Physical Enrichment Triggers Brain Plasticity and Influences Blood Plasma Circulating miRNA in Rainbow Trout (Oncorhynchus mykiss)
Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animal Handling
2.2. Fish and Experimental Design
2.3. Sampling Protocol
2.4. Gene Expression Measurement by Real-Time Quantitative PCR
2.5. Primer Design
2.6. RNA Extraction, Pre-Processing, and Illumina Small RNA Deep Sequencing for Plasma Samples
2.7. c-miRNA Annotation, Quantification and Differential Expression Analysis
2.8. Statistical Analysis
3. Results
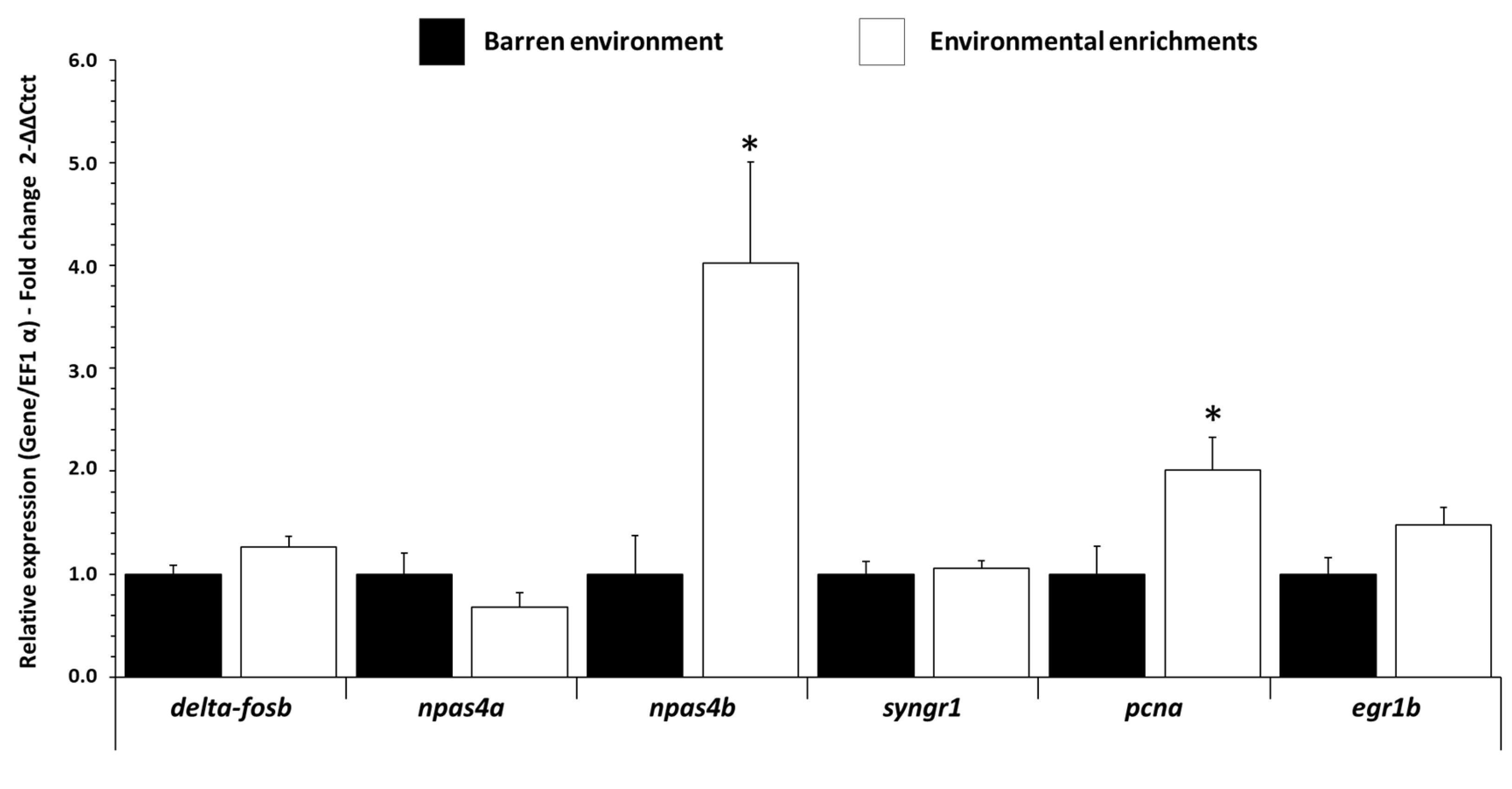
3.1. Influence of EE on mRNA Expression Levels of Neuronal Activity Gene in Trout Brain
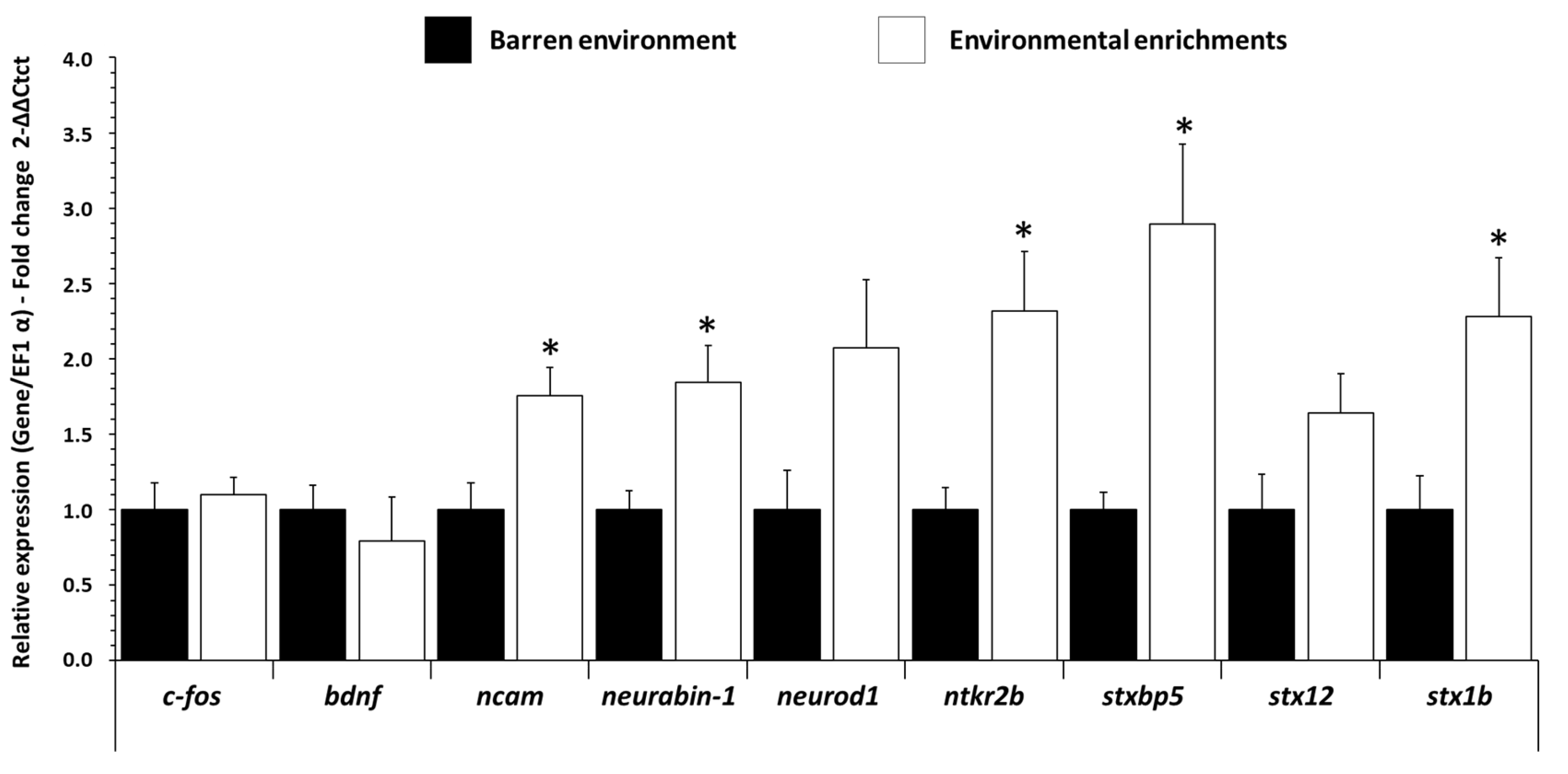
3.2. Influence of EE on mRNA Expression Levels of Neurotrophic and Synaptogenesis Gene in Trout Brain
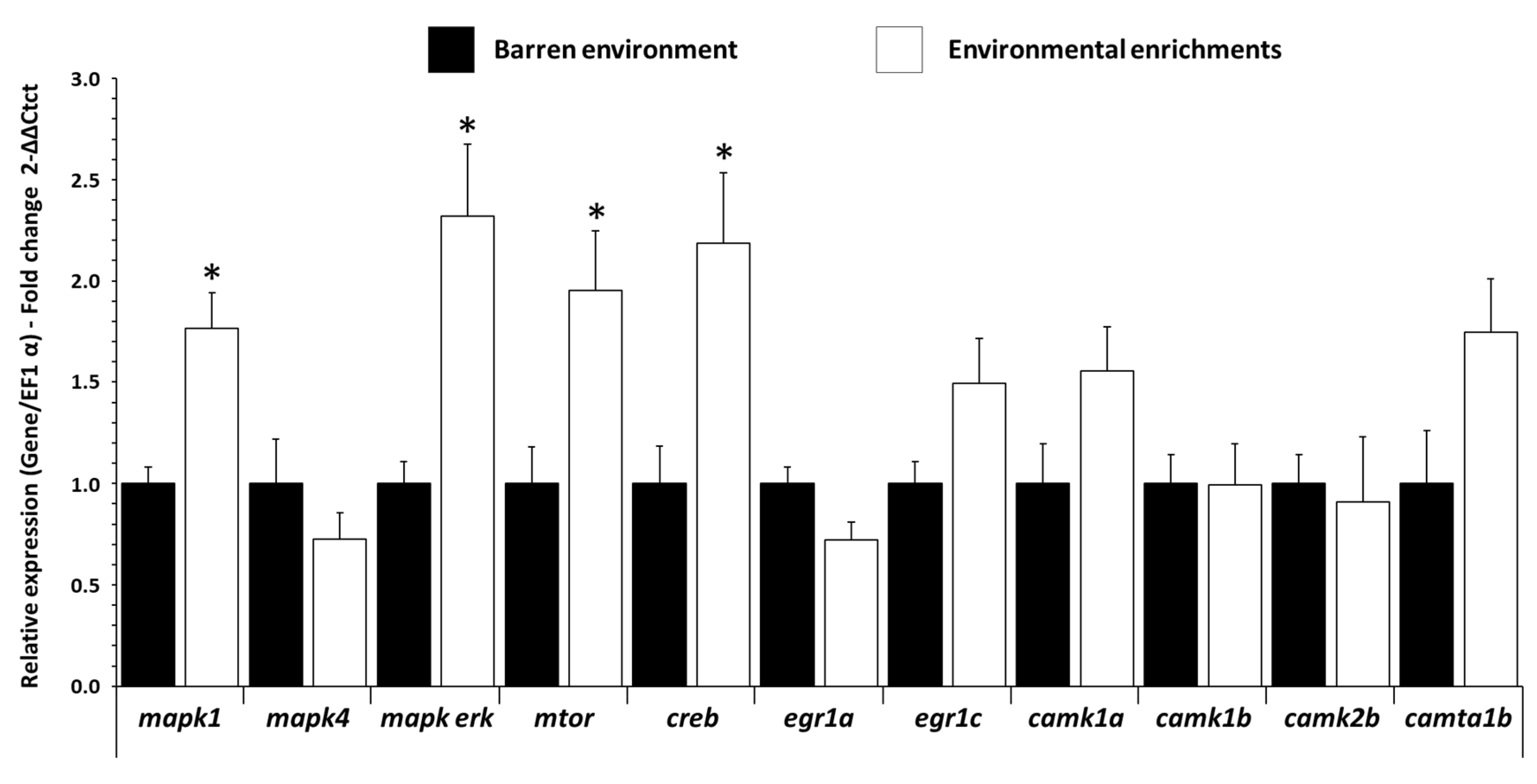
3.3. Influence of EE on mRNA Expression Levels of Plasticity Gene in Trout Brain
3.4. Influence of EE on mRNA Expression Levels of Neurotransmitters Pathways Gene in Trout Brain
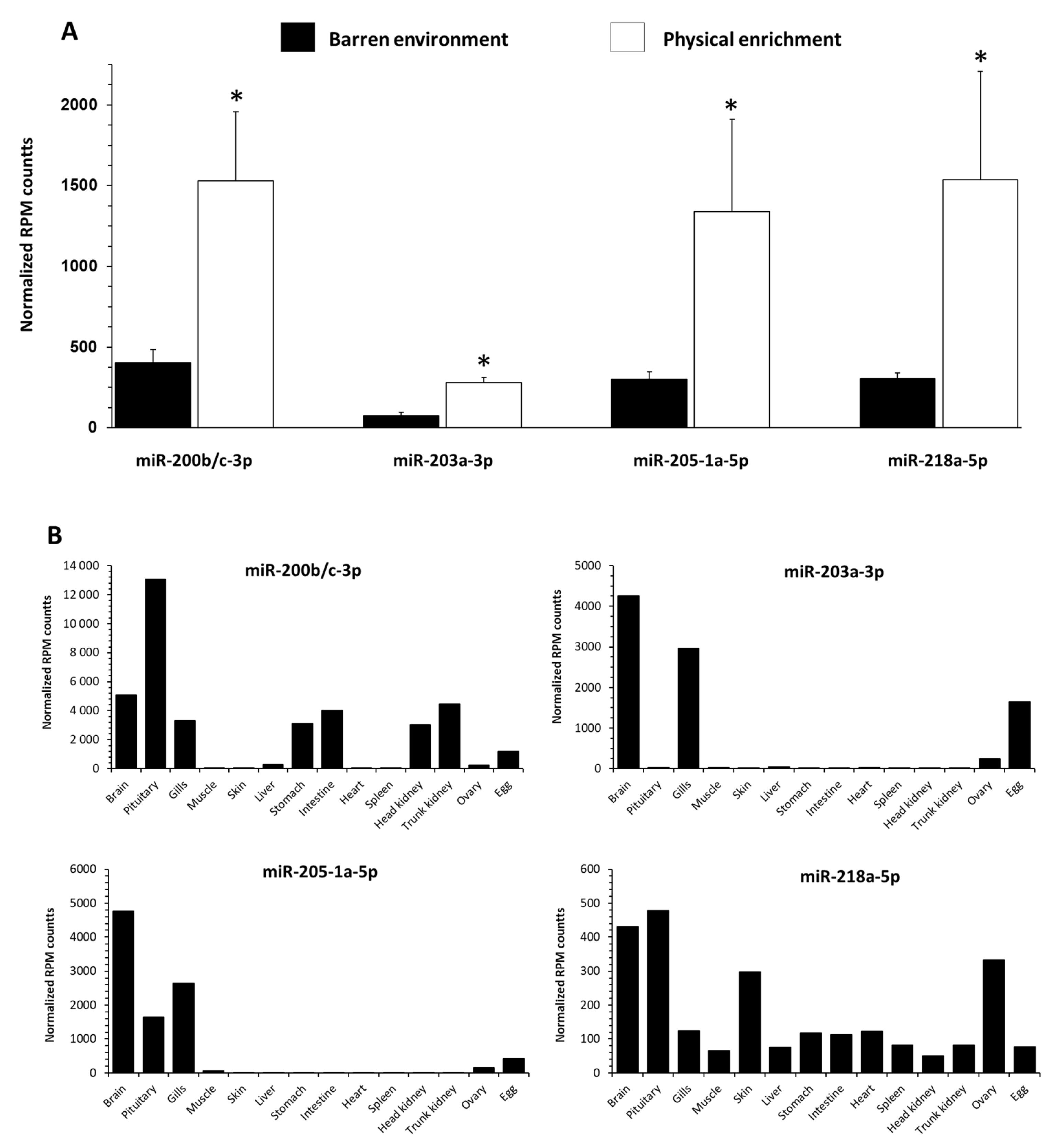
3.5. Influence of EE on Selected c-miRNAs Abundance in Blood-Plasma Fluid in RT
4. Discussion
4.1. EE Affects Cerebral Function in Telencephalon of Trout
4.2. EE Influences Circulating miRNAs Profiles of Plasma in Trout
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hebb, D.O. The effects of early experience on problem-solving at maturity. Am. Psychol. 1947, 2, 306–307. [Google Scholar]
- Bayne, K. Environmental enrichment and mouse models: Current perspectives. Anim. Model Exp. Med. 2018, 1, 82–90. [Google Scholar] [CrossRef]
- Newberry, R.C. Environmental enrichment: Increasing the biological relevance of captive environments. Appl. Anim. Behav. Sci. 1995, 44, 229–243. [Google Scholar] [CrossRef]
- Arechavala-Lopez, P.; Cabrera-Álvarez, M.J.; Maia, C.M.; Saraiva, J.L. Environmental enrichment in fish aquaculture: A review of fundamental and practical aspects. Rev. Aquac. 2022, 14, 704–728. [Google Scholar] [CrossRef]
- Corcoran, M. Environmental enrichment for aquatic animals. Vet. Clin. N. Am. Exot. Anim. Pract. 2015, 18, 305–321. [Google Scholar] [CrossRef] [PubMed]
- Näslund, J.; Johnsson, J.I. Environmental enrichment for fish in captive environments: Effects of physical structures and substrates. Fish Fish. 2016, 17, 1–30. [Google Scholar] [CrossRef]
- Salvanes, A.G.V.; Moberg, O.; Ebbesson, L.O.E.; Nilsen, T.O.; Jensen, K.H.; Braithwaite, V.A. Environmental enrichment promotes neural plasticity and cognitive ability in fish. Proc. Biol. Sci. 2013, 280, 20131331. [Google Scholar] [CrossRef]
- Kadry, V.O.; Barreto, R.E. Environmental enrichment reduces aggression of pearl cichlid, Geophagus brasiliensis, during resident-intruder interactions. Neotrop. Ichthyol. 2010, 8, 329–332. [Google Scholar] [CrossRef]
- Reiser, S.; Pohlmann, D.M.; Blancke, T.; Koops, U.; Trautner, J. Environmental enrichment during early rearing provokes epigenetic changes in the brain of a salmonid fish. Comp. Biochem. Physiol. Part D Genom. Proteom. 2021, 39, 100838. [Google Scholar] [CrossRef]
- Brydges, N.M.; Braithwaite, V.A. Does environmental enrichment affect the behaviour of fish commonly used in laboratory work? Appl. Anim. Behav. Sci. 2009, 118, 137–143. [Google Scholar] [CrossRef]
- Naslund, J.; Rosengren, M.; Johnsson, J. Fish density, but not environmental enrichment, affects the size of cerebellum in the brain of juvenile hatchery-reared Atlantic salmon. Environ. Biol. Fishes 2019, 102, 705–712. [Google Scholar] [CrossRef]
- Toli, E.A.; Noreikiene, K.; DeFaveri, J.; Merilä, J. Environmental enrichment, sexual dimorphism, and brain size in sticklebacks. Ecol. Evol. 2017, 7, 1691–1698. [Google Scholar] [CrossRef] [PubMed]
- Jones, N.A.R.; Webster, M.M.; Salvanes, A.G.V. Physical enrichment research for captive fish: Time to focus on the DETAILS. J. Fish Biol. 2021, 99, 704–725. [Google Scholar] [CrossRef] [PubMed]
- Brunet, V.; Kleiber, A.; Patinote, A.; Sudan, P.-L.; Duret, C.; Gourmelen, G.; Moreau, E.; Fournel, C.; Pineau, L.; Calvez, S.; et al. Positive welfare effects of physical enrichments from the nature-, functions- and feeling- based approaches in farmed rainbow trout (Oncorhynchus mykiss). Aquaculture 2022, 550, 737825. [Google Scholar] [CrossRef]
- Shettleworth, S.J. Clever animals and killjoy explanations in comparative psychology. Trends Cogn. Sci. 2010, 14, 477–481. [Google Scholar] [CrossRef]
- Ebbesson, L.O.E.; Braithwaite, V.A. Environmental effects on fish neural plasticity and cognition. J. Fish Biol. 2012, 81, 2151–2174. [Google Scholar] [CrossRef]
- Kihslinger, R.L.; Nevitt, G.A. Early rearing environment impacts cerebellar growth in juvenile salmon. J. Exp. Biol. 2006, 209, 504–509. [Google Scholar] [CrossRef]
- DePasquale, C.; Neuberger, T.; Hirrlinger, A.M.; Braithwaite, V.A. The influence of complex and threatening environments in early life on brain size and behaviour. Proc. Biol. Sci. 2016, 283, 20152564. [Google Scholar] [CrossRef]
- Turschwell, M.P.; White, C.R. The effects of laboratory housing and spatial enrichment on brain size and metabolic rate in the eastern mosquitofish, Gambusia holbrooki. Biol. Open 2016, 5, 205–210. [Google Scholar] [CrossRef]
- Von Krogh, K.; Sørensen, C.; Nilsson, G.E.; Øverli, Ø. Forebrain cell proliferation, behavior, and physiology of zebrafish, Danio rerio, kept in enriched or barren environments. Physiol. Behav. 2010, 101, 32–39. [Google Scholar] [CrossRef]
- Dunlap, K.D.; Silva, A.C.; Chung, M. Environmental complexity, seasonality and brain cell proliferation in a weakly electric fish, Brachyhypopomus gauderio. J. Exp. Biol. 2011, 214, 794–805. [Google Scholar] [CrossRef] [PubMed]
- Schouten, M.; Aschrafi, A.; Bielefeld, P.; Doxakis, E.; Fitzsimons, C.P. microRNAs and the regulation of neuronal plasticity under stress conditions. Neuroscience 2013, 241, 188–205. [Google Scholar] [CrossRef] [PubMed]
- Fiorenza, A.; Barco, A. Role of Dicer and the miRNA system in neuronal plasticity and brain function. Neurobiol. Learn. Mem. 2016, 135, 3–12. [Google Scholar] [CrossRef]
- Bao, T.; Miao, W.; Han, J.; Yin, M.; Yan, Y.; Wang, W.; Zhu, Y. Spontaneous running wheel improves cognitive functions of mouse associated with miRNA expressional alteration in hippocampus following traumatic brain injury. J. Mol. Neurosci. 2014, 54, 622–629. [Google Scholar] [CrossRef] [PubMed]
- Chandrasekar, V.; Dreyer, J.-L. Regulation of MiR-124, Let-7d, and MiR-181a in the accumbens affects the expression, extinction, and reinstatement of cocaine-induced conditioned place preference. Neuropsychopharmacology 2011, 36, 1149–1164. [Google Scholar] [CrossRef]
- Schaefer, A.; Im, H.-I.; Venø, M.T.; Fowler, C.D.; Min, A.; Intrator, A.; Kjems, J.; Kenny, P.J.; O’Carroll, D.; Greengard, P. Argonaute 2 in dopamine 2 receptor-expressing neurons regulates cocaine addiction. J. Exp. Med. 2010, 207, 1843–1851. [Google Scholar] [CrossRef] [PubMed]
- Greenberg, J.K.; Xia, J.; Zhou, X.; Thatcher, S.R.; Gu, X.; Ament, S.A.; Newman, T.C.; Green, P.J.; Zhang, W.; Robinson, G.E.; et al. Behavioral plasticity in honey bees is associated with differences in brain microRNA transcriptome. Genes Brain Behav. 2012, 11, 660–670. [Google Scholar] [CrossRef]
- Pang, Y.; He, L.; Song, Y.; Song, X.; Lv, J.; Cheng, Y.; Yang, X. Identification and Integrated Analysis of MicroRNA and mRNA Expression Profiles During Agonistic Behavior in Chinese Mitten Crab (Eriocheir sinensis) Using a Deep Sequencing Approach. Front. Genet. 2020, 11, 321. [Google Scholar] [CrossRef]
- Weber, J.A.; Baxter, D.H.; Zhang, S.; Huang, D.Y.; Huang, K.H.; Lee, M.J.; Galas, D.J.; Wang, K. The microRNA spectrum in 12 body fluids. Clin. Chem. 2010, 56, 1733–1741. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Cardona, E.; Guyomar, C.; Desvignes, T.; Montfort, J.; Guendouz, S.; Postlethwait, J.H.; Skiba-Cassy, S.; Bobe, J. Circulating miRNA repertoire as a biomarker of metabolic and reproductive states in rainbow trout. BMC Biol. 2021, 19, 235. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Vigor, C.; Vercauteren, J.; Reversat, G.; Zhou, B.; Surget, A.; Larroquet, L.; Lanuque, A.; Sandres, F.; Terrier, F.; et al. Characterization and modulation of brain lipids content of rainbow trout fed with 100% plant based diet rich in omega-3 long chain polyunsaturated fatty acids DHA and EPA. Biochimie 2020, 178, 137–147. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Terrier, F.; Marchand, M.; Herman, A.; Heraud, C.; Surget, A.; Lanuque, A.; Sandres, F.; Marandel, L. Effects of Low Stocking Densities on Zootechnical Parameters and Physiological Responses of Rainbow Trout (Oncorhynchus mykiss) Juveniles. Biology 2021, 10, 1040. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Larroquet, L.; Surget, A.; Lanuque, A.; Sandres, F.; Terrier, F.; Corraze, G.; Lee, J.C.; Skiba-Cassy, S. Impact on cerebral function in rainbow trout fed with plant based omega-3 long chain polyunsaturated fatty acids enriched with DHA and EPA. Fish Shellfish Immunol. 2020, 103, 409–420. [Google Scholar] [CrossRef] [PubMed]
- Roy, J.; Baranek, E.; Mercier, Y.; Larroquet, L.; Surget, A.; Ganot, A.; Sandres, F.; Lanuque, A.; Terrier, F.; Briand, L. Involvement of Taste Receptors in the Oro-Sensory Perception of Nutrients in Rainbow Trout (Oncorhynchus mikyss) Fed Diets with Different Fatty Acid Profiles. Aquac. Nutr. 2022, 2022, e1152463. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Heraud, C.; Hirschinger, T.; Baranek, E.; Larroquet, L.; Surget, A.; Sandres, F.; Lanuque, A.; Terrier, F.; Roy, J. Detection and Modulation of Olfactory Sensing Receptors in Carnivorous Rainbow Trout (Oncorhynchus mykiss) Fed from First Feeding with Plant-Based Diet. Int. J. Mol. Sci. 2022, 23, 2123. [Google Scholar] [CrossRef] [PubMed]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, 132, 365–386. [Google Scholar] [CrossRef]
- Miandare, H.K.; Farahmand, H.; Akbarzadeh, A.; Ramezanpour, S.; Kaiya, H.; Miyazato, M.; Rytkönen, K.T.; Nikinmaa, M. Developmental transcription of genes putatively associated with growth in two sturgeon species of different growth rate. Gen. Comp. Endocrinol. 2012, 182, 41–47. [Google Scholar] [CrossRef]
- Desvignes, T.; Batzel, P.; Sydes, J.; Eames, B.F.; Postlethwait, J.H. miRNA analysis with Prost! reveals evolutionary conservation of organ-enriched expression and post-transcriptional modifications in three-spined stickleback and zebrafish. Sci Rep. 2019, 9, 3913. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Juanchich, A.; Bardou, P.; Rué, O.; Gabillard, J.-C.; Gaspin, C.; Bobe, J.; Guiguen, Y. Characterization of an extensive rainbow trout miRNA transcriptome by next generation sequencing. BMC Genom. 2016, 17, 164. [Google Scholar] [CrossRef] [PubMed]
- Mes, D.; van Os, R.; Gorissen, M.; Ebbesson, L.O.E.; Finstad, B.; Mayer, I.; Vindas, M.A. Effects of environmental enrichment on forebrain neural plasticity and survival success of stocked Atlantic salmon. J. Exp. Biol. 2019, 222, jeb212258. [Google Scholar] [CrossRef]
- Brockmark, S.; Adriaenssens, B.; Johnsson, J.I. Less is more: Density influences the development of behavioural life skills in trout. Proc. Biol. Sci. 2010, 277, 3035–3043. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.L.; Hori, T.S.; Rise, M.L.; Fleming, I.A. Transcriptomic responses of Atlantic salmon (Salmo salar) to environmental enrichment during juvenile rearing. PLoS ONE 2015, 10, e0118378. [Google Scholar] [CrossRef] [PubMed]
- Salzman, C.D.; Fusi, S. Emotion, cognition, and mental state representation in amygdala and prefrontal cortex. Annu. Rev. Neurosci. 2010, 33, 173–202. [Google Scholar] [CrossRef] [PubMed]
- Northcutt, R.G. Evolution of the telencephalon in nonmammals. Annu. Rev. Neurosci. 1981, 4, 301–350. [Google Scholar] [CrossRef]
- Wullimann, M.F.; Mueller, T. Teleostean and mammalian forebrains contrasted: Evidence from genes to behavior. J. Comp. Neurol. 2004, 475, 143–162. [Google Scholar] [CrossRef]
- Bshary, R.; Brown, C. Fish cognition. Curr. Biol. 2014, 24, R947–R950. [Google Scholar] [CrossRef]
- Kondo, M. Molecular mechanisms of experience-dependent structural and functional plasticity in the brain. Anat. Sci. Int. 2017, 92, 1–17. [Google Scholar] [CrossRef]
- Alwis, D.S.; Rajan, R. Environmental enrichment and the sensory brain: The role of enrichment in remediating brain injury. Front. Syst. Neurosci. 2014, 8, 156. [Google Scholar] [CrossRef] [PubMed]
- Mayer, I.; Meager, J.; Skjæraasen, J.E.; Rodewald, P.; Sverdrup, G.; Fernö, A. Domestication causes rapid changes in heart and brain morphology in Atlantic cod (Gadus morhua). Environ. Biol Fish. 2011, 92, 181–186. [Google Scholar] [CrossRef]
- Switon, K.; Kotulska, K.; Janusz-Kaminska, A.; Zmorzynska, J.; Jaworski, J. Molecular neurobiology of mTOR. Neuroscience 2017, 341, 112–153. [Google Scholar] [CrossRef]
- Bockaert, J.; Marin, P. mTOR in Brain Physiology and Pathologies. Physiol. Rev. 2015, 95, 1157–1187. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Blundell, J.; Ogawa, S.; Kwon, C.-H.; Zhang, W.; Sinton, C.; Powell, C.M.; Parada, L.F. Pharmacological inhibition of mTORC1 suppresses anatomical, cellular, and behavioral abnormalities in neural-specific Pten knock-out mice. J. Neurosci. 2009, 29, 1773–1783. [Google Scholar] [CrossRef] [PubMed]
- Da Peixoto, C.; Parfitt, G.M.; Bruch, G.E.; Cordeiro, M.F.; Almeida, D.V.; Marins, L.F.F.; Barros, D.M. Effects of learning on mTOR pathway gene expression in the brain of zebrafish (Danio rerio) of different ages. Exp. Gerontol. 2017, 89, 8–14. [Google Scholar] [CrossRef]
- Giachello, C.N.G.; Fiumara, F.; Giacomini, C.; Corradi, A.; Milanese, C.; Ghirardi, M.; Benfenati, F.; Montarolo, P.G. MAPK/Erk-dependent phosphorylation of synapsin mediates formation of functional synapses and short-term homosynaptic plasticity. J. Cell Sci. 2010, 123, 881–893. [Google Scholar] [CrossRef]
- Samuels, I.S.; Saitta, S.C.; Landreth, G.E. MAP’ing CNS development and cognition: An ERKsome process. Neuron 2009, 61, 160–167. [Google Scholar] [CrossRef]
- Ciccarelli, A.; Giustetto, M. Role of ERK signaling in activity-dependent modifications of histone proteins. Neuropharmacology 2014, 80, 34–44. [Google Scholar] [CrossRef]
- Guidotti, G.; Calabrese, F.; Auletta, F.; Olivier, J.; Racagni, G.; Homberg, J.; Riva, M.A. Developmental influence of the serotonin transporter on the expression of npas4 and GABAergic markers: Modulation by antidepressant treatment. Neuropsychopharmacol. Publ. Am. Coll. Neuropsychopharmacol. 2012, 37, 746–758. [Google Scholar] [CrossRef]
- Coutellier, L.; Beraki, S.; Ardestani, P.M.; Saw, N.L.; Shamloo, M. Npas4: A neuronal transcription factor with a key role in social and cognitive functions relevant to developmental disorders. PLoS ONE 2012, 7, e46604. [Google Scholar] [CrossRef]
- Jaehne, E.J.; Klarić, T.S.; Koblar, S.A.; Baune, B.T.; Lewis, M.D. Effects of Npas4 deficiency on anxiety, depression-like, cognition and sociability behaviour. Behav. Brain Res. 2015, 281, 276–282. [Google Scholar] [CrossRef] [PubMed]
- Hartz, B.P.; Rønn, L.C.B. NCAM in long-term potentiation and learning. Adv. Exp. Med. Biol. 2010, 663, 257–270. [Google Scholar] [CrossRef]
- Aonurm-Helm, A.; Jurgenson, M.; Zharkovsky, T.; Sonn, K.; Berezin, V.; Bock, E.; Zharkovsky, A. Depression-like behaviour in neural cell adhesion molecule (NCAM)-deficient mice and its reversal by an NCAM-derived peptide, FGL. Eur. J. Neurosci. 2008, 28, 1618–1628. [Google Scholar] [CrossRef]
- Jürgenson, M.; Aonurm-Helm, A.; Zharkovsky, A. Partial reduction in neural cell adhesion molecule (NCAM) in heterozygous mice induces depression-related behaviour without cognitive impairment. Brain Res. 2012, 1447, 106–118. [Google Scholar] [CrossRef] [PubMed]
- Sahún, I.; Delgado-García, J.M.; Amador-Arjona, A.; Giralt, A.; Alberch, J.; Dierssen, M.; Gruart, A. Dissociation between CA3-CA1 synaptic plasticity and associative learning in TgNTRK3 transgenic mice. J. Neurosci. 2007, 27, 2253–2260. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Otnaess, M.K.; Djurovic, S.; Rimol, L.M.; Kulle, B.; Kähler, A.K.; Jönsson, E.G.; Agartz, I.; Sundet, K.; Hall, H.; Timm, S.; et al. Evidence for a possible association of neurotrophin receptor (NTRK-3) gene polymorphisms with hippocampal function and schizophrenia. Neurobiol. Dis. 2009, 34, 518–524. [Google Scholar] [CrossRef] [PubMed]
- Howe, M.W.; Dombeck, D.A. Rapid signalling in distinct dopaminergic axons during locomotion and reward. Nature 2016, 535, 505–510. [Google Scholar] [CrossRef]
- Meneses, A.; Liy-Salmeron, G. Serotonin and emotion, learning and memory. Rev. Neurosci. 2012, 23, 543–553. [Google Scholar] [CrossRef]
- Winberg, S.; Nilsson, G.E. Time Course of Changes in Brain Serotonergic Activity and Brain Tryptophan Levels in Dominant and Subordinate Juvenile Arctic Charr. J. Exp. Biol. 1993, 179, 181–195. [Google Scholar] [CrossRef]
- Whitaker, K.W.; Neumeister, H.; Huffman, L.S.; Kidd, C.E.; Preuss, T.; Hofmann, H.A. Serotonergic modulation of startle-escape plasticity in an African cichlid fish: A single-cell molecular and physiological analysis of a vital neural circuit. J Neurophysiol. 2011, 106, 127–137. [Google Scholar] [CrossRef] [PubMed]
- Ersoy, A.; Waller, M.D. Neurogenesis in adulthood: A possible role in learning. Trends Cogn Sci. 1999, 3, 186–192. [Google Scholar] [CrossRef]
- Wood, L.S.; Desjardins, J.K.; Fernald, R.D. Effects of stress and motivation on performing a spatial task. Neurobiol. Learn. Mem. 2011, 95, 277–285. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Arechavala-Lopez, P.; Caballero-Froilán, J.C.; Jiménez-García, M.; Capó, X.; Tejada, S.; Saraiva, J.L.; Sureda, A.; Moranta, D. Enriched environments enhance cognition, exploratory behaviour and brain physiological functions of Sparus aurata. Sci. Rep. 2020, 10, 11252. [Google Scholar] [CrossRef]
- Makino, H.; Masuda, R.; Tanaka, M. Environmental stimuli improve learning capability in striped knifejaw juveniles: The stage-specific effect of environmental enrichment and the comparison between wild and hatchery-reared fish. Fish. Sci. 2015, 81, 1035–1042. [Google Scholar] [CrossRef]
- Bergendahl, I.A.; Salvanes, A.G.V.; Braithwaite, V.A. Determining the effects of duration and recency of exposure to environmental enrichment. Appl. Anim. Behav. Sci. 2016, 176, 163–169. [Google Scholar] [CrossRef]
- Roy, T.; Bhat, A. Learning and Memory in Juvenile Zebrafish: What makes the Difference—Population or Rearing Environment? Ethology 2016, 122, 308–318. [Google Scholar] [CrossRef]
- Carbia, P.S.; Brown, C. Environmental enrichment influences spatial learning ability in captive-reared intertidal gobies (Bathygobius cocosensis). Anim. Cogn. 2019, 22, 89–98. [Google Scholar] [CrossRef]
- McKibben, L.A.; Dwivedi, Y. MicroRNA Regulates Early-Life Stress–Induced Depressive Behavior via Serotonin Signaling in a Sex-Dependent Manner in the Prefrontal Cortex of Rats. Biol. Psychiatry Glob. Open Sci. 2021, 1, 180–189. [Google Scholar] [CrossRef]
- Ryan, B.; Williams, J.M.; Curtis, M.A. Plasma MicroRNAs Are Altered Early and Consistently in a Mouse Model of Tauopathy. Neuroscience 2019, 411, 164–176. [Google Scholar] [CrossRef]
- Yoshino, Y.; Roy, B.; Dwivedi, Y. Corticosterone-mediated regulation and functions of miR-218-5p in rat brain. Sci. Rep. 2022, 12, 194. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, H.; Yin, H.; Geng, S.; Liu, Y.; Liu, C.; Zhao, J.; Liu, Y.; Wang, X.; Wang, Y. Up-regulated microRNA-218-5p ameliorates the damage of dopaminergic neurons in rats with Parkinson’s disease via suppression of LASP1. Brain Res. Bull. 2021, 166, 92–101. [Google Scholar] [CrossRef] [PubMed]


| Days Post-Fertilization | Average Fish Weight at the Beginning of Each Period | Tank size | Number tank Replicates and Number of Individuals | Number (n) and Type of Structures Used in the Enriched Group | Floor Covering | ||
|---|---|---|---|---|---|---|---|
 |  |  | |||||
| 53–210 | 0.12 g | 100 L (65 × 50 × 30 cm) Covered tanks | 3 tanks/treatment 100 individuals each | n = 1 (10 × 5 × 5 cm) | n = 2 (24 × 15 × 11 cm) | n = 2 (9 × 8 × 8 cm) | ~60% |
| 210–238 | 18.6 g | 72 L (55 × 45 × 29 cm) Uncovered tanks | 1 tank/treatment 30 individuals each | n = 1 (10 × 5 × 5 cm) | n = 2 (24 × 15 × 11 cm) | n = 2 (9 × 8 × 8 cm) | ~70% |
| Days Post-Fertilization | Average Fish Weight at the Beginning of Each Period | Tank Size | Number tank Replicates and Number of Individuals | Number (n) and Type of Structures Used in the Enriched Group | Floor Covering | ||
|---|---|---|---|---|---|---|---|
 |  |  | |||||
| 99–136 | 2.83 g | 72 L (55 × 45 × 29 cm) Uncovered tanks | 2 tanks/treatment 100 individuals each | n = 1 (10 × 5 × 5 cm) | n = 2 (24 × 15 × 11 cm) | n = 2 (9 × 8 × 8 cm) | ~70% |
| 137–189 | 11 g | 3 tanks/treatment 30 individuals each | n = 2 (9 × 8 × 8 cm) | ~70% | |||
| Transcript | Forward Primer | Reverse Primer | Accession Number |
|---|---|---|---|
| Reference | |||
| eef1a1 | TCCTCTTGGTCGTTTCGCTG | ACCCGAGGGACATCCTGTG | AF498320 |
| Neuronal activity | |||
| delta-fosb | TGCAGCCAACTCTCATCTCG | GAGGAATAACTGGGCCCTGG | XM_021587380.1 |
| npas4a | GAGATGGTGTTGCAGGTGGA | TAGTTGTGGCAGCTGATGGG | XM_021618560.2 |
| npas4b | AGCGAGAGAGGGAGGACATT | GTGGGTGGGGTTATTCTGGG | XM_036967710.1 |
| syngr1 | CTTCCCACAGATCAGCTCCG | CAGGAAACAGAACCCCACGA | XP_036805055.1 |
| pcna | GTGGACAAGGAGGAGGAAGC | ACTGTCTTGGAGAGGGGTGT | XM_036936092.1 |
| egr1b | CCCAACATGTCTCTGCCCAT | GCTCTGACACTGGAAAGGCT | XM_021617535.1 |
| Neurotrophic factor | |||
| c-fos | AACAGACTCTCCATGGCAGT | TGCTGATGTGATGACGGTGG | XM_021611391.1 |
| bdnf | GCTGCCGTGGAATAGACAAG | TCCTTATAAACCGCCAGCCA | GU108576.1 |
| ncam | GCTAACGTCACCAAAGCCAA | GGCAGCAGTACAGTTGTAGC | XM_021582629.1 |
| neurabin-1 | AGGAGAGAGAGGAGACAGCA | TTCCTCCGCCTGTTTCTCAT | XM_021572886.1 |
| neurod1 | AACCATGAGTAAGGACGGCG | TTCTCCCGACCCTCCTTCTT | XM_021608264.2 |
| ntkr2b | ACTATCCTGGAGCTGCTGGA | CTGTTCGTGGAGCTCTTGGT | XM_021602993.2 |
| stxbp5 | GTCCTCCAAGTCACACCCTG | TCAGATCCCACAACACCACG | XP_021441002.2 |
| stx12 | ACAACTTCCAGGCCGTACAG | CAGAGGCACCATCTTCAGCA | XM_036970689.1 |
| stx1b | TGACCGAGTACAACACCACG | GCATATCCTCCAGCTCCTCG | NP_001117929.1 |
| Plasticity/synaptogenesis factor | |||
| mapk1 | CCTGCTCATCAACACCACCT | AGCCACGTACTCTGTCAGGA | XM_036937960.1 |
| mapk4 | GGCAGGCTCTAAACCCTTGT | AGAGAGGAGTGGGAGTGGTC | XM_021620877.2 |
| mapk-erk | AGTCCATCTCCACGACCATC | GAAAGCCTCCAGACGTTTCC | NM_001124424.1 |
| mtor | CCAAGGACTTCGCTCACAAG | GCTCCTTGATGTCTTGCTGG | XM_021615845.1 |
| creb | CAGATTGACAGCTGCCCCTA | TGGTGTTCTGGTGTAGTGCT | MG310160.1 |
| egr1a | CGATCACCTGACCACACACA | TCAGGTGGATCTTGGTGTGC | ENSOMYT00000041426.1 |
| egr1c | CTCGTACCCCTCTCCCTCAA | AGATGGAGGCTACGGAGGAG | ENSOMYT00000036541.1 |
| camk1a | AGAGGACGGGAATGGATGGA | CTAGCACCACCTCCGAGAAC | XM_036950122.1 |
| camk1b | GTTGGCCCAGAAACCCTACA | AGAGCTTGGCGTCATTCTCA | NM_001124638.1 |
| camk2b | GCCGCTGTGTCAAACTTTGT | GCTTCCCTCTCCAGCTTCTG | XM_036976944.1 |
| camta1b | CTACTGCCCTGCCCATGAAA | GGGCAAGTCTCGAGCTTTA | XM_036947092.1 |
| Dopamine markers | |||
| th | ACGCTCTCTCAAGGTGTTCG | AAAGTACTCCAGCCCCTCCA | XM_021564247.1 |
| drd1 | GGAGGAGCTGCAGAAGAAGG | TTTCCAGTGACACATCGGCA | XM_021617454.1 |
| drd2 | CCTCCAGTCCACCACCAATT | CCACTCTCCCACCACCTCTA | NM_001124372.2 |
| dat | CTACCTCAGCGTCGACTTCC | TAGCACACCAAACCCGACTC | XM_021592557.1 |
| Serotonin markers | |||
| tph1a | ACACCAGAGCCAGACACATG | TCATCTGAAGCTCCGAGGGA | XM_021598622.1 |
| tph1b | AGCGTCCGTTTACAGTGAGG | GCCCACGATGTCCAGTTCAT | XM_021598622.1 |
| tph2 | AGCACCTCAAAGACCACGTC | ACTGGTCGAGCTCTGCAATC | MG015698 |
| 5ht1aa | CCCAACACTCCACAGTCCTC | ACCGAGCGTCTTTACCGTTT | XM_021622104 |
| 5ht1ab | GAGGACCAACGGGGACCCGA | AATCGCCGTGCTTGACCGCA | CCAF0100015582 |
| sert | CCTGCTGCCCTACATGTTGA | GGGGCAGATGTGTTTCCAGA | M_021582096.1 |
| Glutamatergic markers | |||
| grin1a | AACAAGCGAGGACCTAAGGC | CTGGCGGAGAGGATGATGAC | XM_021602512.2 |
| grin2ca | ACCCTCTGCCTTTCTTGAGC | CACAGGGCTGCAGTACTCAA | XM_036938636.1 |
| grik5a | GCAGATCAGGGTCCAGTCAC | AGTCAAAATACCCTCCCGCG | XM_036969398.1 |
| grik5b | TGAAGAGGAGGTGGTGGGAA | GATGATGAGGCCGCAGATCA | XM_036952756.1 |
| grin3bb1 | CTACTTCAGTGAGCGTGCCA | TACTCGAAGCGCATGTCCTC | XM_021614590.2 |
| grin3bb2 | GGATCCAGAATAGGCCTGCC | GAACACCCTCTTCCCACAGG | XM_036977945.1 |
| gria1b1 | GCCTTTCAGAACCTCCGGAA | CTGGATGTCGATACCCTGGC | XM_036989628.1 |
| gria1b2 | GCGTATTGACATTTCCCGGC | CCTCAATCCGAACCTGCTGT | XR_005035026.1 |
| grm1a | GCTGATCGAAAGTGTGGGGA | ATGTTGGGGAGCAGGAAAGG | XM_021600740.2 |
| grim2a | TGCATCGCCACTTCAGCTAA | TGCGTGTGAAGAGGATGACC | XM_036950357.1 |
| grim2b | GTGAGGGGAAGTGAGACAGC | GGGGTTCCGTGTGTTAGTGT | XM_021568769.2 |
| grm4a | CCATTTCATCTGGGTGGGCT | CCTCTGATGGACTGGCGTTT | XM_021609206.2 |
| grm4b | GTGCCAGAGACCTTCAACGA | GACTGCGAGGTCCCAAAGAA | XM_036988394.1 |
| grim5b | GGGCATCCTGTTTGACGAGA | TGTCCCAGCTCCCTACGTTA | XM_036961649.1 |
| Gabaergic markers | |||
| gabarap | CAGATGCACTTTCCCTCCCC | TCAACCGAAATCCCCATCTCG | NM_001165091.1 |
| gbrl2 | AGAGAGAGATGGGGATGGCT | AGGATGCAAGGGTTGTGTCA | NM_001165109.1 |
| gbrap | CTCACAGTGGGCCAGTTCTA | GAGGTGGGAGGAATGACGTT | NM_001165091.1 |
| gabat1 | GGTGATGGAGTTTTGGGAGC | TAAACCAGGACCCAAGCGAT | XM_021615563.1 |
| chat | CATCATCGTGGCATGCAAGA | AGTTCTCCGCCATCTTCACT | XM_021581165 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cardona, E.; Brunet, V.; Baranek, E.; Milhade, L.; Skiba-Cassy, S.; Bobe, J.; Calandreau, L.; Roy, J.; Colson, V. Physical Enrichment Triggers Brain Plasticity and Influences Blood Plasma Circulating miRNA in Rainbow Trout (Oncorhynchus mykiss). Biology 2022, 11, 1093. https://doi.org/10.3390/biology11081093
Cardona E, Brunet V, Baranek E, Milhade L, Skiba-Cassy S, Bobe J, Calandreau L, Roy J, Colson V. Physical Enrichment Triggers Brain Plasticity and Influences Blood Plasma Circulating miRNA in Rainbow Trout (Oncorhynchus mykiss). Biology. 2022; 11(8):1093. https://doi.org/10.3390/biology11081093
Chicago/Turabian StyleCardona, Emilie, Valentin Brunet, Elodie Baranek, Léo Milhade, Sandrine Skiba-Cassy, Julien Bobe, Ludovic Calandreau, Jérôme Roy, and Violaine Colson. 2022. "Physical Enrichment Triggers Brain Plasticity and Influences Blood Plasma Circulating miRNA in Rainbow Trout (Oncorhynchus mykiss)" Biology 11, no. 8: 1093. https://doi.org/10.3390/biology11081093
APA StyleCardona, E., Brunet, V., Baranek, E., Milhade, L., Skiba-Cassy, S., Bobe, J., Calandreau, L., Roy, J., & Colson, V. (2022). Physical Enrichment Triggers Brain Plasticity and Influences Blood Plasma Circulating miRNA in Rainbow Trout (Oncorhynchus mykiss). Biology, 11(8), 1093. https://doi.org/10.3390/biology11081093







