A. Joint Transcriptomic Analysis of Lung Cancer and Other Lung Diseases. Front. Genet. 2019, 10, 1260. [Google Scholar] [CrossRef]
- Leal, L.G.; López, C.; López-Kleine, L. Construction and Comparison of Gene Co-Expression Networks Shows Complex Plant Immune Responses. PeerJ 2014, 2, e610. [Google Scholar] [CrossRef]
- Oshlack, A.; Robinson, M.D.; Young, M.D. From RNA-Seq Reads to Differential Expression Results. Genome Biol. 2010, 11, 220. [Google Scholar] [CrossRef]
- Lu, T.P.; Tsai, M.H.; Lee, J.M.; Hsu, C.P.; Chen, P.C.; Lin, C.W.; Shih, J.Y.; Yang, P.C.; Hsiao, C.K.; Lai, L.C.; et al. Identification of a Novel Biomarker, SEMA5A, for Non-Small Cell Lung Carcinoma in Nonsmoking Women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 2590–2597. [Google Scholar] [CrossRef]
- Landi, M.T.; Dracheva, T.; Rotunno, M.; Figueroa, J.D.; Liu, H.; Dasgupta, A.; Mann, F.E.; Fukuoka, J.; Hames, M.; Bergen, A.W.; et al. Gene Expression Signature of Cigarette Smoking and Its Role in Lung Adenocarcinoma Development and Survival. PLoS ONE 2008, 3, e1651. [Google Scholar] [CrossRef] [PubMed]
- Wachi, S.; Yoneda, K.; Wu, R. Interactome-Transcriptome Analysis Reveals the High Centrality of Genes Differentially Expressed in Lung Cancer Tissues. Bioinformatics 2005, 21, 4205–4208. [Google Scholar] [CrossRef] [PubMed]
- Asiedu, M.K.; Thomas, C.F.; Dong, J.; Schulte, S.C.; Khadka, P.; Sun, Z.; Kosari, F.; Jen, J.; Molina, J.; Vasmatzis, G.; et al. Pathways Impacted by Genomic Alterations in Pulmonary Carcinoid Tumors. Clin. Cancer Res. 2018, 24, 1691–1704. [Google Scholar] [CrossRef] [PubMed]
- Willuda, J.; Linden, L.; Lerchen, H.-G.; Kopitz, C.; Stelte-Ludwig, B.; Pena, C.; Lange, C.; Golfier, S.; Kneip, C.; Carrigan, P.E.; et al. Preclinical Antitumor Efficacy of BAY 1129980—A Novel Auristatin-Based Anti-C4.4A (LYPD3) Antibody–Drug Conjugate for the Treatment of Non–Small Cell Lung Cancer. Mol. Cancer Ther. 2017, 16, 893–904. [Google Scholar] [CrossRef]
- Koper, A.; Zeef, L.A.H.; Joseph, L.; Kerr, K.; Gosney, J.; Lindsay, M.A.; Booton, R. Whole Transcriptome Analysis of Pre-Invasive and Invasive Early Squamous Lung Carcinoma in Archival Laser Microdissected Samples. Respir. Res. 2017, 18, 12. [Google Scholar] [CrossRef]
- Morton, M.L.; Bai, X.; Merry, C.R.; Linden, P.A.; Khalil, A.M.; Leidner, R.S.; Thompson, C.L. Identification of MRNAs and LincRNAs Associated with Lung Cancer Progression Using Next-Generation RNA Sequencing from Laser Micro-Dissected Archival FFPE Tissue Specimens. Lung Cancer Amst. Neth. 2014, 85, 31–39. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; Salz, T.; Hansen, K.D.; Feinberg, A. Whole-Genome Analysis of the Methylome and Hydroxymethylome in Normal and Malignant Lung and Liver. Genome Res. 2016, 26, 1730–1741. [Google Scholar] [CrossRef]
- Mezheyeuski, A.; Bergsland, C.H.; Backman, M.; Djureinovic, D.; Sjöblom, T.; Bruun, J.; Micke, P. Multispectral Imaging for Quantitative and Compartment-Specific Immune Infiltrates Reveals Distinct Immune Profiles That Classify Lung Cancer Patients. J. Pathol. 2018, 244, 421–431. [Google Scholar] [CrossRef]
- Nazarov, P.V.; Muller, A.; Kaoma, T.; Nicot, N.; Maximo, C.; Birembaut, P.; Tran, N.L.; Dittmar, G.; Vallar, L. RNA Sequencing and Transcriptome Arrays Analyses Show Opposing Results for Alternative Splicing in Patient Derived Samples. BMC Genom. 2017, 18, 443. [Google Scholar] [CrossRef]
- Casey, T.; Bond, J.; Tighe, S.; Hunter, T.; Lintault, L.; Patel, O.; Eneman, J.; Crocker, A.; White, J.; Tessitore, J.; et al. Molecular Signatures Suggest a Major Role for Stromal Cells in Development of Invasive Breast Cancer. Breast Cancer Res. Treat. 2008, 114, 47–62. [Google Scholar] [CrossRef]
- Kretschmer, C.; Sterner-Kock, A.; Siedentopf, F.; Schoenegg, W.; Schlag, P.M.; Kemmner, W. Identification of Early Molecular Markers for Breast Cancer. Mol. Cancer 2011, 10, 15. [Google Scholar] [CrossRef]
- Planche, A.; Bacac, M.; Provero, P.; Fusco, C.; Delorenzi, M.; Stehle, J.-C.; Stamenkovic, I. Identification of Prognostic Molecular Features in the Reactive Stroma of Human Breast and Prostate Cancer. PLoS ONE 2011, 6, e18640. [Google Scholar] [CrossRef] [PubMed]
- Alimonti, A.; Carracedo, A.; Clohessy, J.G.; Trotman, L.C.; Nardella, C.; Egia, A.; Salmena, L.; Sampieri, K.; Haveman, W.J.; Brogi, E.; et al. Subtle Variations in Pten Dose Determine Cancer Susceptibility. Nat. Genet. 2010, 42, 454–458. [Google Scholar] [CrossRef] [PubMed]
- Richardson, A.L.; Wang, Z.C.; De Nicolo, A.; Lu, X.; Brown, M.; Miron, A.; Liao, X.; Iglehart, J.D.; Livingston, D.M.; Ganesan, S. X Chromosomal Abnormalities in Basal-like Human Breast Cancer. Cancer Cell 2006, 9, 121–132. [Google Scholar] [CrossRef] [PubMed]
- Turashvili, G.; Bouchal, J.; Baumforth, K.; Wei, W.; Dziechciarkova, M.; Ehrmann, J.; Klein, J.; Fridman, E.; Skarda, J.; Srovnal, J.; et al. Novel Markers for Differentiation of Lobular and Ductal Invasive Breast Carcinomas by Laser Microdissection and Microarray Analysis. BMC Cancer 2007, 7, 55. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, A.; Tschumper, R.C.; Wu, X.; Shanafelt, T.D.; Eckel-Passow, J.; Huddleston, P.M.; Slager, S.L.; Kay, N.E.; Jelinek, D.F. LEF-1 Is a Prosurvival Factor in Chronic Lymphocytic Leukemia and Is Expressed in the Preleukemic State of Monoclonal B-Cell Lymphocytosis. Blood 2010, 116, 2975–2983. [Google Scholar] [CrossRef]
- Vargova, K.; Curik, N.; Burda, P.; Basova, P.; Kulvait, V.; Pospisil, V.; Savvulidi, F.; Kokavec, J.; Necas, E.; Berkova, A.; et al. MYB Transcriptionally Regulates the MiR-155 Host Gene in Chronic Lymphocytic Leukemia. Blood 2011, 117, 3816–3825. [Google Scholar] [CrossRef]
- Dürig, J.; Bug, S.; Klein-Hitpass, L.; Boes, T.; Jöns, T.; Martin-Subero, J.I.; Harder, L.; Baudis, M.; Dührsen, U.; Siebert, R. Combined Single Nucleotide Polymorphism-Based Genomic Mapping and Global Gene Expression Profiling Identifies Novel Chromosomal Imbalances, Mechanisms and Candidate Genes Important in the Pathogenesis of T-Cell Prolymphocytic Leukemia with Inv(14)(Q11q32). Leukemia 2007, 21, 2153–2163. [Google Scholar] [CrossRef]
- Gutiérrez, N.C.; Ocio, E.M.; de las Rivas, J.; Maiso, P.; Delgado, M.; Fermiñán, E.; Arcos, M.J.; Sánchez, M.L.; Hernández, J.M.; San Miguel, J.F. Gene Expression Profiling of B Lymphocytes and Plasma Cells from Waldenström’s Macroglobulinemia: Comparison with Expression Patterns of the Same Cell Counterparts from Chronic Lymphocytic Leukemia, Multiple Myeloma and Normal Individuals. Leukemia 2007, 21, 541–549. [Google Scholar] [CrossRef]
- Stirewalt, D.L.; Meshinchi, S.; Kopecky, K.J.; Fan, W.; Pogosova-Agadjanyan, E.L.; Engel, J.H.; Cronk, M.R.; Dorcy, K.S.; McQuary, A.R.; Hockenbery, D.; et al. Identification of Genes with Abnormal Expression Changes in Acute Myeloid Leukemia. Genes. Chromosomes Cancer 2008, 47, 8–20. [Google Scholar] [CrossRef] [PubMed]
- Dennis, G.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003, 4, R60. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B Methodol. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Ibragimov, R.; Malek, M.; Guo, J.; Baumbach, J. GEDEVO: An Evolutionary Graph Edit Distance Algorithm for Biological Network Alignment; Schloss Dagstuhl-Leibniz-Zentrum fuer Informatik GmbH, Wadern/Saarbruecken, Germany. In Proceedings of the German Conference on Bioinformatics, Göttingen, Germany, 10–13 September 2013; pp. 68–79. [Google Scholar]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A Software Environment for Integrated Models. Genome Res. 2003, 13, 2498–2503. [Google Scholar] [CrossRef]
- Janky, R.; Verfaillie, A.; Imrichová, H.; Van de Sande, B.; Standaert, L.; Christiaens, V.; Hulselmans, G.; Herten, K.; Naval Sanchez, M.; Potier, D.; et al. IRegulon: From a Gene List to a Gene Regulatory Network Using Large Motif and Track Collections. PLoS Comput. Biol. 2014, 10, e1003731. [Google Scholar] [CrossRef] [PubMed]
- Nicolle, R.; Radvanyi, F.; Elati, M. CoRegNet: Reconstruction and Integrated Analysis of Co-Regulatory Networks. Bioinformatics 2015, 31, 3066–3068. [Google Scholar] [CrossRef] [PubMed]
- Stalpers, L.J.A.; Kaplan, E.L. Edward L. Kaplan and the Kaplan-Meier Survival Curve. BSHM Bull. J. Br. Soc. Hist. Math. 2018, 33, 109–135. [Google Scholar] [CrossRef]
- Otálora-Otálora, B.A. Identificación de Factores de Transcripción Asociados al Establecimiento y Progresión Del Cáncer de Pulmón Mediante Análisis Bioinformático: Validación Experimental de RUNX2 En Cáncer de Pulmón. Univ. Nac. Colomb. 2020, 1–225. [Google Scholar]
- Papetti, M.; Augenlicht, L.H. MYBL2, a Link between Proliferation and Differentiation in Maturing Colon Epithelial Cells. J. Cell. Physiol. 2011, 226, 785–791. [Google Scholar] [CrossRef]
- Jin, Y.; Nenseth, H.Z.; Saatcioglu, F. Role of PLZF as a Tumor Suppressor in Prostate Cancer. Oncotarget 2017, 8, 71317–71324. [Google Scholar] [CrossRef]
- Xiao, G.-Q.; Li, F.; Findeis-Hosey, J.; Hyrien, O.; Unger, P.D.; Xiao, L.; Dunne, R.; Kim, E.S.; Yang, Q.; McMahon, L.; et al. Down-Regulation of Cytoplasmic PLZF Correlates with High Tumor Grade and Tumor Aggression in Non–Small Cell Lung Carcinoma. Hum. Pathol. 2015, 46, 1607–1615. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Wu, M.; Xiong, L.; Gong, Y.; Yu, R.; Peng, W.; Li, L.; Li, L.; Tian, S.; Wang, Y.; et al. BTB/POZ Zinc Finger Protein ZBTB16 Inhibits Breast Cancer Proliferation and Metastasis through Upregulating ZBTB28 and Antagonizing BCL6/ZBTB27. Clin. Epigenetics 2020, 12, 82. [Google Scholar] [CrossRef] [PubMed]
- Porcher, C.; Chagraoui, H.; Kristiansen, M.S. SCL/TAL1: A Multifaceted Regulator from Blood Development to Disease. Blood 2017, 129, 2051–2060. [Google Scholar] [CrossRef]
- Shivdasani, R.A.; Mayer, E.L.; Orkin, S.H. Absence of Blood Formation in Mice Lacking the T-Cell Leukaemia Oncoprotein Tal-1/SCL. Nature 1995, 373, 432–434. [Google Scholar] [CrossRef]
- Meng, X.; Lu, P.; Bai, H.; Xiao, P.; Fan, Q. Transcriptional Regulatory Networks in Human Lung Adenocarcinoma. Mol. Med. Rep. 2012, 6, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Evans, P.M.; Liu, C. Role of Krüppel-like Factor 4 in Normal Homeostasis, Cancer, and Stem Cells. Acta Biochim. Biophys. Sin. 2008, 40, 554–564. [Google Scholar] [CrossRef]
- Evans, P.M.; Zhang, W.; Chen, X.; Yang, J.; Bhakat, K.K.; Liu, C. Krüppel-like Factor 4 Is Acetylated by P300 and Regulates Gene Transcription via Modulation of Histone Acetylation. J. Biol. Chem. 2007, 282, 33994–34002. [Google Scholar] [CrossRef]
- Cho, Y.G.; Song, J.H.; Kim, C.J.; Nam, S.W.; Yoo, N.J.; Lee, J.Y.; Park, W.S. Genetic and Epigenetic Analysis of the KLF4 Gene in Gastric Cancer. APMIS Acta Pathol. Microbiol. Immunol. Scand. 2007, 115, 802–808. [Google Scholar] [CrossRef]
- Kalathil, D.; John, S.; Nair, A.S. FOXM1 and Cancer: Faulty Cellular Signaling Derails Homeostasis. Front. Oncol. 2021, 10, 626836. [Google Scholar] [CrossRef]
- Kopanja, D.; Pandey, A.; Kiefer, M.; Wang, Z.; Chandan, N.; Carr, J.R.; Franks, R.; Yu, D.-Y.; Guzman, G.; Maker, A.; et al. Essential Roles of FoxM1 in Ras-Induced Liver Cancer Progression and in Cancer Cells with Stem Cell Features. J. Hepatol. 2015, 63, 429–436. [Google Scholar] [CrossRef]
- Blanchard, T.; Czinn, S.; Banerjee, V.; Sharda, N.; Bafford, A.; Mubariz, F.; Morozov, D.; Passaniti, A.; Ahmed, A.; Banerjee, A. Identification of Cross Talk between FoxM1 and RASSF1A as a Therapeutic Target of Colon Cancer. Cancers 2019, 11, 199. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.; Xia, T.; Xie, D.; Gao, Y.; Jia, Z.; Wei, D.; Wang, L.; Huang, S.; Quan, M.; Xie, K. HGF/Met and FOXM1 Form a Positive Feedback Loop and Render Pancreatic Cancer Cells Resistance to Met Inhibition and Aggressive Phenotypes. Oncogene 2016, 35, 4708–4718. [Google Scholar] [CrossRef] [PubMed]
- Francica, P.; Nisa, L.; Aebersold, D.; Langer, R.; Bladt, F.; Blaukat, A.; Stroka, D.; Rodríguez Martínez, M.; Zimmer, Y.; Medova, M. Depletion of FOXM1 via MET Targeting Underlies Establishment of a DNA Damage-Induced Senescence Program in Gastric Cancer. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2016, 22, 5322–5336. [Google Scholar] [CrossRef]
- Bektas, N.; ten Haaf, A.; Veeck, J.; Wild, P.J.; Lüscher-Firzlaff, J.; Hartmann, A.; Knüchel, R.; Dahl, E. Tight Correlation between Expression of the Forkhead Transcription Factor FOXM1 and HER2 in Human Breast Cancer. BMC Cancer 2008, 8, 42. [Google Scholar] [CrossRef] [PubMed]
- Katoh, M. Molecular Cloning and Characterization of Human SOX17. Int. J. Mol. Med. 2002, 9, 153–157. [Google Scholar] [CrossRef] [PubMed]
- Fu, D.-Y.; Wang, Z.-M.; Li-Chen; Wang, B.-L.; Shen, Z.-Z.; Huang, W.; Shao, Z.-M. Sox17, the Canonical Wnt Antagonist, Is Epigenetically Inactivated by Promoter Methylation in Human Breast Cancer. Breast Cancer Res. Treat. 2010, 119, 601–612. [Google Scholar] [CrossRef]
- Ansari, A.; Pillarisetty, L.S. Embryology, Ectoderm. In StatPearls; StatPearls Publishing: Treasure Island, FL, USA, 2022. Available online: http://www.ncbi.nlm.nih.gov/books/NBK539836/ (accessed on 26 May 2022).
- Xu, X.-H.; Bao, Y.; Wang, X.; Yan, F.; Guo, S.; Ma, Y.; Xu, D.; Jin, L.; Xu, J.; Wang, J. Hypoxic-Stabilized EPAS1 Proteins Transactivate DNMT1 and Cause Promoter Hypermethylation and Transcription Inhibition of EPAS1 in Non-Small Cell Lung Cancer. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2018, 32, 6694–6705. [Google Scholar] [CrossRef]
- Putra, A.C.; Eguchi, H.; Lee, K.L.; Yamane, Y.; Gustine, E.; Isobe, T.; Nishiyama, M.; Hiyama, K.; Poellinger, L.; Tanimoto, K. The A Allele at Rs13419896 of EPAS1 Is Associated with Enhanced Expression and Poor Prognosis for Non-Small Cell Lung Cancer. PLoS ONE 2015, 10, e0134496. [Google Scholar] [CrossRef]
- Wang, Z.; Wei, Y.; Zhang, R.; Su, L.; Gogarten, S.M.; Liu, G.; Brennan, P.; Field, J.K.; McKay, J.D.; Lissowska, J.; et al. Multi-Omics Analysis Reveals a HIF Network and Hub Gene EPAS1 Associated with Lung Adenocarcinoma. eBioMedicine 2018, 32, 93–101. [Google Scholar] [CrossRef]
- Song, Y.; Zhang, M.; Lu, M.M.; Qu, L.Y.; Xu, S.G.; Li, Y.Z.; Wang, M.Y.; Zhu, H.F.; Zhang, Z.Y.; He, G.Y.; et al. EPAS1 Targeting by MiR-152-3p in Paclitaxel-Resistant Breast Cancer. J. Cancer 2020, 11, 5822–5830. [Google Scholar] [CrossRef]
- Wang, F.; Zhu, Y.; Huang, Y.; McAvoy, S.; Johnson, J.; Th, C.; Tk, C.; Kw, L.; Sf, Y.; Mm, Y.; et al. Transcriptional Repression of WEE1 by Kruppel-like Factor 2 Is Involved in DNA Damage-Induced Apoptosis. Oncogene 2005, 24, 3875–3885. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, R.; Senbanerjee, S.; Lin, Z.; Mir, S.; Hamik, A.; Wang, P.; Mukherjee, P.; Mukhopadhyay, D.; Jain, M.K. Inhibition of Vascular Permeability Factor/Vascular Endothelial Growth Factor-Mediated Angiogenesis by the Kruppel-like Factor KLF2. J. Biol. Chem. 2005, 280, 28848–28851. [Google Scholar] [CrossRef]
- Wu, J.; Lingrel, J.B. KLF2 Inhibits Jurkat T Leukemia Cell Growth via Upregulation of Cyclin-Dependent Kinase Inhibitor P21WAF1/CIP1. Oncogene 2004, 23, 8088–8096. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Xu, X.; Deng, S.; Luo, J.; Xu, H.; Wang, C.; Sun, T.; Lei, G.; Zhang, F.; Yang, C.; et al. Methylation of Kruppel-like Factor 2 (KLF2) Associates with Its Expression and Non-Small Cell Lung Cancer Progression. Am. J. Transl. Res. 2017, 9, 2024–2037. [Google Scholar] [PubMed]
- Zhang, W.; Levi, L.; Banerjee, P.; Jain, M.; Noy, N. Kruppel-like Factor 2 Suppresses Mammary Carcinoma Growth by Regulating Retinoic Acid Signaling. Oncotarget 2015, 6, 35830–35842. [Google Scholar] [CrossRef]
- Pan, S.-H.; Hsu, Y.L.; Hung, P.-F.; Wang, C.-J.; Wang, C.-C. Abstract 1431: Id4 Inhibits Cancer Metastasis through EMT Regulation in Lung Cancer. Cancer Res. 2015, 75, 1431. [Google Scholar] [CrossRef]
- Qi, K.; Li, Y.; Li, X.; Lei, X.; Wang, B.; Zhang, L.; Chu, X. Id4 Promotes Cisplatin Resistance in Lung Cancer through the P38 MAPK Pathway. Anticancer Drugs 2016, 27, 970–978. [Google Scholar] [CrossRef]
- Mullican, S.E.; Zhang, S.; Konopleva, M.; Ruvolo, V.; Andreeff, M.; Milbrandt, J.; Conneely, O.M. Abrogation of Nuclear Receptors Nr4a3 and Nr4a1 Leads to Development of Acute Myeloid Leukemia. Nat. Med. 2007, 13, 730–735. [Google Scholar] [CrossRef]
- Fedorova, O.; Petukhov, A.; Daks, A.; Shuvalov, O.; Leonova, T.; Vasileva, E.; Aksenov, N.; Melino, G.; Barlev, N.A. Orphan Receptor NR4A3 Is a Novel Target of P53 That Contributes to Apoptosis. Oncogene 2019, 38, 2108–2122. [Google Scholar] [CrossRef]
- Iavarone, A.; Garg, P.; Lasorella, A.; Hsu, J.; Israel, M.A. The Helix-Loop-Helix Protein Id-2 Enhances Cell Proliferation and Binds to the Retinoblastoma Protein. Genes Dev. 1994, 8, 1270–1284. [Google Scholar] [CrossRef]
- Coma, S.; Amin, D.N.; Shimizu, A.; Lasorella, A.; Iavarone, A.; Klagsbrun, M. Id2 Promotes Tumor Cell Migration and Invasion through Transcriptional Repression of Semaphorin 3F. Cancer Res. 2010, 70, 3823–3832. [Google Scholar] [CrossRef] [PubMed]
- Rätze, M.A.K.; Koorman, T.; Sijnesael, T.; Bassey-Archibong, B.; van de Ven, R.; Enserink, L.; Visser, D.; Jaksani, S.; Viciano, I.; Bakker, E.R.M.; et al. Loss of E-Cadherin Leads to Id2-Dependent Inhibition of Cell Cycle Progression in Metastatic Lobular Breast Cancer. Oncogene 2022, 41, 2932–2944. [Google Scholar] [CrossRef] [PubMed]
- Lasorella, A.; Iavarone, A.; Israel, M.A. Id2 Specifically Alters Regulation of the Cell Cycle by Tumor Suppressor Proteins. Mol. Cell. Biol. 1996, 16, 2570–2578. [Google Scholar] [CrossRef]
- Rollin, J.; Bléchet, C.; Régina, S.; Tenenhaus, A.; Guyétant, S.; Gidrol, X. The Intracellular Localization of ID2 Expression Has a Predictive Value in Non Small Cell Lung Cancer. PLoS ONE 2009, 4, e4158. [Google Scholar] [CrossRef] [PubMed]
- López-Kleine, L.; Leal, L.; López, C. Biostatistical Approaches for the Reconstruction of Gene Co-Expression Networks Based on Transcriptomic Data. Brief. Funct. Genom. 2013, 12, 457–467. [Google Scholar] [CrossRef] [PubMed]
- Lo Iacono, M.; Monica, V.; Saviozzi, S.; Ceppi, P.; Bracco, E.; Papotti, M.; Scagliotti, G.V. Aurora Kinase A Expression Is Associated with Lung Cancer Histological-Subtypes and with Tumor de-Differentiation. J. Transl. Med. 2011, 9, 100. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dos Santos, E.O.; Carneiro-Lobo, T.C.; Aoki, M.N.; Levantini, E.; Bassères, D.S. Aurora Kinase Targeting in Lung Cancer Reduces KRAS-Induced Transformation. Mol. Cancer 2016, 15, 12. [Google Scholar] [CrossRef]
- Zhang, X.; Xiao, D.; Wang, Z.; Zou, Y.; Huang, L.; Lin, W.; Deng, Q.; Pan, H.; Zhou, J.; Liang, C.; et al. MicroRNA-26a/b Regulate DNA Replication Licensing, Tumorigenesis, and Prognosis by Targeting CDC6 in Lung Cancer. Mol. Cancer Res. 2014, 12, 1535–1546. [Google Scholar] [CrossRef]
- Hateboer, G.; Wobst, A.; Petersen, B.O.; Le Cam, L.; Vigo, E.; Sardet, C.; Helin, K. Cell Cycle-Regulated Expression of Mammalian CDC6 Is Dependent on E2F. Mol. Cell. Biol. 1998, 18, 6679–6697. [Google Scholar] [CrossRef]
- Yan, Z.; DeGregori, J.; Shohet, R.; Leone, G.; Stillman, B.; Nevins, J.R.; Williams, R.S. Cdc6 Is Regulated by E2F and Is Essential for DNA Replication in Mammalian Cells. Proc. Natl. Acad. Sci. USA 1998, 95, 3603–3608. [Google Scholar] [CrossRef]
- Sideridou, M.; Zakopoulou, R.; Evangelou, K.; Liontos, M.; Kotsinas, A.; Rampakakis, E.; Gagos, S.; Kahata, K.; Grabusic, K.; Gkouskou, K.; et al. Cdc6 Expression Represses E-Cadherin Transcription and Activates Adjacent Replication Origins. J. Cell Biol. 2011, 195, 1123–1140. [Google Scholar] [CrossRef]
- Chen, X.; Chen, S.; Pei, N.; Mao, Y.; Wang, S.; Yan, R.; Bai, N.; Li, A.; Zhang, Y.; Du, H.; et al. AAV-Mediated Angiotensin 1-7 Overexpression Inhibits Tumor Growth of Lung Cancer in Vitro and in Vivo. Oncotarget 2017, 8, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Guo, Y.; Zhang, X.; Yang, M.; Miao, X.; Shi, Y.; Yao, J.; Tan, W.; Sun, T.; Zhao, D.; Yu, D.; et al. Functional Evaluation of Missense Variations in the Human MAD1L1 and MAD2L1 Genes and Their Impact on Susceptibility to Lung Cancer. J. Med. Genet. 2010, 47, 616–622. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.Y.; Lee, Y.-S.; Huang, J.-J.; Chang, C.; Chang, J.-M.; Chuang, S.-H.; Kao, K.-J.; Tsai, Y.-J.; Tsai, P.-Y.; Liu, C.-W.; et al. Characterization of the Biological Activity of a Potent Small Molecule Hec1 Inhibitor TAI-1. J. Exp. Clin. Cancer Res. 2014, 33, 6. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, K.; Sato, M.; Hase, T.; Elshazley, M.; Yamashita, R.; Usami, N.; Taniguchi, T.; Yokoi, K.; Nakamura, S.; Kondo, M.; et al. TIMELESS Is Overexpressed in Lung Cancer and Its Expression Correlates with Poor Patient Survival. Cancer Sci. 2013, 104, 171–177. [Google Scholar] [CrossRef] [PubMed]
- Peng, F.; Li, Q.; Niu, S.-Q.; Shen, G.-P.; Luo, Y.; Chen, M.; Bao, Y. ZWINT Is the next Potential Target for Lung Cancer Therapy. J. Cancer Res. Clin. Oncol. 2019, 145. [Google Scholar] [CrossRef]
- Boucherat, O.; Vitry, G.; Trinh, I.; Paulin, R.; Provencher, S.; Bonnet, S. The Cancer Theory of Pulmonary Arterial Hypertension. Pulm. Circ. 2017, 7, 285–299. [Google Scholar] [CrossRef]
- Ness, S.A. Myb Protein Specificity: Evidence of a Context-Specific Transcription Factor Code. Blood Cells. Mol. Dis. 2003, 31, 192–200. [Google Scholar] [CrossRef]
- Manak, J.R.; Mitiku, N.; Lipsick, J.S. Mutation of the Drosophila Homologue of the Myb Protooncogene Causes Genomic Instability. Proc. Natl. Acad. Sci. USA 2002, 99, 7438–7443. [Google Scholar] [CrossRef]
- Iltzsche, F.; Simon, K.; Stopp, S.; Pattschull, G.; Francke, S.; Wolter, P.; Hauser, S.; Murphy, D.J.; Garcia, P.; Rosenwald, A.; et al. An Important Role for Myb-MuvB and Its Target Gene KIF23 in a Mouse Model of Lung Adenocarcinoma. Oncogene 2017, 36, 110–121. [Google Scholar] [CrossRef]
- Cao, Y.; Zhu, W.; Chen, W.; Wu, J.; Hou, G.; Li, Y. Prognostic Value of BIRC5 in Lung Adenocarcinoma Lacking EGFR, KRAS, and ALK Mutations by Integrated Bioinformatics Analysis. Dis. Markers 2019, 2019, 5451290. [Google Scholar] [CrossRef] [PubMed]
- Otálora-Otálora, B.A.; Henríquez, B.; López-Kleine, L.; Rojas, A. RUNX Family: Oncogenes or Tumor Suppressors (Review). Oncol. Rep. 2019, 42, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Bolte, C.; Flood, H.M.; Ren, X.; Jagannathan, S.; Barski, A.; Kalin, T.V.; Kalinichenko, V.V. FOXF1 Transcription Factor Promotes Lung Regeneration after Partial Pneumonectomy. Sci. Rep. 2017, 7, 10690. [Google Scholar] [CrossRef] [PubMed]
- Wei, H.-J.; Nickoloff, J.A.; Chen, W.-H.; Liu, H.-Y.; Lo, W.-C.; Chang, Y.-T.; Yang, P.-C.; Wu, C.-W.; Williams, D.F.; Gelovani, J.G.; et al. FOXF1 Mediates Mesenchymal Stem Cell Fusion-Induced Reprogramming of Lung Cancer Cells. Oncotarget 2014, 5, 9514–9529. [Google Scholar] [CrossRef]
- Herrera-Merchan, A.; Cuadros, M.; Rodriguez, M.I.; Rodriguez, S.; Torres, R.; Estecio, M.; Coira, I.F.; Loidi, C.; Saiz, M.; Carmona-Saez, P.; et al. The Value of LncRNA FENDRR and FOXF1 as a Prognostic Factor for Survival of Lung Adenocarcinoma. Oncotarget 2020, 11, 1172–1185. [Google Scholar] [CrossRef]
- Miao, L.; Huang, Z.; Zengli, Z.; Li, H.; Chen, Q.; Yao, C.; Cai, H.; Xiao, Y.; Xia, H.; Wang, Y. Loss of Long Noncoding RNA FOXF1-AS1 Regulates Epithelial-Mesenchymal Transition, Stemness and Metastasis of Non-Small Cell Lung Cancer Cells. Oncotarget 2016, 7, 68339–68349. [Google Scholar] [CrossRef]
- Xu, R.; Han, Y. Long Non-Coding RNA FOXF1 Adjacent Non-Coding Developmental Regulatory RNA Inhibits Growth and Chemotherapy Resistance in Non-Small Cell Lung Cancer. Arch. Med. Sci. 2019, 15, 1539–1546. [Google Scholar] [CrossRef]
- Zito, G.; Naselli, F.; Saieva, L.; Raimondo, S.; Calabrese, G.; Guzzardo, C.; Forte, S.; Rolfo, C.; Parenti, R.; Alessandro, R. Retinoic Acid Affects Lung Adenocarcinoma Growth by Inducing Differentiation via GATA6 Activation and EGFR and Wnt Inhibition. Sci. Rep. 2017, 7, 4770. [Google Scholar] [CrossRef]
- Cheung, W.K.C.; Zhao, M.; Liu, Z.; Stevens, L.E.; Cao, P.D.; Fang, J.E.; Westbrook, T.F.; Nguyen, D.X. Control of Alveolar Differentiation by the Lineage Transcription Factors GATA6 and HOPX Inhibits Lung Adenocarcinoma Metastasis. Cancer Cell 2013, 23, 725–738. [Google Scholar] [CrossRef]
- Yang, Y.; Tang, X.; Song, X.; Tang, L.; Cao, Y.; Liu, X.; Wang, X.; Li, Y.; Yu, M.; Wan, H.; et al. Evidence for an Oncogenic Role of HOXC6 in Human Non-Small Cell Lung Cancer. PeerJ 2019, 7, e6629. [Google Scholar] [CrossRef]
- Taniwaki, M.; Daigo, Y.; Ishikawa, N.; Takano, A.; Tsunoda, T.; Yasui, W.; Inai, K.; Kohno, N.; Nakamura, Y. Gene Expression Profiles of Small-Cell Lung Cancers: Molecular Signatures of Lung Cancer. Int. J. Oncol. 2006, 29, 567–575. [Google Scholar] [CrossRef] [PubMed]
- Jiang, T.; Hou, C.-C.; She, Z.-Y.; Yang, W.-X. The SOX Gene Family: Function and Regulation in Testis Determination and Male Fertility Maintenance. Mol. Biol. Rep. 2013, 40, 2187–2194. [Google Scholar] [CrossRef]
- Kumar, P.; Mistri, T.K. Transcription Factors in SOX Family: Potent Regulators for Cancer Initiation and Development in the Human Body. Semin. Cancer Biol. 2020, 67, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Fischer, M.; Grossmann, P.; Padi, M.; DeCaprio, J.A. Integration of TP53, DREAM, MMB-FOXM1 and RB-E2F Target Gene Analyses Identifies Cell Cycle Gene Regulatory Networks. Nucleic Acids Res. 2016, 44, 6070–6086. [Google Scholar] [CrossRef]
- Suliman, B.; Xu, D.; Williams, B. The Promyelocytic Leukemia Zinc Finger Protein: Two Decades of Molecular Oncology. Front. Oncol. 2012, 2, 74. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Z.-Y.; He, T.-T.; Gao, X.-M.; Zhao, Y.; Wang, J. ZBTB Transcription Factors: Key Regulators of the Development, Differentiation and Effector Function of T Cells. Front. Immunol. 2021, 12, 713294. [Google Scholar] [CrossRef]
- Chen, Z.; Guidez, F.; Rousselot, P.; Agadir, A.; Chen, S.J.; Wang, Z.Y.; Degos, L.; Zelent, A.; Waxman, S.; Chomienne, C. PLZF-RAR Alpha Fusion Proteins Generated from the Variant t(11;17)(Q23;Q21) Translocation in Acute Promyelocytic Leukemia Inhibit Ligand-Dependent Transactivation of Wild-Type Retinoic Acid Receptors. Proc. Natl. Acad. Sci. USA 1994, 91, 1178–1182. [Google Scholar] [CrossRef]
- Chagraoui, H.; Kristiansen, M.S.; Ruiz, J.P.; Serra-Barros, A.; Richter, J.; Hall-Ponselé, E.; Gray, N.; Waithe, D.; Clark, K.; Hublitz, P.; et al. SCL/TAL1 Cooperates with Polycomb RYBP-PRC1 to Suppress Alternative Lineages in Blood-Fated Cells. Nat. Commun. 2018, 9, 5375. [Google Scholar] [CrossRef]
- Khalil, A.M.; Guttman, M.; Huarte, M.; Garber, M.; Raj, A.; Rivea Morales, D.; Thomas, K.; Presser, A.; Bernstein, B.E.; van Oudenaarden, A.; et al. Many Human Large Intergenic Noncoding RNAs Associate with Chromatin-Modifying Complexes and Affect Gene Expression. Proc. Natl. Acad. Sci. USA 2009, 106, 11667–11672. [Google Scholar] [CrossRef]
- Pradhan, A.; Ustiyan, V.; Zhang, Y.; Kalin, T.V.; Kalinichenko, V.V. Forkhead Transcription Factor FoxF1 Interacts with Fanconi Anemia Protein Complexes to Promote DNA Damage Response. Oncotarget 2015, 7, 1912–1926. [Google Scholar] [CrossRef][Green Version]
- Bikeye, S.-N.N.; Colin, C.; Marie, Y.; Vampouille, R.; Ravassard, P.; Rousseau, A.; Boisselier, B.; Idbaih, A.; Calvo, C.F.; Leuraud, P.; et al. ASPM-Associated Stem Cell Proliferation Is Involved in Malignant Progression of Gliomas and Constitutes an Attractive Therapeutic Target. Cancer Cell Int. 2010, 10, 1. [Google Scholar] [CrossRef] [PubMed]
- Al-Khafaji, A.S.K.; Marcus, M.W.; Davies, M.P.A.; Risk, J.M.; Shaw, R.J.; Field, J.K.; Liloglou, T. AURKA MRNA Expression Is an Independent Predictor of Poor Prognosis in Patients with Non-Small Cell Lung Cancer. Oncol. Lett. 2017, 13, 4463–4468. [Google Scholar] [CrossRef]
- Shah, K.N.; Bhatt, R.; Rotow, J.; Rohrberg, J.; Olivas, V.; Wang, V.E.; Hemmati, G.; Martins, M.M.; Maynard, A.; Kuhn, J.; et al. Aurora Kinase A Drives the Evolution of Resistance to Third-Generation EGFR Inhibitors in Lung Cancer. Nat. Med. 2019, 25, 111–118. [Google Scholar] [CrossRef]
- Zhang, M.-Y.; Liu, X.-X.; Li, H.; Li, R.; Liu, X.; Qu, Y.-Q. Elevated MRNA Levels of AURKA, CDC20 and TPX2 Are Associated with Poor Prognosis of Smoking Related Lung Adenocarcinoma Using Bioinformatics Analysis. Int. J. Med. Sci. 2018, 15, 1676–1685. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.; Du, J.; Parsons, S.H.; Merzoug, F.F.; Webster, Y.; Iversen, P.W.; Chio, L.-C.; Van Horn, R.D.; Lin, X.; Blosser, W.; et al. Aurora A Kinase Inhibition Is Synthetic Lethal with Loss of the RB1 Tumor Suppressor Gene. Cancer Discov. 2019, 9, 248–263. [Google Scholar] [CrossRef] [PubMed]
- Jasinski, P.; Zwolak, P.; Terai, K.; Vogel, R.I.; Borja-Cacho, D.; Dudek, A.Z. MT477 Acts in Tumor Cells as an AURKA Inhibitor and Strongly Induces NRF-2 Signaling. Anticancer Res. 2011, 31, 1181–1187. [Google Scholar] [PubMed]
- Dar, A.A.; Goff, L.W.; Majid, S.; Berlin, J.; El-Rifai, W. Aurora Kinase Inhibitors--Rising Stars in Cancer Therapeutics? Mol. Cancer Ther. 2010, 9, 268–278. [Google Scholar] [CrossRef]
- Bertran-Alamillo, J.; Cattan, V.; Schoumacher, M.; Codony-Servat, J.; Giménez-Capitán, A.; Cantero, F.; Burbridge, M.; Rodríguez, S.; Teixidó, C.; Roman, R.; et al. AURKB as a Target in Non-Small Cell Lung Cancer with Acquired Resistance to Anti-EGFR Therapy. Nat. Commun. 2019, 10, 1812. [Google Scholar] [CrossRef]
- Hirano, H.; Maeda, H.; Yamaguchi, T.; Yokota, S.; Mori, M.; Sakoda, S. Survivin Expression in Lung Cancer: Association with Smoking, Histological Types and Pathological Stages. Oncol. Lett. 2015, 10, 1456–1462. [Google Scholar] [CrossRef]
- Chen, X.-Q.; Yang, S.; Kang, M.-Q.; Li, Z.-Y.; Lu, H.-S.; Lin, T.-Y. Survivin Expression in Human Lung Cancer and the Influence of Its Downregulation on the Biological Behavior of Human Lung Cancer Cells. Exp. Ther. Med. 2012, 3, 1010–1014. [Google Scholar] [CrossRef][Green Version]
- Falleni, M.; Pellegrini, C.; Marchetti, A.; Oprandi, B.; Buttitta, F.; Barassi, F.; Santambrogio, L.; Coggi, G.; Bosari, S. Survivin Gene Expression in Early-Stage Non-Small Cell Lung Cancer. J. Pathol. 2003, 200, 620–626. [Google Scholar] [CrossRef] [PubMed]
- Haruki, N.; Saito, H.; Harano, T.; Nomoto, S.; Takahashi, T.; Osada, H.; Fujii, Y.; Takahashi, T. Molecular Analysis of the Mitotic Checkpoint Genes BUB1, BUBR1 and BUB3 in Human Lung Cancers. Cancer Lett. 2001, 162, 201–205. [Google Scholar] [CrossRef]
- Rio Frio, T.; Lavoie, J.; Hamel, N.; Geyer, F.C.; Kushner, Y.B.; Novak, D.J.; Wark, L.; Capelli, C.; Reis-Filho, J.S.; Mai, S.; et al. Homozygous BUB1B Mutation and Susceptibility to Gastrointestinal Neoplasia. N. Engl. J. Med. 2010, 363, 2628–2637. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.; Lee, J.; Kljavin, N.M.; Haley, B.; Daemen, A.; Johnson, L.; Liang, Y. Requirement for BUB1B/BUBR1 in Tumor Progression of Lung Adenocarcinoma. Genes Cancer 2015, 6, 106–118. [Google Scholar] [CrossRef]
- Arsic, N.; Bendris, N.; Peter, M.; Begon-Pescia, C.; Rebouissou, C.; Gadéa, G.; Bouquier, N.; Bibeau, F.; Lemmers, B.; Blanchard, J.M. A Novel Function for Cyclin A2: Control of Cell Invasion via RhoA Signaling. J. Cell Biol. 2012, 196, 147–162. [Google Scholar] [CrossRef]
- Bendris, N.; Arsic, N.; Lemmers, B.; Blanchard, J.M. Cyclin A2, Rho GTPases and EMT. Small GTPases 2012, 3, 225–228. [Google Scholar] [CrossRef]
- Ruan, J.S.; Zhou, H.; Yang, L.; Wang, L.; Jiang, Z.S.; Wang, S.M. CCNA2 Facilitates Epithelial-to-Mesenchymal Transition via the Integrin Avβ3 Signaling in NSCLC. Int. J. Clin. Exp. Pathol. 2017, 10, 8324–8333. [Google Scholar]
- Soria, J.C.; Jang, S.J.; Khuri, F.R.; Hassan, K.; Liu, D.; Hong, W.K.; Mao, L. Overexpression of Cyclin B1 in Early-Stage Non-Small Cell Lung Cancer and Its Clinical Implication. Cancer Res. 2000, 60, 4000–4004. [Google Scholar] [PubMed]
- Winters, Z.E.; Hunt, N.C.; Bradburn, M.J.; Royds, J.A.; Turley, H.; Harris, A.L.; Norbury, C.J. Subcellular Localisation of Cyclin B, Cdc2 and P21(WAF1/CIP1) in Breast Cancer. Association with Prognosis. Eur. J. Cancer Oxf. Engl. 1990 2001, 37, 2405–2412. [Google Scholar] [CrossRef]
- Müllers, E.; Silva Cascales, H.; Burdova, K.; Macurek, L.; Lindqvist, A. Residual Cdk1/2 Activity after DNA Damage Promotes Senescence. Aging Cell 2017, 16, 575–584. [Google Scholar] [CrossRef]
- Ni, Z.; Wang, X.; Zhang, T.; Li, L.; Li, J. Comprehensive Analysis of Differential Expression Profiles Reveals Potential Biomarkers Associated with the Cell Cycle and Regulated by P53 in Human Small Cell Lung Cancer. Exp. Ther. Med. 2018, 15, 3273–3282. [Google Scholar] [CrossRef] [PubMed]
- Pines, J.; Hunter, T. Isolation of a Human Cyclin CDNA: Evidence for Cyclin MRNA and Protein Regulation in the Cell Cycle and for Interaction with P34cdc2. Cell 1989, 58, 833–846. [Google Scholar] [CrossRef]
- Brandeis, M.; Rosewell, I.; Carrington, M.; Crompton, T.; Jacobs, M.A.; Kirk, J.; Gannon, J.; Hunt, T. Cyclin B2-Null Mice Develop Normally and Are Fertile Whereas Cyclin B1-Null Mice Die in Utero. Proc. Natl. Acad. Sci. USA 1998, 95, 4344–4349. [Google Scholar] [CrossRef] [PubMed]
- Qian, X.; Song, X.; He, Y.; Yang, Z.; Sun, T.; Wang, J.; Zhu, G.; Xing, W.; You, C. CCNB2 Overexpression Is a Poor Prognostic Biomarker in Chinese NSCLC Patients. Biomed. Pharmacother. Biomed. Pharmacother. 2015, 74, 222–227. [Google Scholar] [CrossRef]
- Kato, T.; Daigo, Y.; Aragaki, M.; Ishikawa, K.; Sato, M.; Kaji, M. Overexpression of CDC20 Predicts Poor Prognosis in Primary Non-Small Cell Lung Cancer Patients. J. Surg. Oncol. 2012, 106, 423–430. [Google Scholar] [CrossRef]
- Zhang, W.; Gong, W.; Ai, H.; Tang, J.; Shen, C. Gene Expression Analysis of Lung Adenocarcinoma and Matched Adjacent Non-Tumor Lung Tissue. Tumori J. 2014, 100, 338–345. [Google Scholar] [CrossRef]
- Kidokoro, T.; Tanikawa, C.; Furukawa, Y.; Katagiri, T.; Nakamura, Y.; Matsuda, K. CDC20, a Potential Cancer Therapeutic Target, Is Negatively Regulated by P53. Oncogene 2008, 27, 1562–1571. [Google Scholar] [CrossRef]
- Shi, R.; Sun, Q.; Sun, J.; Wang, X.; Xia, W.; Dong, G.; Wang, A.; Jiang, F.; Xu, L. Cell Division Cycle 20 Overexpression Predicts Poor Prognosis for Patients with Lung Adenocarcinoma. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2017, 39, 1010428317692233. [Google Scholar] [CrossRef]
- Veena, V.; Rajan, K.; Saritha, V.; George, G.; Chandramohan, K.; Jayasree, K.; Thara, S.; Sujathan, K. DNA Replication Licensing Proteins for Early Detection of Lung Cancer. Asian Pac. J. Cancer Prev. APJCP 2017, 18, 3041–3047. [Google Scholar] [CrossRef]
- Wu, X.; Li, S.; Hu, X.; Xiang, X.; Halloran, M.; Yang, L.; Williams, T.M.; Houghton, P.J.; Shen, C.; He, Z. MTOR Signaling Upregulates CDC6 via Suppressing MiR-3178 and Promotes the Loading of DNA Replication Helicase. Sci. Rep. 2019, 9, 9805. [Google Scholar] [CrossRef]
- Allera-Moreau, C.; Rouquette, I.; Lepage, B.; Oumouhou, N.; Walschaerts, M.; Leconte, E.; Schilling, V.; Gordien, K.; Brouchet, L.; Delisle, M.B.; et al. DNA Replication Stress Response Involving PLK1, CDC6, POLQ, RAD51 and CLASPIN Upregulation Prognoses the Outcome of Early/Mid-Stage Non-Small Cell Lung Cancer Patients. Oncogenesis 2012, 1, e30. [Google Scholar] [CrossRef] [PubMed]
- Gamell, C.; Gulati, T.; Solomon, B.; Haupt, S.; Haupt, Y. Uncovering a Novel Pathway for P16 Silencing: Therapeutic Implications for Lung Cancer. Mol. Cell. Oncol. 2017, 4, e1299273. [Google Scholar] [CrossRef] [PubMed]
- Putkey, F.R.; Cramer, T.; Morphew, M.K.; Silk, A.D.; Johnson, R.S.; McIntosh, J.R.; Cleveland, D.W. Unstable Kinetochore-Microtubule Capture and Chromosomal Instability Following Deletion of CENP-E. Dev. Cell 2002, 3, 351–365. [Google Scholar] [CrossRef]
- Weng, M.-T.; Lee, J.-H.; Wei, S.-C.; Li, Q.; Shahamatdar, S.; Hsu, D.; Schetter, A.J.; Swatkoski, S.; Mannan, P.; Garfield, S.; et al. Evolutionarily Conserved Protein ERH Controls CENP-E MRNA Splicing and Is Required for the Survival of KRAS Mutant Cancer Cells. Proc. Natl. Acad. Sci. USA 2012, 109, E3659–E3667. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-J.; Tan, J.; Gao, X.-H.; Wang, L.-X. Integrated Analysis Reveals Key Genes with Prognostic Value in Lung Adenocarcinoma. Cancer Manag. Res. 2018, 10, 6097–6108. [Google Scholar] [CrossRef] [PubMed]
- Huang, R.; Gao, L. Identification of Potential Diagnostic and Prognostic Biomarkers in Non-Small Cell Lung Cancer Based on Microarray Data. Oncol. Lett. 2018, 15, 6436–6442. [Google Scholar] [CrossRef]
- Hsu, N.-Y.; Wang, H.-C.; Wang, C.-H.; Chiu, C.-F.; Tseng, H.-C.; Liang, S.-Y.; Tsai, C.-W.; Lin, C.-C.; Bau, D.-T. Lung Cancer Susceptibility and Genetic Polymorphisms of Exo1 Gene in Taiwan. Anticancer Res. 2009, 29, 725–730. [Google Scholar] [PubMed]
- Jin, G.; Wang, H.; Hu, Z.; Liu, H.; Sun, W.; Ma, H.; Chen, D.; Miao, R.; Tian, T.; Jin, L.; et al. Potentially Functional Polymorphisms of EXO1 and Risk of Lung Cancer in a Chinese Population: A Case-Control Analysis. Lung Cancer Amst. Neth. 2008, 60, 340–346. [Google Scholar] [CrossRef]
- Tang, C.; Jiang, Y.; Shao, W.; Shi, W.; Gao, X.; Qin, W.; Jiang, T.; Wang, F.; Feng, S. Abnormal Expression of FOSB Correlates with Tumor Progression and Poor Survival in Patients with Gastric Cancer. Int. J. Oncol. 2016, 49, 1489–1496. [Google Scholar] [CrossRef]
- Duan, F.; Song, C.; Dai, L.; Cui, S.; Zhang, X.; Zhao, X. The Significance of Exo1 K589E Polymorphism on Cancer Susceptibility: Evidence Based on a Meta-Analysis. PLoS ONE 2014, 9, e96764. [Google Scholar] [CrossRef]
- Yang, S.-Y.; Hsiung, C.-N.; Li, Y.-J.; Chang, G.-C.; Tsai, Y.-H.; Chen, K.-Y.; Huang, M.-S.; Su, W.-C.; Chen, Y.-M.; Hsiung, C.A.; et al. Fanconi Anemia Genes in Lung Adenocarcinoma- a Pathway-Wide Study on Cancer Susceptibility. J. Biomed. Sci. 2016, 23, 23. [Google Scholar] [CrossRef] [PubMed]
- Smogorzewska, A.; Matsuoka, S.; Vinciguerra, P.; McDonald, E.R.; Hurov, K.E.; Luo, J.; Ballif, B.A.; Gygi, S.P.; Hofmann, K.; D’Andrea, A.D.; et al. Identification of the FANCI Protein, a Monoubiquitinated FANCD2 Paralog Required for DNA Repair. Cell 2007, 129, 289–301. [Google Scholar] [CrossRef] [PubMed]
- Somyajit, K.; Subramanya, S.; Nagaraju, G. RAD51C: A Novel Cancer Susceptibility Gene Is Linked to Fanconi Anemia and Breast Cancer. Carcinogenesis 2010, 31, 2031–2038. [Google Scholar] [CrossRef] [PubMed]
- Howlett, N.G.; Harney, J.A.; Rego, M.A.; Kolling, F.W.; Glover, T.W. Functional Interaction between the Fanconi Anemia D2 Protein and Proliferating Cell Nuclear Antigen (PCNA) via a Conserved Putative PCNA Interaction Motif. J. Biol. Chem. 2009, 284, 28935–28942. [Google Scholar] [CrossRef] [PubMed]
- Stevens, L.E.; Cheung, W.K.C.; Adua, S.J.; Arnal-Estapé, A.; Zhao, M.; Liu, Z.; Brewer, K.; Herbst, R.S.; Nguyen, D.X. Extracellular Matrix Receptor Expression in Subtypes of Lung Adenocarcinoma Potentiates Outgrowth of Micrometastases. Cancer Res. 2017, 77, 1905–1917. [Google Scholar] [CrossRef]
- Stevens, L.E.; Zhao, M.; Liu, Z.Z.; Nguyen, D.X. Abstract 2269: A Novel Molecular Subset of Metastatic Lung Adenocarcinoma Is Defined by the Function of the Proteoglycan Receptor HMMR. Cancer Res. 2015, 75, 2269. [Google Scholar] [CrossRef]
- Blangy, A.; Lane, H.A.; d’Hérin, P.; Harper, M.; Kress, M.; Nigg, E.A. Phosphorylation by P34cdc2 Regulates Spindle Association of Human Eg5, a Kinesin-Related Motor Essential for Bipolar Spindle Formation in Vivo. Cell 1995, 83, 1159–1169. [Google Scholar] [CrossRef]
- Koffa, M.D.; Casanova, C.M.; Santarella, R.; Köcher, T.; Wilm, M.; Mattaj, I.W. HURP Is Part of a Ran-Dependent Complex Involved in Spindle Formation. Curr. Biol. 2006, 16, 743–754. [Google Scholar] [CrossRef]
- Schneider, M.A.; Christopoulos, P.; Muley, T.; Warth, A.; Klingmueller, U.; Thomas, M.; Herth, F.J.F.; Dienemann, H.; Mueller, N.S.; Theis, F.; et al. AURKA, DLGAP5, TPX2, KIF11 and CKAP5: Five Specific Mitosis-Associated Genes Correlate with Poor Prognosis for Non-Small Cell Lung Cancer Patients. Int. J. Oncol. 2017, 50, 365–372. [Google Scholar] [CrossRef]
- Kato, T.; Lee, D.; Huang, H.; Cruz, W.; Ujiie, H.; Fujino, K.; Wada, H.; Patel, P.; Hu, H.-P.; Hirohashi, K.; et al. Personalized SiRNA-Nanoparticle Systemic Therapy Using Metastatic Lymph Node Specimens Obtained with EBUS-TBNA in Lung Cancer. Mol. Cancer Res. MCR 2018, 16, 47–57. [Google Scholar] [CrossRef]
- Kato, T.; Lee, D.; Wu, L.; Patel, P.; Young, J.A.; Wada, H.; Hu, H.-P.; Ujiie, H.; Kaji, M.; Kano, S.; et al. Kinesin Family Members KIF11 and KIF23 as Potential Therapeutic Targets in Malignant Pleural Mesothelioma. Int. J. Oncol. 2016, 49, 448–456. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Wada, H.; Patel, P.; Hu, H.-P.; Lee, D.; Ujiie, H.; Hirohashi, K.; Nakajima, T.; Sato, M.; Kaji, M.; et al. Overexpression of KIF23 Predicts Clinical Outcome in Primary Lung Cancer Patients. Lung Cancer Amst. Neth. 2016, 92, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Vikberg, A.-L.; Vooder, T.; Lokk, K.; Annilo, T.; Golovleva, I. Mutation Analysis and Copy Number Alterations of KIF23 in Non-Small-Cell Lung Cancer Exhibiting KIF23 over-Expression. OncoTargets Ther. 2017, 10, 4969–4979. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Gu, J.; Xu, F.; Zhu, Q.; Ge, D.; Lu, C. Transcriptomic and Functional Network Features of Lung Squamous Cell Carcinoma through Integrative Analysis of GEO and TCGA Data. Sci. Rep. 2018, 8, 15834. [Google Scholar] [CrossRef]
- Bai, Y.; Xiong, L.; Zhu, M.; Yang, Z.; Zhao, J.; Tang, H. Co-Expression Network Analysis Identified KIF2C in Association with Progression and Prognosis in Lung Adenocarcinoma. Cancer Biomark. Sect. Dis. Markers 2019, 24, 371–382. [Google Scholar] [CrossRef]
- Shi, Y.-X.; Zhu, T.; Zou, T.; Zhuo, W.; Chen, Y.-X.; Huang, M.-S.; Zheng, W.; Wang, C.-J.; Li, X.; Mao, X.-Y.; et al. Prognostic and Predictive Values of CDK1 and MAD2L1 in Lung Adenocarcinoma. Oncotarget 2016, 7, 85235–85243. [Google Scholar] [CrossRef]
- Wei, X.; Zhang, K.; Qin, H.; Zhu, J.; Qin, Q.; Yu, Y.; Wang, H. GMDS Knockdown Impairs Cell Proliferation and Survival in Human Lung Adenocarcinoma. BMC Cancer 2018, 18, 600. [Google Scholar] [CrossRef]
- Mincheva, A.; Todorov, I.; Werner, D.; Fink, T.M.; Lichter, P. The Human Gene for Nuclear Protein BM28 (CDCL1), a New Member of the Early S-Phase Family of Proteins, Maps to Chromosome Band 3q21. Cytogenet. Cell Genet. 1994, 65, 276–277. [Google Scholar] [CrossRef]
- Yabuta, N.; Kajimura, N.; Mayanagi, K.; Sato, M.; Gotow, T.; Uchiyama, Y.; Ishimi, Y.; Nojima, H. Mammalian Mcm2/4/6/7 Complex Forms a Toroidal Structure. Genes Cells 2003, 8, 413–421. [Google Scholar] [CrossRef]
- Cheung, C.H.Y.; Hsu, C.-L.; Chen, K.-P.; Chong, S.-T.; Wu, C.-H.; Huang, H.-C.; Juan, H.-F. MCM2-Regulated Functional Networks in Lung Cancer by Multi-Dimensional Proteomic Approach. Sci. Rep. 2017, 7, 13302. [Google Scholar] [CrossRef]
- Yang, J.; Ramnath, N.; Moysich, K.B.; Asch, H.L.; Swede, H.; Alrawi, S.J.; Huberman, J.; Geradts, J.; Brooks, J.S.; Tan, D. Prognostic Significance of MCM2, Ki-67 and Gelsolin in Non-Small Cell Lung Cancer. BMC Cancer 2006, 6, 203. [Google Scholar] [CrossRef] [PubMed]
- Tan, D.F.; Huberman, J.A.; Hyland, A.; Loewen, G.M.; Brooks, J.S.; Beck, A.F.; Todorov, I.T.; Bepler, G. MCM2—A Promising Marker for Premalignant Lesions of the Lung: A Cohort Study. BMC Cancer 2001, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Kikuchi, J.; Kinoshita, I.; Shimizu, Y.; Kikuchi, E.; Takeda, K.; Aburatani, H.; Oizumi, S.; Konishi, J.; Kaga, K.; Matsuno, Y.; et al. Minichromosome Maintenance (MCM) Protein 4 as a Marker for Proliferation and Its Clinical and Clinicopathological Significance in Non-Small Cell Lung Cancer. Lung Cancer Amst. Neth. 2011, 72, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Choy, B.; LaLonde, A.; Que, J.; Wu, T.; Zhou, Z. MCM4 and MCM7, Potential Novel Proliferation Markers, Significantly Correlated with Ki-67, Bmi1, and Cyclin E Expression in Esophageal Adenocarcinoma, Squamous Cell Carcinoma, and Precancerous Lesions. Hum. Pathol. 2016, 57, 126–135. [Google Scholar] [CrossRef]
- Warth, A.; Cortis, J.; Soltermann, A.; Meister, M.; Budczies, J.; Stenzinger, A.; Goeppert, B.; Thomas, M.; Herth, F.J.F.; Schirmacher, P.; et al. Tumour Cell Proliferation (Ki-67) in Non-Small Cell Lung Cancer: A Critical Reappraisal of Its Prognostic Role. Br. J. Cancer 2014, 111, 1222–1229. [Google Scholar] [CrossRef]
- Sadasivam, S.; DeCaprio, J.A. The DREAM Complex: Master Coordinator of Cell Cycle Dependent Gene Expression. Nat. Rev. Cancer 2013, 13, 585. [Google Scholar] [CrossRef]
- Fan, X.; Wang, Y.; Jiang, T.; Cai, W.; Jin, Y.; Niu, Y.; Zhu, H.; Bu, Y. B-Myb Mediates Proliferation and Migration of Non-Small-Cell Lung Cancer via Suppressing IGFBP3. Int. J. Mol. Sci. 2018, 19, 1479. [Google Scholar] [CrossRef]
- Zhang, S.; Li, M.; Ji, H.; Fang, Z. Landscape of Transcriptional Deregulation in Lung Cancer. BMC Genom. 2018, 19, 435. [Google Scholar] [CrossRef]
- Furth, P.A.; St Onge, L.; Böger, H.; Gruss, P.; Gossen, M.; Kistner, A.; Bujard, H.; Hennighausen, L. Temporal Control of Gene Expression in Transgenic Mice by a Tetracycline-Responsive Promoter. Proc. Natl. Acad. Sci. USA 1994, 91, 9302–9306. [Google Scholar] [CrossRef]
- Zhou, W.; Yang, Y.; Xia, J.; Wang, H.; Salama, M.E.; Xiong, W.; Xu, H.; Shetty, S.; Chen, T.; Zeng, Z.; et al. NEK2 Induces Drug-Resistance Mainly through Activation of Efflux Drug Pumps and Is Associated with Poor Prognosis in Myeloma and Other Cancers. Cancer Cell 2013, 23, 48–62. [Google Scholar] [CrossRef]
- Bidkhori, G.; Narimani, Z.; Hosseini Ashtiani, S.; Moeini, A.; Nowzari-Dalini, A.; Masoudi-Nejad, A. Reconstruction of an Integrated Genome-Scale Co-Expression Network Reveals Key Modules Involved in Lung Adenocarcinoma. PLoS ONE 2013, 8, e67552. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Guan, X.; Liu, W.; Zhang, L. Aberrant Expression of NEK2 and Its Clinical Significance in Non-Small Cell Lung Cancer. Oncol. Lett. 2014, 8, 1470–1476. [Google Scholar] [CrossRef] [PubMed]
- Kaowinn, S.; Oh, S.; Moon, J.; Yoo, A.Y.; Kang, H.Y.; Lee, M.R.; Kim, J.E.; Hwang, D.Y.; Youn, S.E.; Koh, S.S.; et al. CGK062, a Small Chemical Molecule, Inhibits Cancer Upregulated Gene 2-Induced Oncogenesis through NEK2 and β-Catenin. Int. J. Oncol. 2019, 54, 1295–1305. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Zhan, X.; Furqan, M.; Abu Hejleh, T.; Clamon, G.H.; Ren, S.; Zhan, F. NEK2 as a Prognostic Marker and Therapeutic Target in Adenocarcinoma of the Lung. J. Clin. Oncol. 2016, 34, e23282. [Google Scholar] [CrossRef]
- Das, T.K.; Dana, D.; Paroly, S.S.; Perumal, S.K.; Singh, S.; Jhun, H.; Pendse, J.; Cagan, R.L.; Talele, T.T.; Kumar, S. Centrosomal Kinase Nek2 Cooperates with Oncogenic Pathways to Promote Metastasis. Oncogenesis 2013, 2, e69. [Google Scholar] [CrossRef]
- Zhong, X.; Guan, X.; Dong, Q.; Yang, S.; Liu, W.; Zhang, L. Examining Nek2 as a Better Proliferation Marker in Non-Small Cell Lung Cancer Prognosis. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014, 35, 7155–7162. [Google Scholar] [CrossRef]
- Wen, P.; Chidanguro, T.; Shi, Z.; Gu, H.; Wang, N.; Wang, T.; Li, Y.; Gao, J. Identification of Candidate Biomarkers and Pathways Associated with SCLC by Bioinformatics Analysis. Mol. Med. Rep. 2018, 18, 1538–1550. [Google Scholar] [CrossRef]
- Lu, C.; Chen, H.; Shan, Z.; Yang, L. Identification of Differentially Expressed Genes between Lung Adenocarcinoma and Lung Squamous Cell Carcinoma by Gene Expression Profiling. Mol. Med. Rep. 2016, 14, 1483–1490. [Google Scholar] [CrossRef]
- Erdogan, E.; Klee, E.W.; Thompson, E.A.; Fields, A.P. Meta-Analysis of Oncogenic Protein Kinase Cι Signaling in Lung Adenocarcinoma. Clin. Cancer Res. 2009, 15, 1527–1533. [Google Scholar] [CrossRef]
- Song, L.; Dai, Z.; Zhang, S.; Zhang, H.; Liu, C.; Ma, X.; Liu, D.; Zan, Y.; Yin, X. MicroRNA-1179 Suppresses Cell Growth and Invasion by Targeting Sperm-Associated Antigen 5-Mediated Akt Signaling in Human Non-Small Cell Lung Cancer. Biochem. Biophys. Res. Commun. 2018, 504, 164–170. [Google Scholar] [CrossRef]
- Wang, T.; Li, K.; Song, H.; Xu, D.; Liao, Y.; Jing, B.; Guo, W.; Hu, M.; Kuang, Y.; Sun, B.; et al. P53 Suppression Is Essential for Oncogenic SPAG5 Upregulation in Lung Adenocarcinoma. Biochem. Biophys. Res. Commun. 2019, 513, 319–325. [Google Scholar] [CrossRef] [PubMed]
- Välk, K.; Vooder, T.; Kolde, R.; Reintam, M.-A.; Petzold, C.; Vilo, J.; Metspalu, A. Gene Expression Profiles of Non-Small Cell Lung Cancer: Survival Prediction and New Biomarkers. Oncology 2010, 79, 283–292. [Google Scholar] [CrossRef] [PubMed]
- Dingemans, A.C.; van Ark-Otte, J.; Span, S.; Scagliotti, G.V.; van der Valk, P.; Postmus, P.E.; Giaccone, G. Topoisomerase IIalpha and Other Drug Resistance Markers in Advanced Non-Small Cell Lung Cancer. Lung Cancer Amst. Neth. 2001, 32, 117–128. [Google Scholar] [CrossRef]
- Hou, G.-X.; Liu, P.; Yang, J.; Wen, S. Mining Expression and Prognosis of Topoisomerase Isoforms in Non-Small-Cell Lung Cancer by Using Oncomine and Kaplan–Meier Plotter. PLoS ONE 2017, 12, e0174515. [Google Scholar] [CrossRef]
- Neubauer, E.; Wirtz, R.M.; Kaemmerer, D.; Athelogou, M.; Schmidt, L.; Sänger, J.; Lupp, A. Comparative Evaluation of Three Proliferation Markers, Ki-67, TOP2A, and RacGAP1, in Bronchopulmonary Neuroendocrine Neoplasms: Issues and Prospects. Oncotarget 2016, 7, 41959–41973. [Google Scholar] [CrossRef][Green Version]
- Van Gijn, S.E.; Wierenga, E.; van den Tempel, N.; Kok, Y.P.; Heijink, A.M.; Spierings, D.C.J.; Foijer, F.; van Vugt, M.A.T.M.; Fehrmann, R.S.N. TPX2/Aurora Kinase A Signaling as a Potential Therapeutic Target in Genomically Unstable Cancer Cells. Oncogene 2019, 38, 852–867. [Google Scholar] [CrossRef] [PubMed]
- Du, L.; Zhao, Z.; Suraokar, M.; Shelton, S.S.; Ma, X.; Hsiao, T.-H.; Minna, J.D.; Wistuba, I.; Pertsemlidis, A. LMO1 Functions as an Oncogene by Regulating TTK Expression and Correlates with Neuroendocrine Differentiation of Lung Cancer. Oncotarget 2018, 9, 29601–29618. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Yu, C.; Gao, J.; Zhu, H.; Cui, B.; Zhang, T.; Zhou, Y.; Liu, Q.; He, H.; Xiao, R.; et al. A Novel USP9X Substrate TTK Contributes to Tumorigenesis in Non-Small-Cell Lung Cancer. Theranostics 2018, 8, 2348–2360. [Google Scholar] [CrossRef]
- Zheng, L.; Chen, Z.; Kawakami, M.; Chen, Y.; Roszik, J.; Mustachio, L.M.; Kurie, J.M.; Villalobos, P.; Lu, W.; Behrens, C.; et al. Tyrosine Threonine Kinase Inhibition Eliminates Lung Cancers by Augmenting Apoptosis and Polyploidy. Mol. Cancer Ther. 2019, 18, 1775–1786. [Google Scholar] [CrossRef]
- Mills, G.B.; Schmandt, R.; McGill, M.; Amendola, A.; Hill, M.; Jacobs, K.; May, C.; Rodricks, A.M.; Campbell, S.; Hogg, D. Expression of TTK, a Novel Human Protein Kinase, Is Associated with Cell Proliferation. J. Biol. Chem. 1992, 267, 16000–16006. [Google Scholar] [CrossRef] [PubMed]
- Yuan, W.; Xie, S.; Wang, M.; Pan, S.; Huang, X.; Xiong, M.; Xiao, R.; Xiong, J.; Zhang, Q.; Shao, L. Bioinformatic Analysis of Prognostic Value of ZW10 Interacting Protein in Lung Cancer. OncoTargets Ther. 2018, 11, 1683–1695. [Google Scholar] [CrossRef] [PubMed]
- Choi, K.H.; Shin, C.H.; Lee, W.J.; Ji, H.; Kim, H.H. Dual-Strand Tumor Suppressor MiR-193b-3p and -5p Inhibit Malignant Phenotypes of Lung Cancer by Suppressing Their Common Targets. Biosci. Rep. 2019, 39, BSR20190634. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhao, L.; Zhao, P.; Liu, Z. Long Non-Coding RNA LINC00641 Suppresses Non-Small-Cell Lung Cancer by Sponging MiR-424-5p to Upregulate PLSCR4. Cancer Biomark. Sect. Dis. Markers 2019, 26, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Hsiao, C.-C.; Chen, K.-D.; Hung, Y.-C.; Wu, C.-Y.; Lie, C.-H.; Liu, S.-F.; Sung, M.-T.; Chen, C.-J.; Wang, T.-Y.; et al. Peripheral Immune Cell Gene Expression Changes in Advanced Non-Small Cell Lung Cancer Patients Treated with First Line Combination Chemotherapy. PLoS ONE 2013, 8, e57053. [Google Scholar] [CrossRef] [PubMed]
- Lin, J.; Marquardt, G.; Mullapudi, N.; Wang, T.; Han, W.; Shi, M.; Keller, S.; Zhu, C.; Locker, J.; Spivack, S.D. Lung Cancer Transcriptomes Refined with Laser Capture Microdissection. Am. J. Pathol. 2014, 184, 2868–2884. [Google Scholar] [CrossRef] [PubMed]
- Orvis, T.; Hepperla, A.; Walter, V.; Song, S.; Simon, J.; Parker, J.; Wilkerson, M.D.; Desai, N.; Major, M.B.; Hayes, D.N.; et al. BRG1/SMARCA4 Inactivation Promotes Non-Small Cell Lung Cancer Aggressiveness by Altering Chromatin Organization. Cancer Res. 2014, 74, 6486–6498. [Google Scholar] [CrossRef]
- Yang, Z.; Zhuan, B.; Yan, Y.; Jiang, S.; Wang, T. Integrated Analyses of Copy Number Variations and Gene Differential Expression in Lung Squamous-Cell Carcinoma. Biol. Res. 2015, 48, 47. [Google Scholar] [CrossRef]
- Wang, W.; Dong, M.; Cui, J.; Xu, F.; Yan, C.; Ma, C.; Yi, L.; Tang, W.; Dong, J.; Wei, Y. NME4 May Enhance Non-small Cell Lung Cancer Progression by Overcoming Cell Cycle Arrest and Promoting Cellular Proliferation. Mol. Med. Rep. 2019, 20, 1629–1636. [Google Scholar] [CrossRef]
- Lu, Q.; Xie, Z.; Yan, C.; Ding, Y.; Ma, Z.; Wu, S.; Qiu, Y.; Cossette, S.M.; Bordas, M.; Ramchandran, R.; et al. SNRK (Sucrose Nonfermenting 1-Related Kinase) Promotes Angiogenesis In Vivo. Arterioscler. Thromb. Vasc. Biol. 2018, 38, 373–385. [Google Scholar] [CrossRef]
- Zheng, Y.; Miyamoto, D.T.; Wittner, B.S.; Sullivan, J.P.; Aceto, N.; Jordan, N.V.; Yu, M.; Karabacak, N.M.; Comaills, V.; Morris, R.; et al. Expression of β-Globin by Cancer Cells Promotes Cell Survival during Blood-Borne Dissemination. Nat. Commun. 2017, 8, 14344. [Google Scholar] [CrossRef]
- Casciaro, M.; Cardia, R.; Di Salvo, E.; Tuccari, G.; Ieni, A.; Gangemi, S. Interleukin-33 Involvement in Nonsmall Cell Lung Carcinomas: An Update. Biomolecules 2019, 9, 203. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Chen, Z.; Bu, X.; Han, Y.; Shan, S.; Ren, T.; Song, W. IL-33 Signaling Fuels Outgrowth and Metastasis of Human Lung Cancer. Biochem. Biophys. Res. Commun. 2016, 479, 461–468. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.S.; Kim, E.; Heo, J.-S.; Bae, D.-J.; Lee, J.-U.W.; Lee, T.-H.; Lee, H.J.; Chang, H.S.; Park, J.S.; Jang, A.S.; et al. Circulating IL-33 Level Is Associated with the Progression of Lung Cancer. Lung Cancer Amst. Neth. 2015, 90, 346–351. [Google Scholar] [CrossRef]
- Wang, K.; Shan, S.; Yang, Z.; Gu, X.; Wang, Y.; Wang, C.; Ren, T. IL-33 Blockade Suppresses Tumor Growth of Human Lung Cancer through Direct and Indirect Pathways in a Preclinical Model. Oncotarget 2017, 8, 68571–68582. [Google Scholar] [CrossRef]
- Hu, L.-A.; Fu, Y.; Zhang, D.-N.; Zhang, J. Serum IL-33 as a Diagnostic and Prognostic Marker in Non-Small Cell Lung Cancer. Asian Pac. J. Cancer Prev. 2013, 14, 2563–2566. [Google Scholar] [CrossRef] [PubMed]
- Jo, U.; Whang, Y.M.; Kim, H.K.; Kim, Y.H. AKAP12alpha Is Associated with Promoter Methylation in Lung Cancer. Cancer Res. Treat. 2006, 38, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Tessema, M.; Willink, R.; Do, K.; Yu, Y.Y.; Yu, W.; Machida, E.O.; Brock, M.; Van Neste, L.; Stidley, C.A.; Baylin, S.B.; et al. Promoter Methylation of Genes in and around the Candidate Lung Cancer Susceptibility Locus 6q23-25. Cancer Res. 2008, 68, 1707–1714. [Google Scholar] [CrossRef] [PubMed]
- Chen, K.-Y.; Hsiao, C.-F.; Chang, G.-C.; Tsai, Y.-H.; Su, W.-C.; Chen, Y.-M.; Huang, M.-S.; Tsai, F.-Y.; Jiang, S.-S.; Chang, I.-S.; et al. Estrogen Receptor Gene Polymorphisms and Lung Adenocarcinoma Risk in Never-Smoking Women. J. Thorac. Oncol. 2015, 10, 1413–1420. [Google Scholar] [CrossRef]
- Kim, W.; Kim, E.; Lee, S.; Kim, D.; Chun, J.; Park, K.H.; Youn, H.; Youn, B. TFAP2C-Mediated Upregulation of TGFBR1 Promotes Lung Tumorigenesis and Epithelial-Mesenchymal Transition. Exp. Mol. Med. 2016, 48, e273. [Google Scholar] [CrossRef]
- Kao, Y.-C.; Jiang, S.-J.; Pan, W.-A.; Wang, K.-C.; Chen, P.-K.; Wei, H.-J.; Chen, W.-S.; Chang, B.-I.; Shi, G.-Y.; Wu, H.-L. The Epidermal Growth Factor-like Domain of CD93 Is a Potent Angiogenic Factor. PLoS ONE 2012, 7, e51647. [Google Scholar] [CrossRef]
- Overbeck, T.R.; Arnemann, J.; Waldmann-Beushausen, R.; Trümper, L.; Schöndube, F.A.; Reuter-Jessen, K.; Danner, B.C. ABCA3 Phenotype in Non-Small Cell Lung Cancer Indicates Poor Outcome. Oncology 2017, 93, 270–278. [Google Scholar] [CrossRef] [PubMed]
- Beers, M.F.; Mulugeta, S. The Biology of the ABCA3 Lipid Transporter in Lung Health and Disease. Cell Tissue Res. 2017, 367, 481–493. [Google Scholar] [CrossRef] [PubMed]
- Arnemann, J.; Overbeck, T.; Ort, K.; Emmert, A.; Waldmann-Beushausen, R.; Trümper, L.; Wulf, G.; Schöndube, F.A.; Danner, B. Expression Patterns of ABCA3 and TTF-1 in Non-Small Cell Lung Cancer. Thorac. Cardiovasc. Surg. 2015, 63, ePP71. [Google Scholar] [CrossRef]
- Zhou, W.; Zhuang, Y.; Sun, J.; Wang, X.; Zhao, Q.; Xu, L.; Wang, Y. Variants of the ABCA3 Gene Might Contribute to Susceptibility to Interstitial Lung Diseases in the Chinese Population. Sci. Rep. 2017, 7, 4097. [Google Scholar] [CrossRef]
- Arkova, O.V.; Drachkova, I.A.; Arshinova, T.V.; Rasskazov, D.A.; Suslov, V.V.; Ponomarenko, P.M.; Ponomarenko, M.P.; Kolchanov, N.A.; Savinkova, L.K. Prediction and Verification of the Influence of the Rs367781716 SNP on the Interaction of the TATA-Binding Protein with the Promoter of the Human АВСА9 Gene. Russ. J. Genet. Appl. Res. 2016, 6, 785–791. [Google Scholar] [CrossRef]
- Piehler, A.; Kaminski, W.E.; Wenzel, J.J.; Langmann, T.; Schmitz, G. Molecular Structure of a Novel Cholesterol-Responsive A Subclass ABC Transporter, ABCA9. Biochem. Biophys. Res. Commun. 2002, 295, 408–416. [Google Scholar] [CrossRef]
- Fang, X.; Yin, Z.; Li, X.; Xia, L.; Quan, X.; Zhao, Y.; Zhou, B. Multiple Functional SNPs in Differentially Expressed Genes Modify Risk and Survival of Non-Small Cell Lung Cancer in Chinese Female Non-Smokers. Oncotarget 2017, 8, 18924–18934. [Google Scholar] [CrossRef]
- Selamat, S.A.; Chung, B.S.; Girard, L.; Zhang, W.; Zhang, Y.; Campan, M.; Siegmund, K.D.; Koss, M.N.; Hagen, J.A.; Lam, W.L.; et al. Genome-Scale Analysis of DNA Methylation in Lung Adenocarcinoma and Integration with MRNA Expression. Genome Res. 2012, 22, 1197–1211. [Google Scholar] [CrossRef]
- Marchitti, S.A.; Orlicky, D.J.; Brocker, C.; Vasiliou, V. Aldehyde Dehydrogenase 3B1 (ALDH3B1): Immunohistochemical Tissue Distribution and Cellular-Specific Localization in Normal and Cancerous Human Tissues. J. Histochem. Cytochem. Off. J. Histochem. Soc. 2010, 58, 765–783. [Google Scholar] [CrossRef]
- Deng, T.; Lin, D.; Zhang, M.; Zhao, Q.; Li, W.; Zhong, B.; Deng, Y.; Fu, X. Differential Expression of Bone Morphogenetic Protein 5 in Human Lung Squamous Cell Carcinoma and Adenocarcinoma. Acta Biochim. Biophys. Sin. 2015, 47, 557–563. [Google Scholar] [CrossRef]
- Tian, X.-P.; Jin, X.-H.; Li, M.; Huang, W.-J.; Xie, D.; Zhang, J.-X. The Depletion of PinX1 Involved in the Tumorigenesis of Non-Small Cell Lung Cancer Promotes Cell Proliferation via P15/Cyclin D1 Pathway. Mol. Cancer 2017, 16, 74. [Google Scholar] [CrossRef] [PubMed]
- Al Zeyadi, M.; Dimova, I.; Ranchich, V.; Rukova, B.; Nesheva, D.; Hamude, Z.; Georgiev, S.; Petrov, D.; Toncheva, D. Whole Genome Microarray Analysis in Non-Small Cell Lung Cancer. Biotechnol. Biotechnol. Equip. 2015, 29, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.; Liu, X.; Li, X.; Guan, Y. Integrated Analysis of DNA Methylation and MRNA Expression Profiles Data to Identify Key Genes in Lung Adenocarcinoma. BioMed Res. Int. 2016, 2016, 4369431. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Chen, X.; Wei, K.; Liu, D.; Xu, X.; Zhang, X.; Shi, H. Identification of Key Transcription Factors Associated with Lung Squamous Cell Carcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 172–206. [Google Scholar] [CrossRef][Green Version]
- Faner, R.; Cruz, T.; Casserras, T.; López-Giraldo, A.; Noell, G.; Coca, I.; Tal-Singer, R.; Miller, B.; Rodriguez-Roisin, R.; Spira, A.; et al. Network Analysis of Lung Transcriptomics Reveals a Distinct B-Cell Signature in Emphysema. Am. J. Respir. Crit. Care Med. 2016, 193, 1242–1253. [Google Scholar] [CrossRef]
- Hong, S.-Y.; Kao, Y.-R.; Lee, T.-C.; Wu, C.-W. Upregulation of E3 Ubiquitin Ligase CBLC Enhances EGFR Dysregulation and Signaling in Lung Adenocarcinoma. Cancer Res. 2018, 78, 4984–4996. [Google Scholar] [CrossRef]
- Kim, B.; Lee, H.J.; Choi, H.Y.; Shin, Y.; Nam, S.; Seo, G.; Son, D.-S.; Jo, J.; Kim, J.; Lee, J.; et al. Clinical Validity of the Lung Cancer Biomarkers Identified by Bioinformatics Analysis of Public Expression Data. Cancer Res. 2007, 67, 7431–7438. [Google Scholar] [CrossRef]
- Demidova, A.R.; Aau, M.Y.; Zhuang, L.; Yu, Q. Dual Regulation of Cdc25A by Chk1 and P53-ATF3 in DNA Replication Checkpoint Control *. J. Biol. Chem. 2009, 284, 4132–4139. [Google Scholar] [CrossRef]
- Chiba, I.; Takahashi, T.; Nau, M.M.; D’Amico, D.; Curiel, D.T.; Mitsudomi, T.; Buchhagen, D.L.; Carbone, D.; Piantadosi, S.; Koga, H. Mutations in the P53 Gene Are Frequent in Primary, Resected Non-Small Cell Lung Cancer. Lung Cancer Study Group. Oncogene 1990, 5, 1603–1610. [Google Scholar] [PubMed]
- He, N.; Li, C.; Zhang, X.; Sheng, T.; Chi, S.; Chen, K.; Wang, Q.; Vertrees, R.; Logrono, R.; Xie, J. Regulation of Lung Cancer Cell Growth and Invasiveness by Beta-TRCP. Mol. Carcinog. 2005, 42, 18–28. [Google Scholar] [CrossRef]
- Wu, W.; Fan, Y.H.; Kemp, B.L.; Walsh, G.; Mao, L. Overexpression of Cdc25A and Cdc25B Is Frequent in Primary Non-Small Cell Lung Cancer but Is Not Associated with Overexpression of c-Myc. Cancer Res. 1998, 58, 4082–4085. [Google Scholar] [PubMed]
- Younis, R.H.; Cao, W.; Lin, R.; Xia, R.; Liu, Z.; Edelman, M.J.; Mei, Y.; Mao, L.; Ren, H. CDC25AQ110del: A Novel Cell Division Cycle 25A Isoform Aberrantly Expressed in Non-Small Cell Lung Cancer. PLoS ONE 2012, 7, e46464. [Google Scholar] [CrossRef]
- Cai, W.; Chen, C.; Li, X.; Shi, J.; Sun, Q.; Liu, D.; Sun, Y.; Hou, L.; Zhao, X.; Gu, S.; et al. Association of CDC25 Phosphatase Family Polymorphisms with the Efficacy/Toxicity of Platinum-Based Chemotherapy in Chinese Advanced NSCLC Patients. Future Oncol. Lond. Engl. 2014, 10, 1175–1185. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, M.-H.; Koinuma, J.; Ueda, K.; Ito, T.; Tsuchiya, E.; Nakamura, Y.; Daigo, Y. Phosphorylation and Activation of Cell Division Cycle Associated 5 by Mitogen-Activated Protein Kinase Play a Crucial Role in Human Lung Carcinogenesis. Cancer Res. 2010, 70, 5337–5347. [Google Scholar] [CrossRef] [PubMed]
- Osthus, R.C.; Karim, B.; Prescott, J.E.; Smith, B.D.; McDevitt, M.; Huso, D.L.; Dang, C.V. The Myc Target Gene JPO1/CDCA7 Is Frequently Over-Expressed in Human Tumors and Has Limited Transforming Activity In Vivo. Cancer Res. 2005, 65, 5620–5627. [Google Scholar] [CrossRef]
- Wang, H.; Ye, L.; Xing, Z.; Li, H.; Lv, T.; Liu, H.; Zhang, F.; Song, Y. CDCA7 Promotes Lung Adenocarcinoma Proliferation via Regulating the Cell Cycle. Pathol. Res. Pract. 2019, 215, 152559. [Google Scholar] [CrossRef] [PubMed]
- Cybulski, C.; Masojc, B.; Oszutowska, D.; Jaworowska, E.; Grodzki, T.; Waloszczyk, P.; Serwatowski, P.; Pankowski, J.; Huzarski, T.; Byrski, T.; et al. Constitutional CHEK2 Mutations Are Associated with a Decreased Risk of Lung and Laryngeal Cancers. Carcinogenesis 2008, 29, 762–765. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kukita, Y.; Okami, J.; Yoneda-Kato, N.; Nakamae, I.; Kawabata, T.; Higashiyama, M.; Kato, J.; Kodama, K.; Kato, K. Homozygous Inactivation of CHEK2 Is Linked to a Familial Case of Multiple Primary Lung Cancer with Accompanying Cancers in Other Organs. Cold Spring Harb. Mol. Case Stud. 2016, 2, a001032. [Google Scholar] [CrossRef][Green Version]
- Wang, Y.; McKay, J.D.; Rafnar, T.; Wang, Z.; Timofeeva, M.; Broderick, P.; Zong, X.; Laplana, M.; Wei, Y.; Han, Y.; et al. Rare Variants of Large Effect in BRCA2 and CHEK2 Affect Risk of Lung Cancer. Nat. Genet. 2014, 46, 736–741. [Google Scholar] [CrossRef]
- Cybulski, C.; Górski, B.; Huzarski, T.; Masojć, B.; Mierzejewski, M.; Dębniak, T.; Teodorczyk, U.; Byrski, T.; Gronwald, J.; Matyjasik, J.; et al. CHEK2 Is a Multiorgan Cancer Susceptibility Gene. Am. J. Hum. Genet. 2004, 75, 1131–1135. [Google Scholar] [CrossRef]
- Xu, W.; Liu, D.; Yang, Y.; Ding, X.; Sun, Y.; Zhang, B.; Xu, J.; Su, B. Association of CHEK2 Polymorphisms with the Efficacy of Platinum-Based Chemotherapy for Advanced Non-Small-Cell Lung Cancer in Chinese Never-Smoking Women. J. Thorac. Dis. 2016, 8, 2519–2529. [Google Scholar] [CrossRef] [PubMed]
- Okamoto, T.; Kohno, M.; Ito, K.; Takada, K.; Katsura, M.; Morodomi, Y.; Toyokawa, G.; Shoji, F.; Maehara, Y. Clinical Significance of DNA Damage Response Factors and Chromosomal Instability in Primary Lung Adenocarcinoma. Anticancer Res. 2017, 37, 1729–1735. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wei, H.; Gao, Z.; Chen, G.; Liu, Y.; Gao, X.; Bai, G.; He, S.; Liu, T.; Xu, W.; et al. COL5A1 May Contribute the Metastasis of Lung Adenocarcinoma. Gene 2018, 665, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, H.; Zhang, H.; Li, Y.; Li, S.; Hou, Q.; Wu, S.; Yang, S.-Y. Identification of Curcumin-Inhibited Extracellular Matrix Receptors in Non–Small Cell Lung Cancer A549 Cells by RNA Sequencing. Tumor Biol. 2017, 39, 1010428317705334. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhang, H.; Wang, Y.; Zhou, Y.; Luo, Y.; Cui, Y.; Jiang, N.; Jiang, W.; Wang, H.; Xu, D.; et al. CTHRC1 Induces Non-Small Cell Lung Cancer (NSCLC) Invasion through Upregulating MMP-7/MMP-9. BMC Cancer 2018, 18, 400. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Liu, B.; Cui, Y.; Wang, F.; Sun, H.; Lv, F. Collagen Triple Helix Repeat Containing 1 (Cthrc1) Is an Independently Prognostic Biomarker of Non-Small Cell Lung Cancers with Cigarette Smoke. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2014, 35, 11677–11683. [Google Scholar] [CrossRef]
- Ke, Z.; He, W.; Lai, Y.; Guo, X.; Chen, S.; Li, S.; Wang, Y.; Wang, L. Overexpression of Collagen Triple Helix Repeat Containing 1 (CTHRC1) Is Associated with Tumour Aggressiveness and Poor Prognosis in Human Non-Small Cell Lung Cancer. Oncotarget 2014, 5, 9410–9424. [Google Scholar] [CrossRef]
- Chen, S.; Li, P.; Yang, R.; Cheng, R.; Zhang, F.; Wang, Y.; Chen, X.; Sun, Q.; Zang, W.; Du, Y.; et al. MicroRNA-30b Inhibits Cell Invasion and Migration through Targeting Collagen Triple Helix Repeat Containing 1 in Non-Small Cell Lung Cancer. Cancer Cell Int. 2015, 15, 85. [Google Scholar] [CrossRef]
- Ge, N.; Chu, X.-M.; Xuan, Y.-P.; Ren, D.-Q.; Wang, Y.; Ma, K.; Gao, H.-J.; Jiao, W.-J. Associations between Abnormal Vitamin D Metabolism Pathway Function and Non-Small Cell Lung Cancer. Oncol. Lett. 2017, 14, 7538–7544. [Google Scholar] [CrossRef]
- Kim, S.A.; Miettinen, M. 35 Aberrant Expression of Desmin in Primary Lung Cancer Is Observed Exclusively in Carcinomas With Neuroendocrine Differentiation. Am. J. Clin. Pathol. 2018, 149, S15–S16. [Google Scholar] [CrossRef]
- Terada, T. Pleomorphic Carcinoma of the Lung: A Case Report with Immunohistochemical Studies. Respir. Med. CME 2010, 3, 252–256. [Google Scholar] [CrossRef][Green Version]
- Bahadur, S.; Pujani, M.; Jetley, S.; Khetrapal, S.; Raina, P.K. Large Cell Lung Carcinoma with Rhabdoid Phenotype: Report of a Rare Entity Presenting with Chest Wall Involvement. J. Cancer Res. Ther. 2015, 11, 657. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.-R.; Tai, Y.; Jin, Y.; Hammell, M.C.; Wilkinson, J.E.; Roe, J.-S.; Vakoc, C.R.; Van Aelst, L. TGF-β/Smad Signaling through DOCK4 Facilitates Lung Adenocarcinoma Metastasis. Genes Dev. 2015, 29, 250–261. [Google Scholar] [CrossRef] [PubMed]
- Little, A.C.; Sham, D.; Hristova, M.; Danyal, K.; Heppner, D.E.; Bauer, R.A.; Sipsey, L.M.; Habibovic, A.; van der Vliet, A. DUOX1 Silencing in Lung Cancer Promotes EMT, Cancer Stem Cell Characteristics and Invasive Properties. Oncogenesis 2016, 5, e261. [Google Scholar] [CrossRef]
- Little, A.C.; Hristova, M.; van Lith, L.; Schiffers, C.; Dustin, C.M.; Habibovic, A.; Danyal, K.; Heppner, D.E.; Lin, M.-C.J.; van der Velden, J.; et al. Dysregulated Redox Regulation Contributes to Nuclear EGFR Localization and Pathogenicity in Lung Cancer. Sci. Rep. 2019, 9, 4844. [Google Scholar] [CrossRef]
- Rigutto, S.; Hoste, C.; Grasberger, H.; Milenkovic, M.; Communi, D.; Dumont, J.E.; Corvilain, B.; Miot, F.; De Deken, X. Activation of Dual Oxidases Duox1 and Duox2: Differential Regulation Mediated by Camp-Dependent Protein Kinase and Protein Kinase C-Dependent Phosphorylation. J. Biol. Chem. 2009, 284, 6725–6734. [Google Scholar] [CrossRef]
- Luxen, S.; Belinsky, S.A.; Knaus, U.G. Silencing of DUOX NADPH Oxidases by Promoter Hypermethylation in Lung Cancer. Cancer Res. 2008, 68, 1037–1045. [Google Scholar] [CrossRef]
- Sun, C.-C.; Zhou, Q.; Hu, W.; Li, S.-J.; Zhang, F.; Chen, Z.-L.; Li, G.; Bi, Z.-Y.; Bi, Y.-Y.; Gong, F.-Y.; et al. Transcriptional E2F1/2/5/8 as Potential Targets and Transcriptional E2F3/6/7 as New Biomarkers for the Prognosis of Human Lung Carcinoma. Aging 2018, 10, 973–987. [Google Scholar] [CrossRef]
- Yu, L.; Fang, F.; Lu, S.; Li, X.; Yang, Y.; Wang, Z. LncRNA-HIT Promotes Cell Proliferation of Non-Small Cell Lung Cancer by Association with E2F1. Cancer Gene Ther. 2017, 24, 221–226. [Google Scholar] [CrossRef]
- Eymin, B.; Gazzeri, S.; Brambilla, C.; Brambilla, E. Distinct Pattern of E2F1 Expression in Human Lung Tumours: E2F1 Is Upregulated in Small Cell Lung Carcinoma. Oncogene 2001, 20, 1678–1687. [Google Scholar] [CrossRef]
- Yin, J.; Fu, W.; Dai, L.; Jiang, Z.; Liao, H.; Chen, W.; Pan, L.; Zhao, J. ANKRD22 Promotes Progression of Non-Small Cell Lung Cancer through Transcriptional up-Regulation of E2F1. Sci. Rep. 2017, 7, 4430. [Google Scholar] [CrossRef] [PubMed]
- Busslinger, M. Transcriptional Control of Early B Cell Development. Annu. Rev. Immunol. 2004, 22, 55–79. [Google Scholar] [CrossRef] [PubMed]
- Liao, D. Emerging Roles of the EBF Family of Transcription Factors in Tumor Suppression. Mol. Cancer Res. MCR 2009, 7, 1893–1901. [Google Scholar] [CrossRef] [PubMed]
- Gao, H.; Lukin, K.; Ramírez, J.; Fields, S.; Lopez, D.; Hagman, J. Opposing Effects of SWI/SNF and Mi-2/NuRD Chromatin Remodeling Complexes on Epigenetic Reprogramming by EBF and Pax5. Proc. Natl. Acad. Sci. USA 2009, 106, 11258–11263. [Google Scholar] [CrossRef]
- Decker, T.; Pasca di Magliano, M.; McManus, S.; Sun, Q.; Bonifer, C.; Tagoh, H.; Busslinger, M. Stepwise Activation of Enhancer and Promoter Regions of the B Cell Commitment Gene Pax5 in Early Lymphopoiesis. Immunity 2009, 30, 508–520. [Google Scholar] [CrossRef]
- Shen, A.; Chen, Y.; Liu, L.; Huang, Y.; Chen, H.; Qi, F.; Lin, J.; Shen, Z.; Wu, X.; Wu, M.; et al. EBF1-Mediated Upregulation of Ribosome Assembly Factor PNO1 Contributes to Cancer Progression by Negatively Regulating the P53 Signaling Pathway. Cancer Res. 2019, 79, 2257–2270. [Google Scholar] [CrossRef]
- Welsh, S.J.; Churchman, M.L.; Togni, M.; Mullighan, C.G.; Hagman, J. Deregulation of Kinase Signaling and Lymphoid Development in EBF1-PDGFRB ALL Leukemogenesis. Leukemia 2018, 32, 38–48. [Google Scholar] [CrossRef]
- Richtmann, S.; Wilkens, D.; Warth, A.; Lasitschka, F.; Winter, H.; Christopoulos, P.; Herth, F.J.F.; Muley, T.; Meister, M.; Schneider, M.A. FAM83A and FAM83B as Prognostic Biomarkers and Potential New Therapeutic Targets in NSCLC. Cancers 2019, 11, 652. [Google Scholar] [CrossRef]
- Shi, R.; Jiao, Z.; Yu, A.; Wang, T. Long Noncoding Antisense RNA FAM83A-AS1 Promotes Lung Cancer Cell Progression by Increasing FAM83A. J. Cell. Biochem. 2019, 120, 10505–10512. [Google Scholar] [CrossRef]
- Zhang, J.-T.; Lin, Y.-C.; Xiao, B.-F.; Yu, B.-T. Overexpression of Family with Sequence Similarity 83, Member A (FAM83A) Predicts Poor Clinical Outcomes in Lung Adenocarcinoma. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2019, 25, 4264–4272. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, T.; Wo, Y.; Sun, X.; Li, S.; Miao, S.; Dong, Y.; Leng, X.; Jiao, W. Identification of a Putative Competitive Endogenous RNA Network for Lung Adenocarcinoma Using TCGA Datasets. PeerJ 2019, 7, e6809. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Shen, Y.; Feng, G. Predicting the Survival of Patients with Lung Adenocarcinoma Using a Four-Gene Prognosis Risk Model. Oncol. Lett. 2019, 18, 535–544. [Google Scholar] [CrossRef]
- Marsit, C.J.; Liu, M.; Nelson, H.H.; Posner, M.; Suzuki, M.; Kelsey, K.T. Inactivation of the Fanconi Anemia/BRCA Pathway in Lung and Oral Cancers: Implications for Treatment and Survival. Oncogene 2004, 23, 1000–1004. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Fan, J.; Chen, Q.; Lei, C.; Qiao, B.; Liu, Q. SPP1 and AGER as Potential Prognostic Biomarkers for Lung Adenocarcinoma. Oncol. Lett. 2018, 15, 7028–7036. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.B.; Tan, S.J.; Lim, W.-T.; Lim, C.T. An Extracellular Matrix-Related Prognostic and Predictive Indicator for Early-Stage Non-Small Cell Lung Cancer. Nat. Commun. 2017, 8, 1734. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Jun, Y.; Kim, S.; Kim, E.; Jung, Y.; Park, B.J.; Lee, J.; Kim, J.; Lee, S.; Kim, J. FCN3 Functions as a Tumor Suppressor of Lung Adenocarcinoma through Induction of Endoplasmic Reticulum Stress. Cell Death Dis. 2021, 12, 407. [Google Scholar] [CrossRef]
- Sin, S.; Bonin, F.; Petit, V.; Meseure, D.; Lallemand, F.; Bièche, I.; Bellahcène, A.; Castronovo, V.; de Wever, O.; Gespach, C.; et al. Role of the Focal Adhesion Protein Kindlin-1 in Breast Cancer Growth and Lung Metastasis. J. Natl. Cancer Inst. 2011, 103, 1323–1337. [Google Scholar] [CrossRef]
- Zhan, J.; Zhu, X.; Guo, Y.; Wang, Y.; Wang, Y.; Qiang, G.; Niu, M.; Hu, J.; Du, J.; Li, Z.; et al. Opposite Role of Kindlin-1 and Kindlin-2 in Lung Cancers. PLoS ONE 2012, 7, e50313. [Google Scholar] [CrossRef]
- Gu, Z.; Gao, S.; Zhang, F.; Wang, Z.; Ma, W.; Davis, R.E.; Wang, Z. Protein Arginine Methyltransferase 5 Is Essential for Growth of Lung Cancer Cells. Biochem. J. 2012, 446, 235–241. [Google Scholar] [CrossRef]
- Woenckhaus, M.; Klein-Hitpass, L.; Grepmeier, U.; Merk, J.; Pfeifer, M.; Wild, P.; Bettstetter, M.; Wuensch, P.; Blaszyk, H.; Hartmann, A.; et al. Smoking and Cancer-Related Gene Expression in Bronchial Epithelium and Non-Small-Cell Lung Cancers. J. Pathol. 2006, 210, 192–204. [Google Scholar] [CrossRef]
- Sen, P.; Dharmadhikari, A.V.; Majewski, T.; Mohammad, M.A.; Kalin, T.V.; Zabielska, J.; Ren, X.; Bray, M.; Brown, H.M.; Welty, S.; et al. Comparative Analyses of Lung Transcriptomes in Patients with Alveolar Capillary Dysplasia with Misalignment of Pulmonary Veins and in Foxf1 Heterozygous Knockout Mice. PLoS ONE 2014, 9, e94390. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kong, P.-Z.; Li, G.-M.; Tian, Y.; Song, B.; Shi, R. Decreased Expression of FOXF2 as New Predictor of Poor Prognosis in Stage I Non-Small Cell Lung Cancer. Oncotarget 2016, 7, 55601–55610. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Kundu, S.T.; Byers, L.A.; Peng, D.H.; Roybal, J.D.; Diao, L.; Wang, J.; Tong, P.; Creighton, C.J.; Gibbons, D.L. The MiR-200 Family and the MiR-183~96~182 Cluster Target Foxf2 to Inhibit Invasion and Metastasis in Lung Cancers. Oncogene 2016, 35, 173–186. [Google Scholar] [CrossRef] [PubMed]
- Goldie, S.J.; Mulder, K.W.; Tan, D.W.-M.; Lyons, S.K.; Sims, A.H.; Watt, F.M. FRMD4A Upregulation in Human Squamous Cell Carcinoma Promotes Tumor Growth and Metastasis and Is Associated with Poor Prognosis. Cancer Res. 2012, 72, 3424–3436. [Google Scholar] [CrossRef]
- Velcheti, V.; Thawani, R.; Khunger, M.; Mukhopadhyay, S.; Chute, D.J.; Schrock, A.B.; Ali, S.M. FRMD4A/RET: A Novel RET Oncogenic Fusion Variant in Non–Small Cell Lung Carcinoma. J. Thorac. Oncol. 2017, 12, e15–e16. [Google Scholar] [CrossRef]
- Liao, C.-M.; Mukherjee, S.; Tiyaboonchai, A.; Maguire, J.A.; Cardenas-Diaz, F.L.; French, D.L.; Gadue, P. GATA6 Suppression Enhances Lung Specification from Human Pluripotent Stem Cells. J. Clin. Investig. 2018, 128, 2944–2950. [Google Scholar] [CrossRef]
- Mehta, A.; Cordero, J.; Dobersch, S.; Romero-Olmedo, A.J.; Savai, R.; Bodner, J.; Chao, C.; Fink, L.; Guzmán-Díaz, E.; Singh, I.; et al. Non-invasive Lung Cancer Diagnosis by Detection of GATA6 and NKX2-1 Isoforms in Exhaled Breath Condensate. EMBO Mol. Med. 2016, 8, 1380–1389. [Google Scholar] [CrossRef]
- Li, H.; Feng, C.; Shi, S. MiR-196b Promotes Lung Cancer Cell Migration and Invasion through the Targeting of GATA6. Oncol. Lett. 2018, 16, 247–252. [Google Scholar] [CrossRef]
- Ma, R.; Li, X.; Liu, H.; Jiang, R.; Yang, M.; Zhang, M.; Wang, Y.; Zhao, Y.; Li, H. GATA6-Upregulating Autophagy Promotes TKI Resistance in Nonsmall Cell Lung Cancer. Cancer Biol. Ther. 2019, 20, 1206–1212. [Google Scholar] [CrossRef]
- Mulvihill, M.S.; Kwon, Y.-W.; Lee, S.; Fang, L.T.; Choi, H.; Ray, R.; Kang, H.C.; Mao, J.-H.; Jablons, D.; Kim, I.-J. Gremlin Is Overexpressed in Lung Adenocarcinoma and Increases Cell Growth and Proliferation in Normal Lung Cells. PLoS ONE 2012, 7, e42264. [Google Scholar] [CrossRef]
- Cahill, E.; Costello, C.M.; Rowan, S.C.; Harkin, S.; Howell, K.; Leonard, M.O.; Southwood, M.; Cummins, E.P.; Fitzpatrick, S.F.; Taylor, C.T.; et al. Gremlin Plays a Key Role in the Pathogenesis of Pulmonary Hypertension. Circulation 2012, 125, 920–930. [Google Scholar] [CrossRef] [PubMed]
- Gentles, A.J.; Hui, A.B.-Y.; Feng, W.; Azizi, A.; Nair, R.V.; Knowles, D.A.; Yu, A.; Jeong, Y.; Bejnood, A.; Forgó, E.; et al. Clinically-Relevant Cell Type Cross-Talk Identified from a Human Lung Tumor Microenvironment Interactome. bioRxiv 2019, 637306. [Google Scholar] [CrossRef]
- Landry-Truchon, K.; Houde, N.; Boucherat, O.; Joncas, F.-H.; Dasen, J.S.; Philippidou, P.; Mansfield, J.H.; Jeannotte, L. HOXA5 Plays Tissue-Specific Roles in the Developing Respiratory System. Dev. Camb. Engl. 2017, 144, 3547–3561. [Google Scholar] [CrossRef]
- Zhang, M.; Nie, F.; Sun, M.; Xia, R.; Xie, M.; Lu, K.; Li, W. HOXA5 Indicates Poor Prognosis and Suppresses Cell Proliferation by Regulating P21 Expression in Non Small Cell Lung Cancer. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2015, 36, 3521–3531. [Google Scholar] [CrossRef] [PubMed]
- Chang, C.-J.; Chen, Y.-L.; Hsieh, C.-H.; Liu, Y.-J.; Yu, S.-L.; Chen, J.J.W.; Wang, C.-C. HOXA5 and P53 Cooperate to Suppress Lung Cancer Cell Invasion and Serve as Good Prognostic Factors in Non-Small Cell Lung Cancer. J. Cancer 2017, 8, 1071–1081. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-C.; Su, K.-Y.; Chen, H.-Y.; Chang, S.-Y.; Shen, C.-F.; Hsieh, C.-H.; Hong, Q.-S.; Chiang, C.-C.; Chang, G.-C.; Yu, S.-L.; et al. HOXA5 Inhibits Metastasis via Regulating Cytoskeletal Remodelling and Associates with Prolonged Survival in Non-Small-Cell Lung Carcinoma. PLoS ONE 2015, 10, e0124191. [Google Scholar] [CrossRef]
- Zhu, Q.; Lv, T.; Wu, Y.; Shi, X.; Liu, H.; Song, Y. Long Non-coding RNA 00312 Regulated by HOXA5 Inhibits Tumour Proliferation and Promotes Apoptosis in Non-small Cell Lung Cancer. J. Cell. Mol. Med. 2017, 21, 2184–2198. [Google Scholar] [CrossRef] [PubMed]
- Dong, Y.; Zhang, D.; Cai, M.; Luo, Z.; Zhu, Y.; Gong, L.; Lei, Y.; Tan, X.; Zhu, Q.; Han, S. SPOP Regulates the DNA Damage Response and Lung Adenocarcinoma Cell Response to Radiation. Am. J. Cancer Res. 2019, 9, 1469–1483. [Google Scholar]
- Leithner, K.; Hirschmugl, B.; Li, Y.; Tang, B.; Papp, R.; Nagaraj, C.; Stacher, E.; Stiegler, P.; Lindenmann, J.; Olschewski, A.; et al. TASK-1 Regulates Apoptosis and Proliferation in a Subset of Non-Small Cell Lung Cancers. PLoS ONE 2016, 11, e0157453. [Google Scholar] [CrossRef]
- van der Wekken, A.J.; Kuiper, J.L.; Saber, A.; Terpstra, M.M.; Wei, J.; Hiltermann, T.J.N.; Thunnissen, E.; Heideman, D.a.M.; Timens, W.; Schuuring, E.; et al. Overall Survival in EGFR Mutated Non-Small-Cell Lung Cancer Patients Treated with Afatinib after EGFR TKI and Resistant Mechanisms upon Disease Progression. PLoS ONE 2017, 12, e0182885. [Google Scholar] [CrossRef]
- Salvador, F.; Martin, A.; López-Menéndez, C.; Moreno-Bueno, G.; Santos, V.; Vázquez-Naharro, A.; Santamaria, P.G.; Morales, S.; Dubus, P.R.; Muinelo-Romay, L.; et al. Lysyl Oxidase-like Protein LOXL2 Promotes Lung Metastasis of Breast Cancer. Cancer Res. 2017, 77, 5846–5859. [Google Scholar] [CrossRef]
- Kamikawaji, K.; Seki, N.; Watanabe, M.; Mataki, H.; Kumamoto, T.; Takagi, K.; Mizuno, K.; Inoue, H. Regulation of LOXL2 and SERPINH1 by Antitumor MicroRNA-29a in Lung Cancer with Idiopathic Pulmonary Fibrosis. J. Hum. Genet. 2016, 61, 985–993. [Google Scholar] [CrossRef] [PubMed]
- Leeming, D.J.; Willumsen, N.; Sand, J.M.B.; Holm Nielsen, S.; Dasgupta, B.; Brodmerkel, C.; Curran, M.; Bager, C.L.; Karsdal, M.A. A Serological Marker of the N-Terminal Neoepitope Generated during LOXL2 Maturation Is Elevated in Patients with Cancer or Idiopathic Pulmonary Fibrosis. Biochem. Biophys. Rep. 2018, 17, 38–43. [Google Scholar] [CrossRef] [PubMed]
- Canesin, G.; Cuevas, E.P.; Santos, V.; López-Menéndez, C.; Moreno-Bueno, G.; Huang, Y.; Csiszar, K.; Portillo, F.; Peinado, H.; Lyden, D.; et al. Lysyl Oxidase-like 2 (LOXL2) and E47 EMT Factor: Novel Partners in E-Cadherin Repression and Early Metastasis Colonization. Oncogene 2015, 34, 951–964. [Google Scholar] [CrossRef]
- Jin, H.; Sun, L.; Tang, L.; Yu, W.; Li, H. Expression of GARP Is Increased in Tumor-Infiltrating Regulatory T Cells and Is Correlated to Clinicopathology of Lung Cancer Patients. Front. Immunol. 2017, 8, 138. [Google Scholar] [CrossRef]
- Zhu, H.; Shi, L.; Zhang, Y.; Zhu, Y.; Bai, C.; Wang, X.; Zhou, J. Myocyte Enhancer Factor 2D Provides a Cross-Talk between Chronic Inflammation and Lung Cancer. J. Transl. Med. 2017, 15, 65. [Google Scholar] [CrossRef]
- Zhao, X.; Liu, M.; Li, D. Oleanolic Acid Suppresses the Proliferation of Lung Carcinoma Cells by MiR-122/Cyclin G1/MEF2D Axis. Mol. Cell. Biochem. 2015, 400, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Li, D.; Zhao, Y.; Gu, Y.; Zhao, D.; Li, X.; Bai, X.; Sun, Y.; Zhang, X.; Sun, H.; et al. MiR-218 Suppressed the Growth of Lung Carcinoma by Reducing MEF2D Expression. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2016, 37, 2891–2900. [Google Scholar] [CrossRef] [PubMed]
- Tsubouchi, H.; Roeder, G.S. The Mnd1 Protein Forms a Complex with Hop2 to Promote Homologous Chromosome Pairing and Meiotic Double-Strand Break Repair. Mol. Cell. Biol. 2002, 22, 3078–3088. [Google Scholar] [CrossRef]
- Sparaneo, A.; Fabrizio, F.P.; la Torre, A.; Graziano, P.; Di Maio, M.; Fontana, A.; Bisceglia, M.; Rossi, A.; Pizzolitto, S.; De Maglio, G.; et al. Effects of KEAP1 Silencing on the Regulation of NRF2 Activity in Neuroendocrine Lung Tumors. Int. J. Mol. Sci. 2019, 20, 2531. [Google Scholar] [CrossRef]
- Frank, R.; Scheffler, M.; Merkelbach-Bruse, S.; Ihle, M.A.; Kron, A.; Rauer, M.; Ueckeroth, F.; König, K.; Michels, S.; Fischer, R.; et al. Clinical and Pathological Characteristics of KEAP1- and NFE2L2-Mutated Non-Small Cell Lung Carcinoma (NSCLC). Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2018, 24, 3087–3096. [Google Scholar] [CrossRef]
- Paik, P.K.; Qeriqi, B.; Ahn, L.S.H.; Ginsberg, M.S.; Tandon, N.; McFarland, D.; Doyle, L.A.; de Stanchina, E.; Rudin, C.M. Targeting NFE2L2 Mutations in Advanced Squamous Cell Lung Cancers with the TORC1/2 Inhibitor TAK228. J. Clin. Oncol. 2018, 36, 9098. [Google Scholar] [CrossRef]
- Qian, Z.; Zhou, T.; Gurguis, C.I.; Xu, X.; Wen, Q.; Lv, J.; Fang, F.; Hecker, L.; Cress, A.E.; Natarajan, V.; et al. Nuclear Factor, Erythroid 2-like 2-Associated Molecular Signature Predicts Lung Cancer Survival. Sci. Rep. 2015, 5, 16889. [Google Scholar] [CrossRef] [PubMed]
- Tong, Y.-H.; Zhang, B.; Fan, Y.; Lin, N.-M. Keap1–Nrf2 Pathway: A Promising Target towards Lung Cancer Prevention and Therapeutics. Chronic Dis. Transl. Med. 2015, 1, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Rennhack, J.; Andrechek, E.R.; Rockwell, C.E.; Liby, K.T. Identification of an Unfavorable Immune Signature in Advanced Lung Tumors from Nrf2-Deficient Mice. Antioxid. Redox Signal. 2018, 29, 1535–1552. [Google Scholar] [CrossRef] [PubMed]
- Hellyer, J.A.; Stehr, H.; Das, M.; Padda, S.K.; Ramchandran, K.; Neal, J.W.; Diehn, M.; Wakelee, H.A. Impact of KEAP1/NFE2L2/CUL3 Mutations on Duration of Response to EGFR Tyrosine Kinase Inhibitors in EGFR Mutated Non-Small Cell Lung Cancer. Lung Cancer Amst. Neth. 2019, 134, 42–45. [Google Scholar] [CrossRef]
- Kamizono, S.; Duncan, G.S.; Seidel, M.G.; Morimoto, A.; Hamada, K.; Grosveld, G.; Akashi, K.; Lind, E.F.; Haight, J.P.; Ohashi, P.S.; et al. Nfil3/E4bp4 Is Required for the Development and Maturation of NK Cells in Vivo. J. Exp. Med. 2009, 206, 2977–2986. [Google Scholar] [CrossRef]
- Zimmermann, K.; Opitz, N.; Dedio, J.; Renne, C.; Muller-Esterl, W.; Oess, S. NOSTRIN: A Protein Modulating Nitric Oxide Release and Subcellular Distribution of Endothelial Nitric Oxide Synthase. Proc. Natl. Acad. Sci. USA 2002, 99, 17167–17172. [Google Scholar] [CrossRef] [PubMed]
- Chakraborty, S.; Ain, R. Nitric-Oxide Synthase Trafficking Inducer Is a Pleiotropic Regulator of Endothelial Cell Function and Signaling. J. Biol. Chem. 2017, 292, 6600–6620. [Google Scholar] [CrossRef] [PubMed]
- Ciribilli, Y.; Singh, P.; Inga, A.; Borlak, J. C-Myc Targeted Regulators of Cell Metabolism in a Transgenic Mouse Model of Papillary Lung Adenocarcinoma. Oncotarget 2016, 7, 65514–65539. [Google Scholar] [CrossRef][Green Version]
- Safe, S.; Jin, U.-H.; Hedrick, E.; Reeder, A.; Lee, S.-O. Minireview: Role Of Orphan Nuclear Receptors in Cancer and Potential as Drug Targets. Mol. Endocrinol. 2014, 28, 157–172. [Google Scholar] [CrossRef]
- Safe, S.; Jin, U.-H.; Morpurgo, B.; Abudayyeh, A.; Singh, M.; Tjalkens, R.B. Nuclear Receptor 4A (NR4A) Family - Orphans No More. J. Steroid Biochem. Mol. Biol. 2016, 157, 48–60. [Google Scholar] [CrossRef] [PubMed]
- Beard, J.A.; Tenga, A.; Hills, J.; Hoyer, J.D.; Cherian, M.T.; Wang, Y.-D.; Chen, T. The Orphan Nuclear Receptor NR4A2 Is Part of a P53-MicroRNA-34 Network. Sci. Rep. 2016, 6, 25108. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Mao, J.-H.; Zhu, W.; Jain, A.K.; Liu, K.; Brown, J.B.; Karpen, G.H. Centromere and Kinetochore Gene Misexpression Predicts Cancer Patient Survival and Response to Radiotherapy and Chemotherapy. Nat. Commun. 2016, 7, 12619. [Google Scholar] [CrossRef] [PubMed]
- Rao, C.V.; Farooqui, M.; Zhang, Y.; Kumar, G.; Panneerselvam, J.; Yarla, N.; Asch, A.S.; Yamada, H.Y. Abstract 1199: Identification of Candidate Regulators of Genomic Instability in Human Lung Adenocarcinoma through a New Cross-Species in Silico Analysis. Cancer Res. 2019, 79, 1199. [Google Scholar] [CrossRef]
- Christmann, R.B.; Wooten, A.; Sampaio-Barros, P.; Borges, C.L.; Carvalho, C.R.R.; Kairalla, R.A.; Feghali-Bostwick, C.; Ziemek, J.; Mei, Y.; Goummih, S.; et al. MiR-155 in the Progression of Lung Fibrosis in Systemic Sclerosis. Arthritis Res. Ther. 2016, 18, 155. [Google Scholar] [CrossRef]
- Raja, R.; Sahasrabuddhe, N.A.; Radhakrishnan, A.; Syed, N.; Solanki, H.S.; Puttamallesh, V.N.; Balaji, S.A.; Nanjappa, V.; Datta, K.K.; Babu, N.; et al. Chronic Exposure to Cigarette Smoke Leads to Activation of P21 (RAC1)-Activated Kinase 6 (PAK6) in Non-Small Cell Lung Cancer Cells. Oncotarget 2016, 7, 61229–61245. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, H.; Wang, C. Persistence of Smoking Induced Non-small Cell Lung Carcinogenesis by Decreasing ERBB Pathway-related MicroRNA Expression. Thorac. Cancer 2019, 10, 890–897. [Google Scholar] [CrossRef]
- Li, W.; Huang, K.; Guo, H.; Cui, G.; Zhao, S. Inhibition of Non-Small-Cell Lung Cancer Cell Proliferation by Pbx1. Chin. J. Cancer Res. 2014, 26, 573–578. [Google Scholar] [CrossRef]
- Mo, M.-L.; Chen, Z.; Zhou, H.-M.; Li, H.; Hirata, T.; Jablons, D.M.; He, B. Detection of E2A-PBX1 Fusion Transcripts in Human Non-Small-Cell Lung Cancer. J. Exp. Clin. Cancer Res. CR 2013, 32, 29. [Google Scholar] [CrossRef]
- Wu, X.; Ruan, L.; Yang, Y.; Mei, Q. Analysis of Gene Expression Changes Associated with Human Carcinoma-Associated Fibroblasts in Non-Small Cell Lung Carcinoma. Biol. Res. 2017, 50, 6. [Google Scholar] [CrossRef]
- Bender, A.T.; Beavo, J.A. Cyclic Nucleotide Phosphodiesterases: Molecular Regulation to Clinical Use. Pharmacol. Rev. 2006, 58, 488–520. [Google Scholar] [CrossRef] [PubMed]
- Klutzny, S.; Anurin, A.; Nicke, B.; Regan, J.L.; Lange, M.; Schulze, L.; Parczyk, K.; Steigemann, P. PDE5 Inhibition Eliminates Cancer Stem Cells via Induction of PKA Signaling. Cell Death Dis. 2018, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Wharton, J.; Strange, J.W.; Møller, G.M.O.; Growcott, E.J.; Ren, X.; Franklyn, A.P.; Phillips, S.C.; Wilkins, M.R. Antiproliferative Effects of Phosphodiesterase Type 5 Inhibition in Human Pulmonary Artery Cells. Am. J. Respir. Crit. Care Med. 2005, 172, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Ding, L.; Wang, H.; Lang, W.; Xiao, L. Protein Kinase C-Epsilon Promotes Survival of Lung Cancer Cells by Suppressing Apoptosis through Dysregulation of the Mitochondrial Caspase Pathway. J. Biol. Chem. 2002, 277, 35305–35313. [Google Scholar] [CrossRef]
- Caino, M.C.; Lopez-Haber, C.; Kissil, J.L.; Kazanietz, M.G. Non-Small Cell Lung Carcinoma Cell Motility, Rac Activation and Metastatic Dissemination Are Mediated by Protein Kinase C Epsilon. PLoS ONE 2012, 7, e31714. [Google Scholar] [CrossRef][Green Version]
- Garg, R.; Cooke, M.; Benavides, F.; Abba, M.C.; Cicchini, M.; Feldser, D.M.; Kazanietz, M.G. PKCε Is Required for KRAS-Driven Lung Tumorigenesis. Cancer Res. 2020, 80, 5166–5173. [Google Scholar] [CrossRef]
- Baxter, G.; Oto, E.; Daniel-Issakani, S.; Strulovici, B. Constitutive Presence of a Catalytic Fragment of Protein Kinase C Epsilon in a Small Cell Lung Carcinoma Cell Line. J. Biol. Chem. 1992, 267, 1910–1917. [Google Scholar] [CrossRef] [PubMed]
- Bae, K.-M.; Wang, H.; Jiang, G.; Chen, M.G.; Lu, L.; Xiao, L. Protein Kinase C Epsilon Is Overexpressed in Primary Human Non-Small Cell Lung Cancers and Functionally Required for Proliferation of Non-Small Cell Lung Cancer Cells in a P21/Cip1-Dependent Manner. Cancer Res. 2007, 67, 6053–6063. [Google Scholar] [CrossRef]
- Zhang, N.; Su, Y.; Xu, L. Targeting PKCε by MiR-143 Regulates Cell Apoptosis in Lung Cancer. FEBS Lett. 2013, 587, 3661–3667. [Google Scholar] [CrossRef]
- Tuomi, S.; Mai, A.; Nevo, J.; Laine, J.O.; Vilkki, V.; Öhman, T.J.; Gahmberg, C.G.; Parker, P.J.; Ivaska, J. PKCɛ Regulation of an A5 Integrin–ZO-1 Complex Controls Lamellae Formation in Migrating Cancer Cells. Sci. Signal. 2009, 2, ra32. [Google Scholar] [CrossRef]
- Lemjabbar-Alaoui, H.; Sidhu, S.S.; Mengistab, A.; Gallup, M.; Basbaum, C. TACE/ADAM-17 Phosphorylation by PKC-Epsilon Mediates Premalignant Changes in Tobacco Smoke-Exposed Lung Cells. PLoS ONE 2011, 6, e17489. [Google Scholar] [CrossRef]
- Kim, C.; Yang, H.; Fukushima, Y.; Saw, P.E.; Lee, J.; Park, J.-S.; Park, I.; Jung, J.; Kataoka, H.; Lee, D.; et al. Vascular RhoJ Is an Effective and Selective Target for Tumor Angiogenesis and Vascular Disruption. Cancer Cell 2014, 25, 102–117. [Google Scholar] [CrossRef]
- Mullapudi, N.; Ye, B.; Suzuki, M.; Fazzari, M.; Han, W.; Shi, M.K.; Marquardt, G.; Lin, J.; Wang, T.; Keller, S.; et al. Genome Wide Methylome Alterations in Lung Cancer. PLoS ONE 2015, 10, e0143826. [Google Scholar] [CrossRef] [PubMed]
- Gan, T.; Chen, W.; Qin, H.; Huang, S.; Yang, L.; Fang, Y.; Pan, L.; Li, Z.; Chen, G. Clinical Value and Prospective Pathway Signaling of MicroRNA-375 in Lung Adenocarcinoma: A Study Based on the Cancer Genome Atlas (TCGA), Gene Expression Omnibus (GEO) and Bioinformatics Analysis. Med. Sci. Monit. Int. Med. J. Exp. Clin. Res. 2017, 23, 2453–2464. [Google Scholar] [CrossRef] [PubMed]
- Bell, S.M.; Schreiner, C.M.; Wert, S.E.; Mucenski, M.L.; Scott, W.J.; Whitsett, J.A. R-Spondin 2 Is Required for Normal Laryngeal-Tracheal, Lung and Limb Morphogenesis. Dev. Camb. Engl. 2008, 135, 1049–1058. [Google Scholar] [CrossRef]
- Ramsey, J.; Butnor, K.; Peng, Z.; Leclair, T.; van der Velden, J.; Stein, G.; Lian, J.; Kinsey, C.M. Loss of RUNX1 Is Associated with Aggressive Lung Adenocarcinomas. J. Cell. Physiol. 2018, 233, 3487–3497. [Google Scholar] [CrossRef]
- Matsuo, A. Abstract 3516: RUNX1 Controls EGFR Signaling Pathway in Non-Small Cell Lung Cancer Cells. Cancer Res. 2017, 77, 3516. [Google Scholar] [CrossRef]
- Ishii, T.; Wakabayashi, R.; Kurosaki, H.; Gemma, A.; Kida, K. Association of Serotonin Transporter Gene Variation with Smoking, Chronic Obstructive Pulmonary Disease, and Its Depressive Symptoms. J. Hum. Genet. 2011, 56, 41–46. [Google Scholar] [CrossRef]
- Zhu, Q.; Liang, X.; Dai, J.; Guan, X. Prostaglandin Transporter, SLCO2A1, Mediates the Invasion and Apoptosis of Lung Cancer Cells via PI3K/AKT/MTOR Pathway. Int. J. Clin. Exp. Pathol. 2015, 8, 9175–9181. [Google Scholar]
- Lenka, G.; Tsai, M.-H.; Lin, H.-C.; Hsiao, J.-H.; Lee, Y.-C.; Lu, T.-P.; Lee, J.-M.; Hsu, C.-P.; Lai, L.-C.; Chuang, E.Y. Identification of Methylation-Driven, Differentially Expressed STXBP6 as a Novel Biomarker in Lung Adenocarcinoma. Sci. Rep. 2017, 7, 42573. [Google Scholar] [CrossRef]
- Wang, J.; Duan, Y.; Meng, Q.-H.; Gong, R.; Guo, C.; Zhao, Y.; Zhang, Y. Integrated Analysis of DNA Methylation Profiling and Gene Expression Profiling Identifies Novel Markers in Lung Cancer in Xuanwei, China. PLoS ONE 2018, 13, e0203155. [Google Scholar] [CrossRef] [PubMed]
- de Aberasturi, A.L.; Redrado, M.; Villalba, M.; Larzabal, L.; Pajares, M.J.; Garcia, J.; Evans, S.R.; Garcia-Ros, D.; Bodegas, M.E.; Lopez, L.; et al. TMPRSS4 Induces Cancer Stem Cell-like Properties in Lung Cancer Cells and Correlates with ALDH Expression in NSCLC Patients. Cancer Lett. 2016, 370, 165–176. [Google Scholar] [CrossRef]
- Chikaishi, Y.; Uramoto, H.; Koyanagi, Y.; Yamada, S.; Yano, S.; Tanaka, F. TMPRSS4 Expression as a Marker of Recurrence in Patients with Lung Cancer. Anticancer Res. 2016, 36, 121–127. [Google Scholar] [PubMed]
- Fan, X.; Liang, Y.; Liu, Y.; Bai, Y.; Yang, C.; Xu, S. The Upregulation of TMPRSS4, Partly Ascribed to the Downregulation of MiR-125a-5p, Promotes the Growth of Human Lung Adenocarcinoma via the NF-ΚB Signaling Pathway. Int. J. Oncol. 2018, 53, 148–158. [Google Scholar] [CrossRef] [PubMed]
- Larzabal, L.; Nguewa, P.A.; Pio, R.; Blanco, D.; Sanchez, B.; Rodríguez, M.J.; Pajares, M.J.; Catena, R.; Montuenga, L.M.; Calvo, A. Overexpression of TMPRSS4 in Non-Small Cell Lung Cancer Is Associated with Poor Prognosis in Patients with Squamous Histology. Br. J. Cancer 2011, 105, 1608–1614. [Google Scholar] [CrossRef] [PubMed]
- Hamamoto, J.; Soejima, K.; Naoki, K.; Yasuda, H.; Hayashi, Y.; Yoda, S.; Nakayama, S.; Satomi, R.; Terai, H.; Ikemura, S.; et al. Methylation-Induced Downregulation of TFPI-2 Causes TMPRSS4 Overexpression and Contributes to Oncogenesis in a Subset of Non-Small-Cell Lung Carcinoma. Cancer Sci. 2015, 106, 34–42. [Google Scholar] [CrossRef]
- Valero-Jiménez, A.; Zúñiga, J.; Cisneros, J.; Becerril, C.; Salgado, A.; Checa, M.; Buendía-Roldán, I.; Mendoza-Milla, C.; Gaxiola, M.; Pardo, A.; et al. Transmembrane Protease, Serine 4 (TMPRSS4) Is Upregulated in IPF Lungs and Increases the Fibrotic Response in Bleomycin-Induced Lung Injury. PLoS ONE 2018, 13, e0192963. [Google Scholar] [CrossRef]
- Expósito, F.; Villalba, M.; Pajares, M.J.; Redrado, M.; Sainz, C.; Wistuba, I.; Behrens, C.; Redin, E.; Andrea, C.; Cirauquiz, C.; et al. P1.03-24 TMPRSS4: A Novel Prognostic Biomarker and Therapeutic Target in NSCLC. J. Thorac. Oncol. 2018, 13, S521. [Google Scholar] [CrossRef]
- Link, T.M.; Park, U.; Vonakis, B.M.; Raben, D.M.; Soloski, M.J.; Caterina, M.J. TRPV2 Has a Pivotal Role in Macrophage Particle Binding and Phagocytosis. Nat. Immunol. 2010, 11, 232–239. [Google Scholar] [CrossRef]
- Hao, J.; Xu, A.; Xie, X.; Hao, J.; Tian, T.; Gao, S.; Xiao, X.; He, D. Elevated Expression of UBE2T in Lung Cancer Tumors and Cell Lines. Tumour Biol. J. Int. Soc. Oncodevelopmental Biol. Med. 2008, 29, 195–203. [Google Scholar] [CrossRef]
- Perez-Peña, J.; Corrales-Sánchez, V.; Amir, E.; Pandiella, A.; Ocana, A. Ubiquitin-Conjugating Enzyme E2T (UBE2T) and Denticleless Protein Homolog (DTL) Are Linked to Poor Outcome in Breast and Lung Cancers. Sci. Rep. 2017, 7, 17530. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Liu, X. UBE2T Silencing Inhibited Non-Small Cell Lung Cancer Cell Proliferation and Invasion by Suppressing the Wnt/β-Catenin Signaling Pathway. Int. J. Clin. Exp. Pathol. 2017, 10, 9482. [Google Scholar] [PubMed]
- Madar, S.; Brosh, R.; Buganim, Y.; Ezra, O.; Goldstein, I.; Solomon, H.; Kogan, I.; Goldfinger, N.; Klocker, H.; Rotter, V. Modulated Expression of WFDC1 during Carcinogenesis and Cellular Senescence. Carcinogenesis 2009, 30, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Larsen, J.E.; Nathan, V.; Osborne, J.K.; Farrow, R.K.; Deb, D.; Sullivan, J.P.; Dospoy, P.D.; Augustyn, A.; Hight, S.K.; Sato, M.; et al. ZEB1 Drives Epithelial-to-Mesenchymal Transition in Lung Cancer. J. Clin. Investig. 2016, 126, 3219–3235. [Google Scholar] [CrossRef]
- Zhang, T.; Guo, L.; Creighton, C.J.; Lu, Q.; Gibbons, D.L.; Yi, E.S.; Deng, B.; Molina, J.R.; Sun, Z.; Yang, P.; et al. A Genetic Cell Context-Dependent Role for ZEB1 in Lung Cancer. Nat. Commun. 2016, 7, 12231. [Google Scholar] [CrossRef] [PubMed]
- Yochum, Z.A.; Socinski, M.A.; Burns, T.F. Paradoxical Functions of ZEB1 in EGFR-Mutant Lung Cancer: Tumor Suppressor and Driver of Therapeutic Resistance. J. Thorac. Dis. 2016, 8, E1528–E1531. [Google Scholar] [CrossRef] [PubMed]
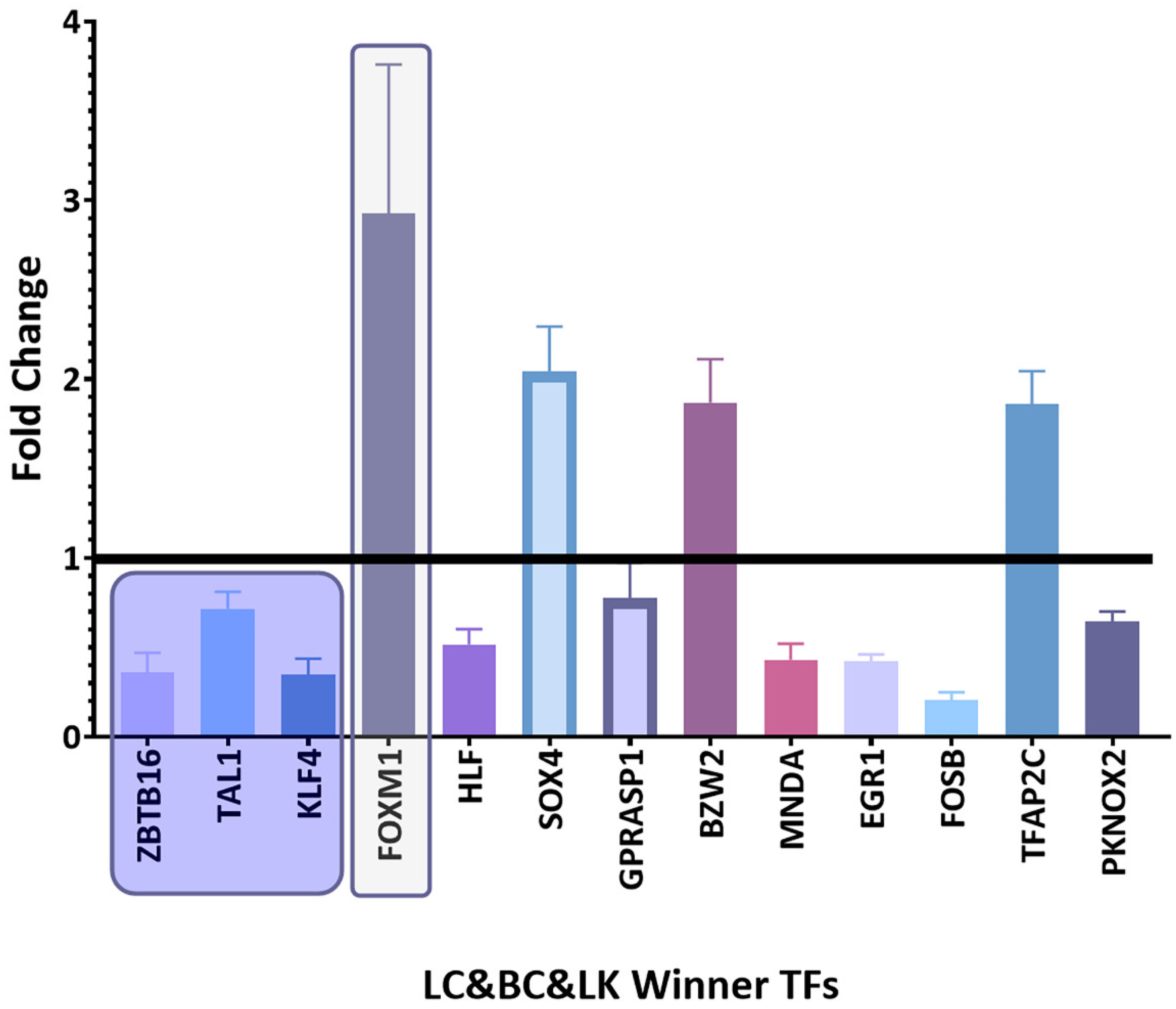
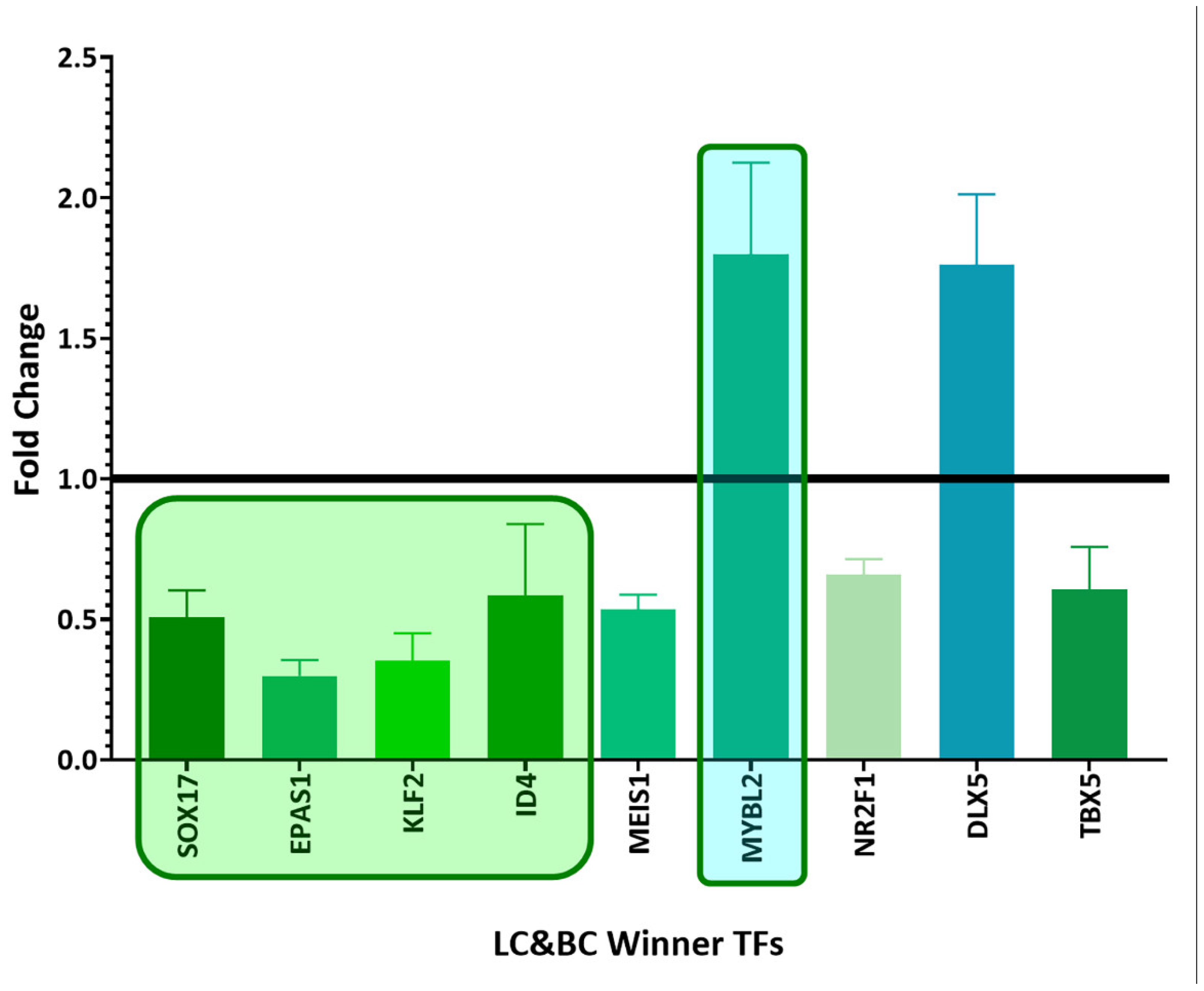

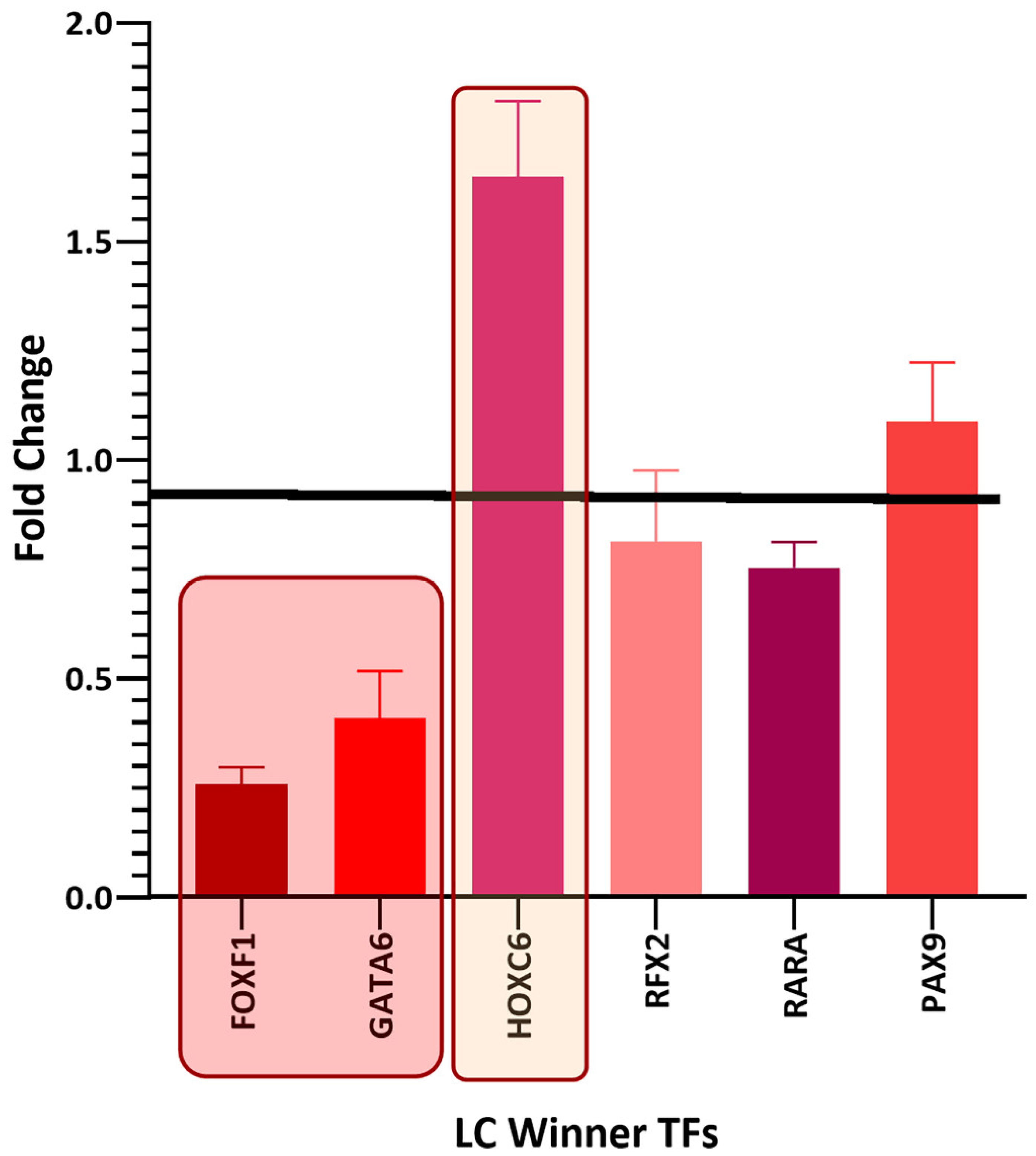
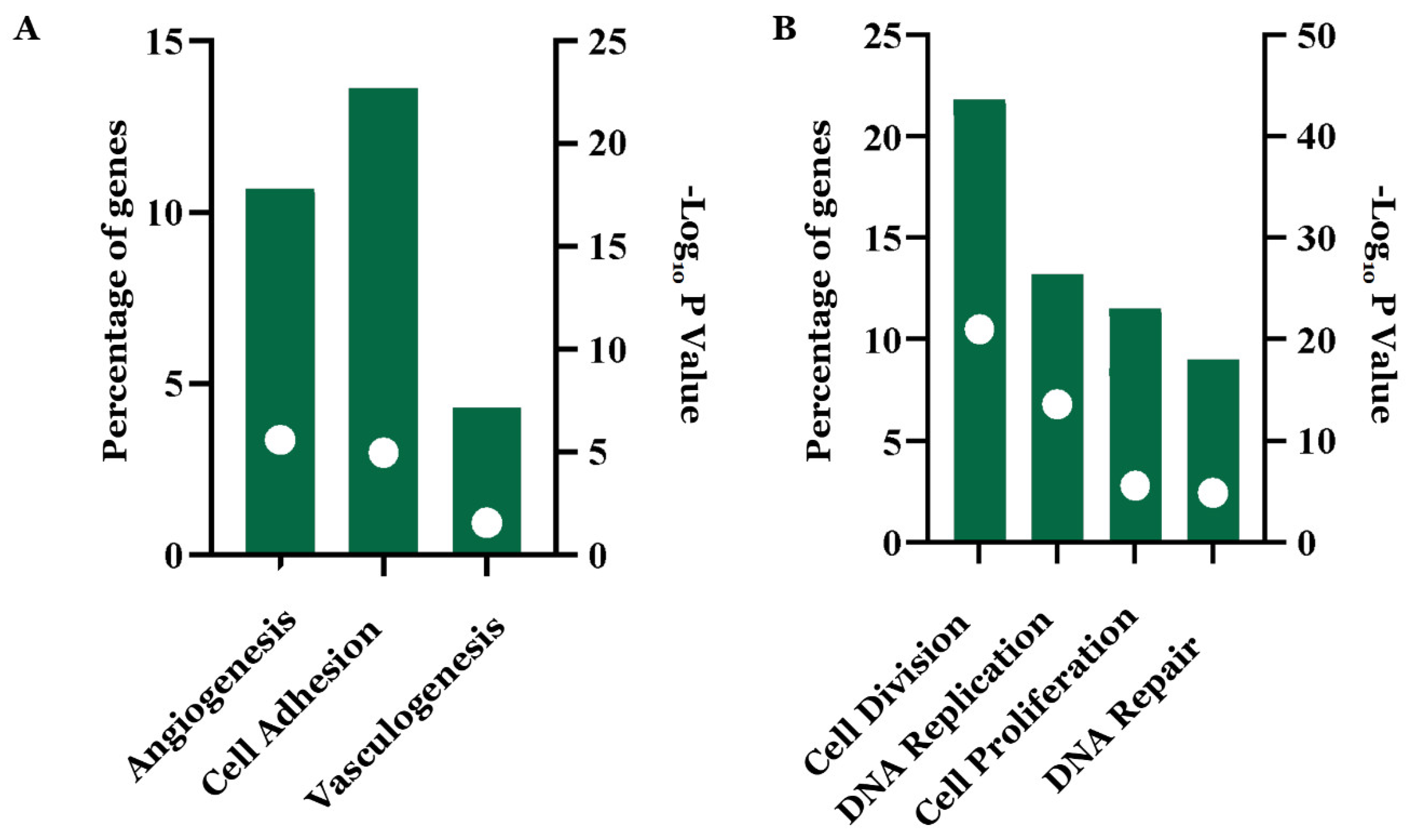
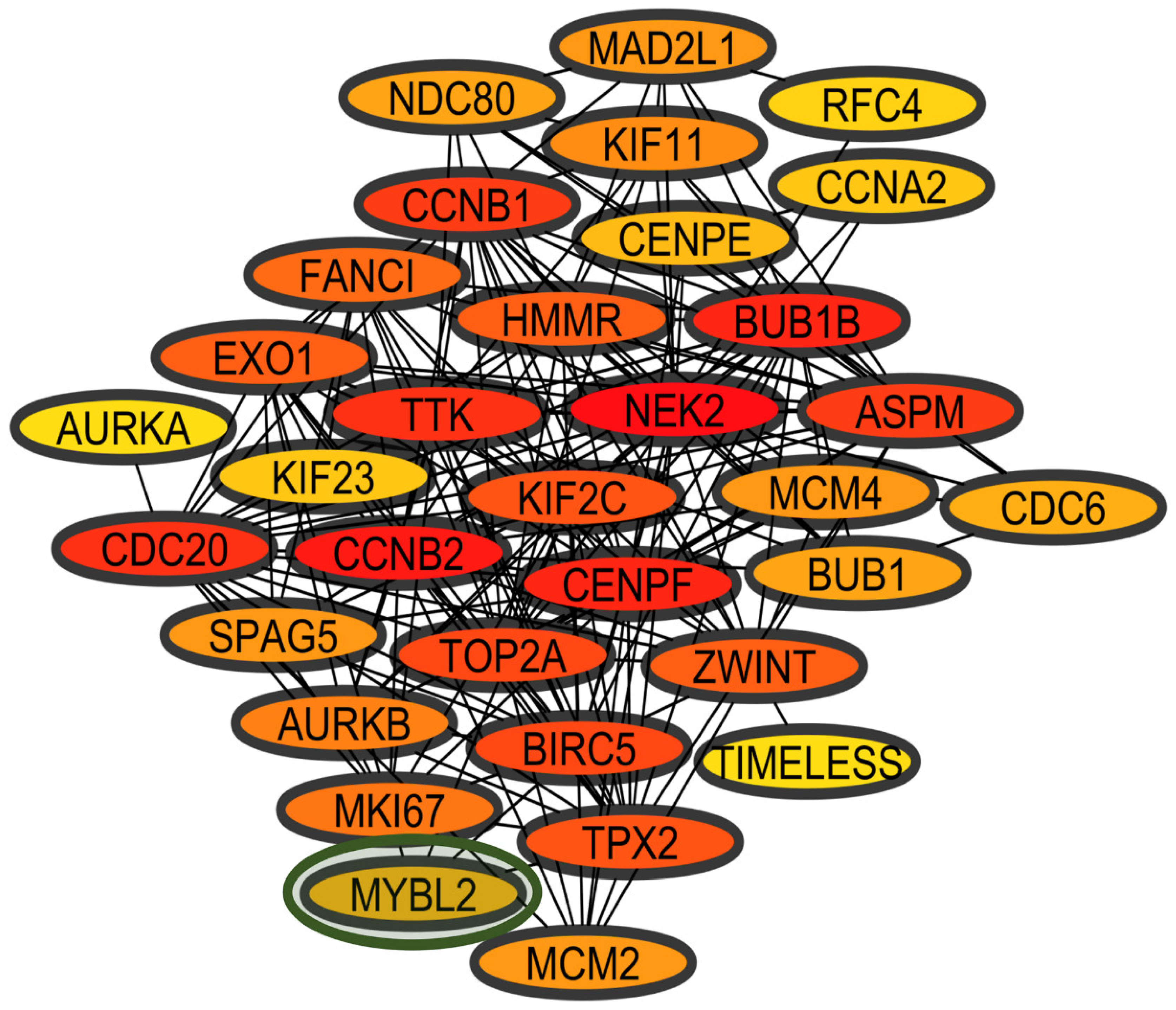
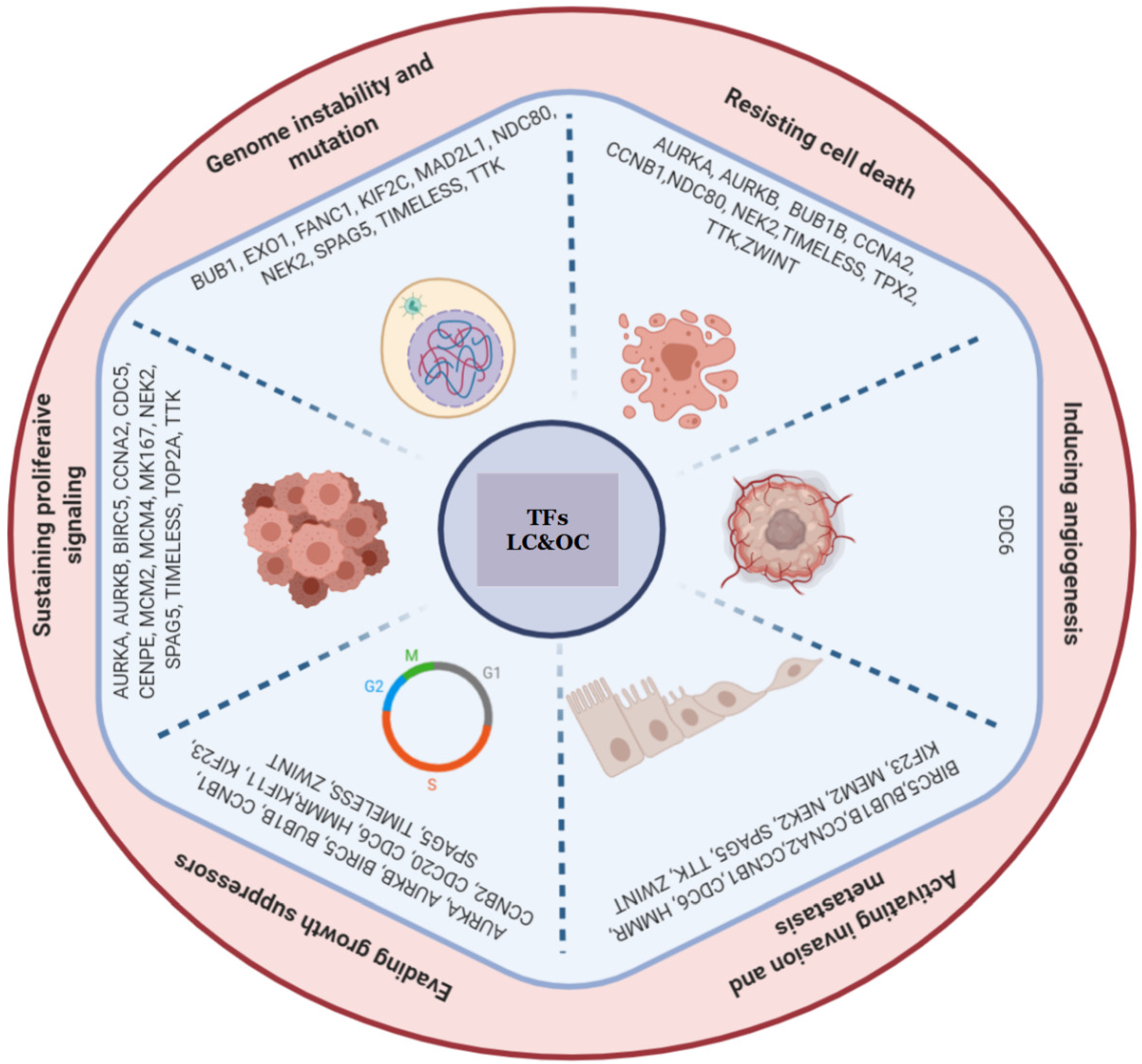

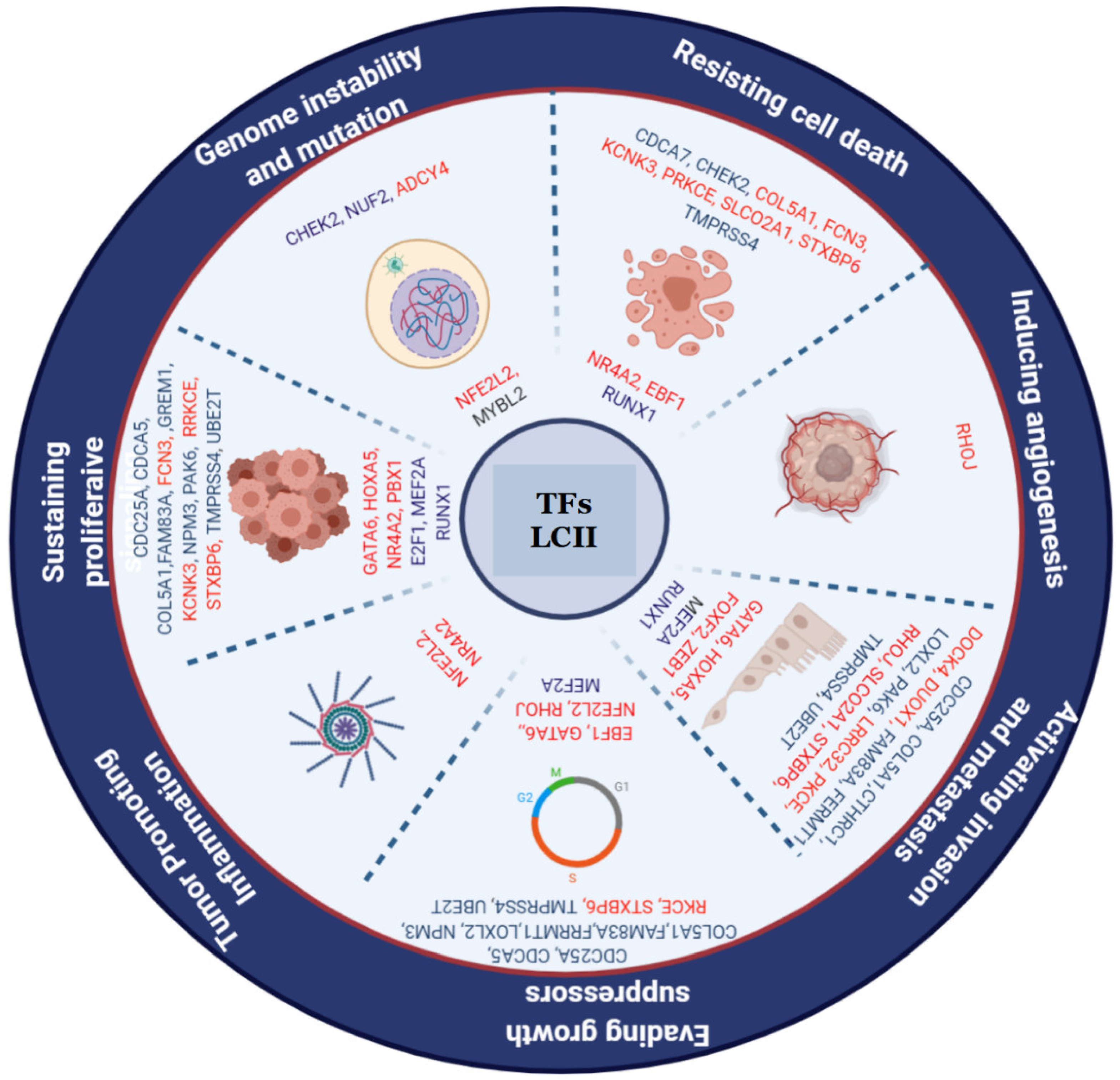
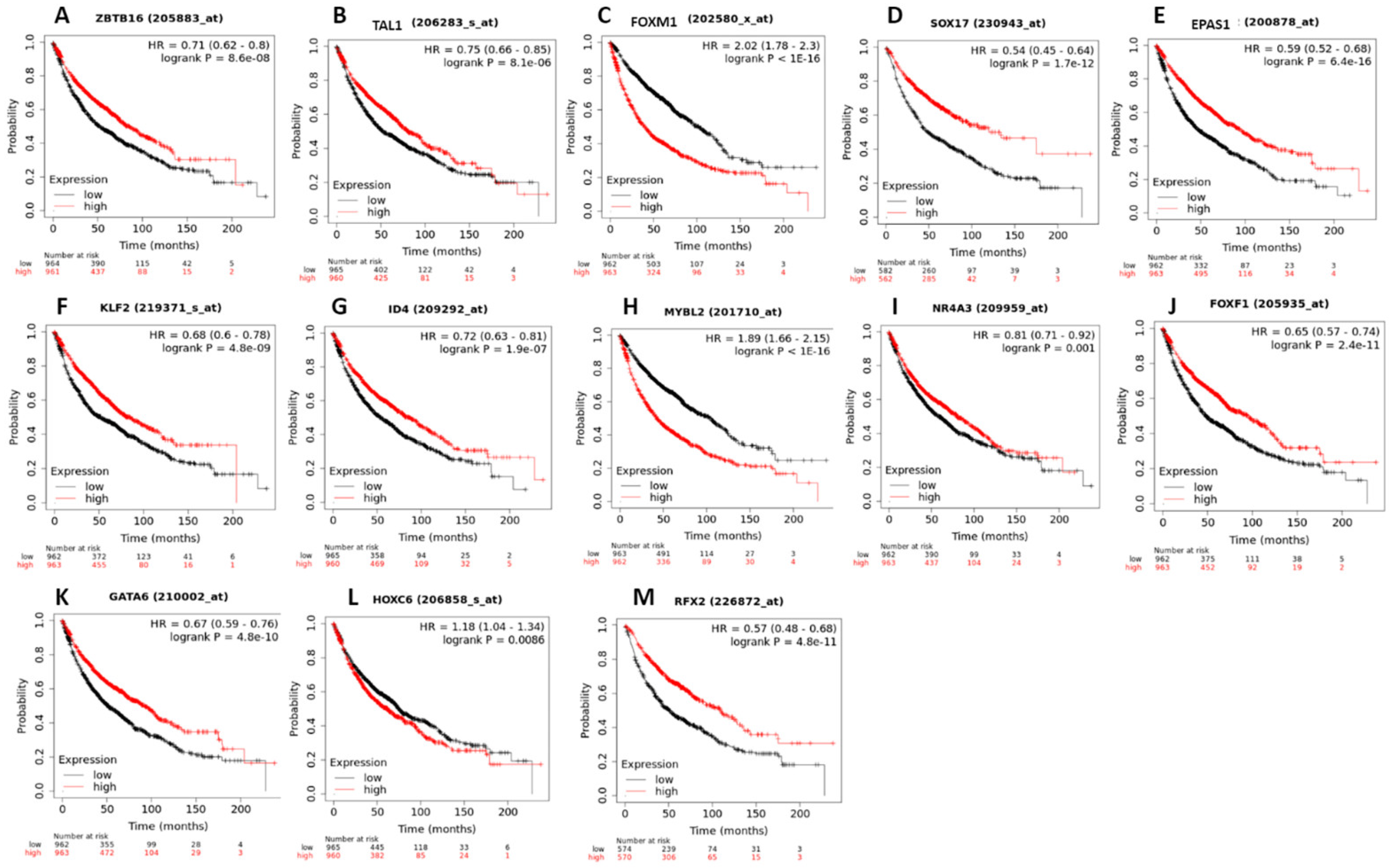

| Study Code | Subjects’ Disease | Samples | Reference |
|---|---|---|---|
| GSE19804 | Non-smoking women with NSCLC | Normal (60) vs. Cancer (60) | [23] |
| GSE10072 | Patients with lung adenocarcinoma | Normal (49) vs. Cancer (58) | [24] |
| GSE3268 | Patients with squamous lung cancer cells | Normal (5) vs. Cancer (5) | [25] |
| GSE108055 | Typical and atypical carcinoid, and SCLC | Normal (9) vs. Cancer (54) | [26] |
| E-MTAB−5231 | Patients with NSCLC | Normal (18) vs. Cancer (SCC = 11 AC = 11) | [27] |
| E-MTAB−3950 | Pre-invasive and invasive early squamous carcinoma | Normal (30) vs. Cancer (30) (SCC) | [28] |
| GSE52248 | Patients with lung adenocarcinoma | Normal (6) vs. Cancer (12) | [29] |
| GSE70089 | Patients with lung carcinoma | Normal (3) vs. Cancer(3) | [30] |
| GSE81089 | Patients with NSCLC | Normal (19) vs. Cancer (199) | [31] |
| GSE84776 | Patients with squamous lung cancer cells | Normal (9) vs. Cancer (9) | [32] |
| GSE10797 | Invasive breast cancer | Normal (10) vs. Cancer (56) | [33] |
| GSE21422 | Ductal carcinoma in situ and invasive breast carcinoma | Normal (5) vs. Cancer (14) | [34] |
| GSE26910 | Invasive breast primary breast cancer | Normal (6) vs. Cancer (6) | [35] |
| GSE3744 | Sporadic basal-like breast cancer | Normal (7) vs. Cancer (40) | [36,37] |
| GSE5764 | Invasive lobular and ductal breast cancer | Normal (10) vs. Cancer (20) | [38] |
| GSE22529 | Chronic lymphocytic leukemia | Normal (11) vs. Cancer (41) | [39] |
| GSE26725 | Chronic lymphocytic leukemia | Normal (5) vs. Cancer (12) | [40] |
| GSE5788 | T-cell prolymphocytic leukemia | Normal (8) vs. Cancer (6) | [41] |
| GSE6691 | Chronic lymphocytic leukemia | Normal (13) vs. Cancer (11) | [42] |
| GSE9476 | Acute myeloid leukemia | Normal (38) vs. Cancer (26) | [43] |
| Transcription Factors (TFs) | Lung Cancer (LC) | Breast Cancer (BC) | Leukemia (LK) | Total |
|---|---|---|---|---|
| ZBTB16 | 9 | 4 | 3 | 16 |
| KLF4 | 9 | 4 | 1 | 14 |
| TAL1 | 9 | 3 | 2 | 14 |
| FOXM1 | 9 | 2 | 1 | 12 |
| BZW2 | 9 | 1 | 1 | 11 |
| HLF | 8 | 3 | 1 | 12 |
| GPRASP1 | 8 | 2 | 2 | 12 |
| MNDA | 8 | 2 | 1 | 11 |
| PKNOX2 | 8 | 1 | 1 | 10 |
| TFAP2C | 8 | 1 | 1 | 10 |
| SOX4 | 10 | 1 | 2 | 13 |
| EGR1 | 7 | 4 | 3 | 14 |
| FOSB | 7 | 4 | 1 | 12 |
| SOX17 | 10 | 3 | 0 | 13 |
| EPAS1 | 8 | 3 | 0 | 11 |
| KLF2 | 8 | 3 | 0 | 11 |
| ID4 | 8 | 3 | 0 | 11 |
| MEIS1 | 8 | 2 | 0 | 10 |
| MYBL2 | 8 | 2 | 0 | 10 |
| NR2F1 | 8 | 2 | 0 | 10 |
| DLX5 | 8 | 1 | 0 | 9 |
| TBX5 | 8 | 1 | 0 | 9 |
| NR4A3 | 8 | 0 | 1 | 9 |
| ID2 | 7 | 0 | 3 | 10 |
| ETV4 | 7 | 0 | 1 | 8 |
| SOX12 | 7 | 0 | 1 | 8 |
| TCF3 | 7 | 0 | 1 | 8 |
| RORA | 6 | 0 | 2 | 8 |
| FOXF1 | 8 | 0 | 0 | 8 |
| HOXC6 | 8 | 0 | 0 | 8 |
| RFX2 | 8 | 0 | 0 | 8 |
| GATA6 | 7 | 0 | 0 | 7 |
| RARA | 7 | 0 | 0 | 7 |
| PAX9 | 7 | 0 | 0 | 7 |
| Alignment LC&BC | Number of Alignments | Percentage | Median |
|---|---|---|---|
| HEG1—HEG1 | 5 | 10.4% | 0.58 |
| PLSCR4—PLSCR4 | 5 | 10.4% | 0.50 |
| GMFG—GMFG | 4 | 8.3% | 0.51 |
| FCGR3B—FCGR3B | 3 | 6.3% | 0.52 |
| NME4—NME4 | 3 | 6.3% | 0.50 |
| Alignment LC&LK | Number of Alignments | Percentage | Median |
|---|---|---|---|
| SNRK—SNRK | 6 | 15.0% | 0.51 |
| BIRC5—BIRC5 | 5 | 12.5% | 0.57 |
| GIMAP5—GIMAP5 | 3 | 7.5% | 0.51 |
| HBB—HBB | 3 | 7.5% | 0.51 |
| IL33—IL33 | 3 | 7.5% | 0.52 |
| AKAP12—AKAP12 | 3 | 7.5% | 0.52 |
| CD93—CD93 | 3 | 7.5% | 0.60 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Otálora-Otálora, B.A.; Osuna-Garzón, D.A.; Carvajal-Parra, M.S.; Cañas, A.; Montecino, M.; López-Kleine, L.; Rojas, A. Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis. Biology 2022, 11, 1082. https://doi.org/10.3390/biology11071082
Otálora-Otálora BA, Osuna-Garzón DA, Carvajal-Parra MS, Cañas A, Montecino M, López-Kleine L, Rojas A. Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis. Biology. 2022; 11(7):1082. https://doi.org/10.3390/biology11071082
Chicago/Turabian StyleOtálora-Otálora, Beatriz Andrea, Daniel Alejandro Osuna-Garzón, Michael Steven Carvajal-Parra, Alejandra Cañas, Martín Montecino, Liliana López-Kleine, and Adriana Rojas. 2022. "Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis" Biology 11, no. 7: 1082. https://doi.org/10.3390/biology11071082
APA StyleOtálora-Otálora, B. A., Osuna-Garzón, D. A., Carvajal-Parra, M. S., Cañas, A., Montecino, M., López-Kleine, L., & Rojas, A. (2022). Identifying General Tumor and Specific Lung Cancer Biomarkers by Transcriptomic Analysis. Biology, 11(7), 1082. https://doi.org/10.3390/biology11071082