Twentieth-Century Paleoproteomics: Lessons from Venta Micena Fossils
Abstract
Simple Summary
Abstract
1. Introduction
2. Short Survival of Fossil DNA, Longer Survival of Fossil Proteins
3. Detection of Fossil Proteins with Immunological Methods: Applications in Paleontological Controversies
4. The Case of Orce Man
4.1. The Orce fossils
4.2. Methods to Investigate Proteins in Fossils
4.2.1. Preparation of Fossil Extracts
4.2.2. Immunoassays for the Detection of Albumin
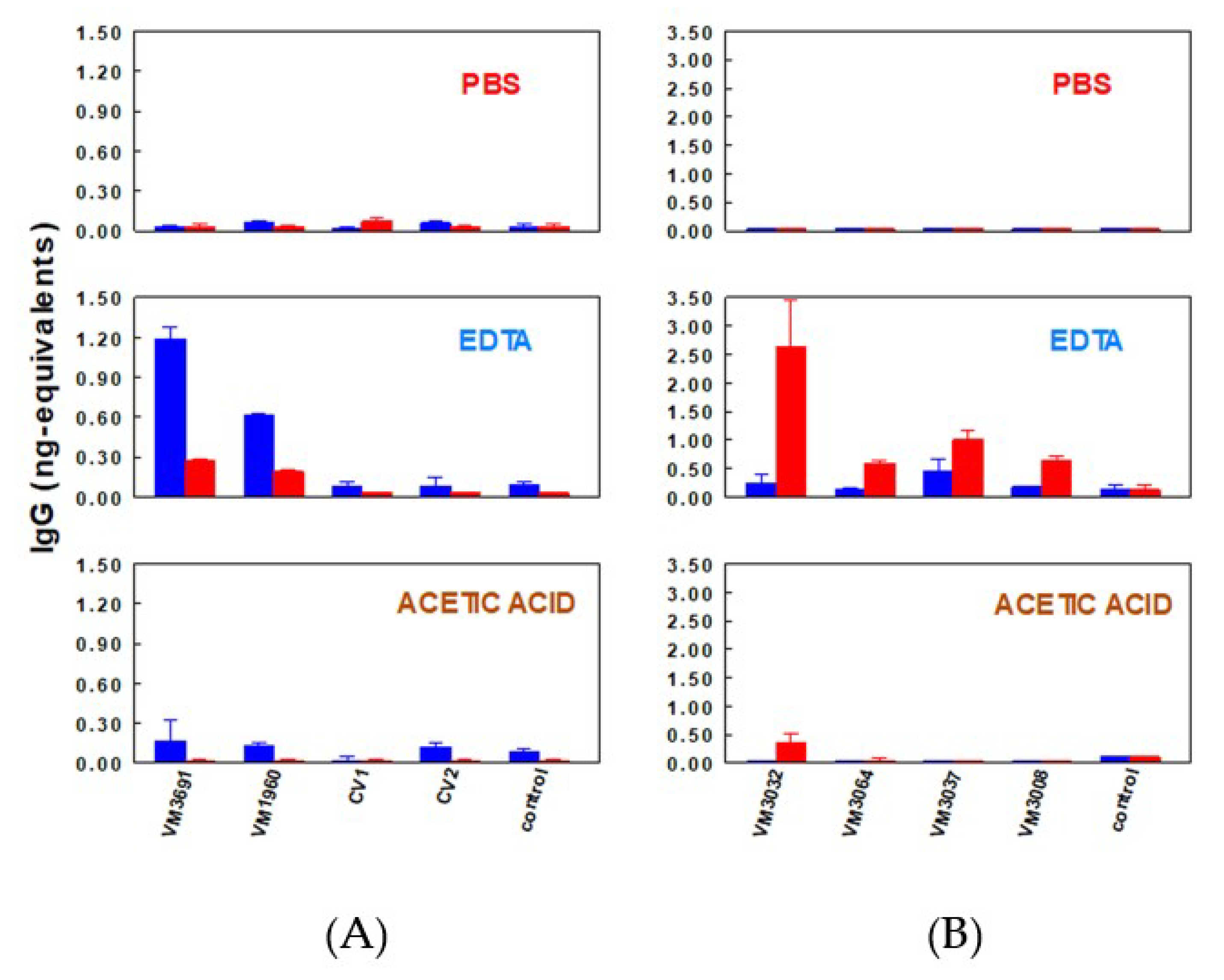
4.2.3. Detection of IgG by Quantitative Dot-Blotting
4.3. Detection of Albumin in Fossil Bones
4.4. Fossil Proteins or Contamination
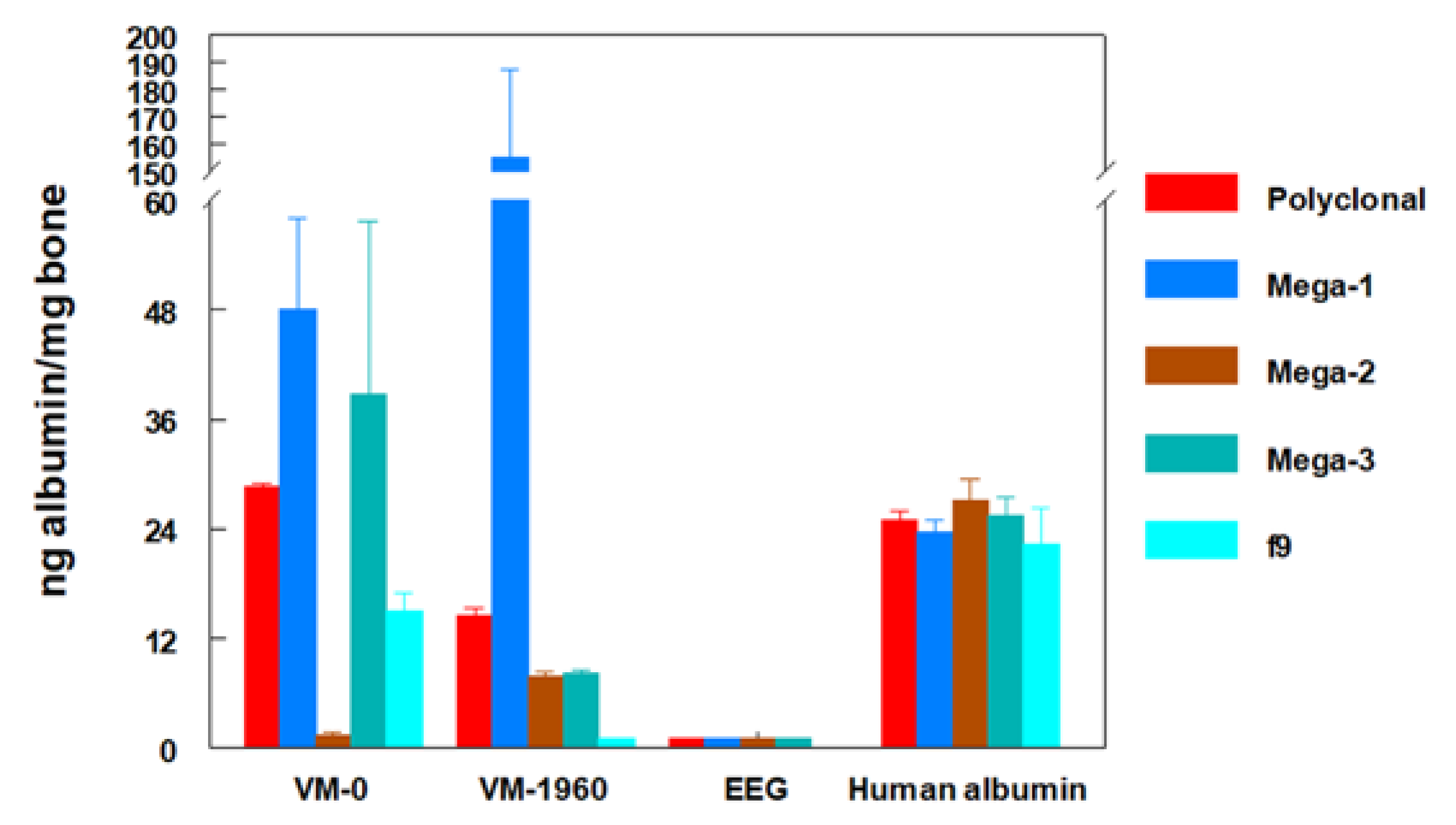
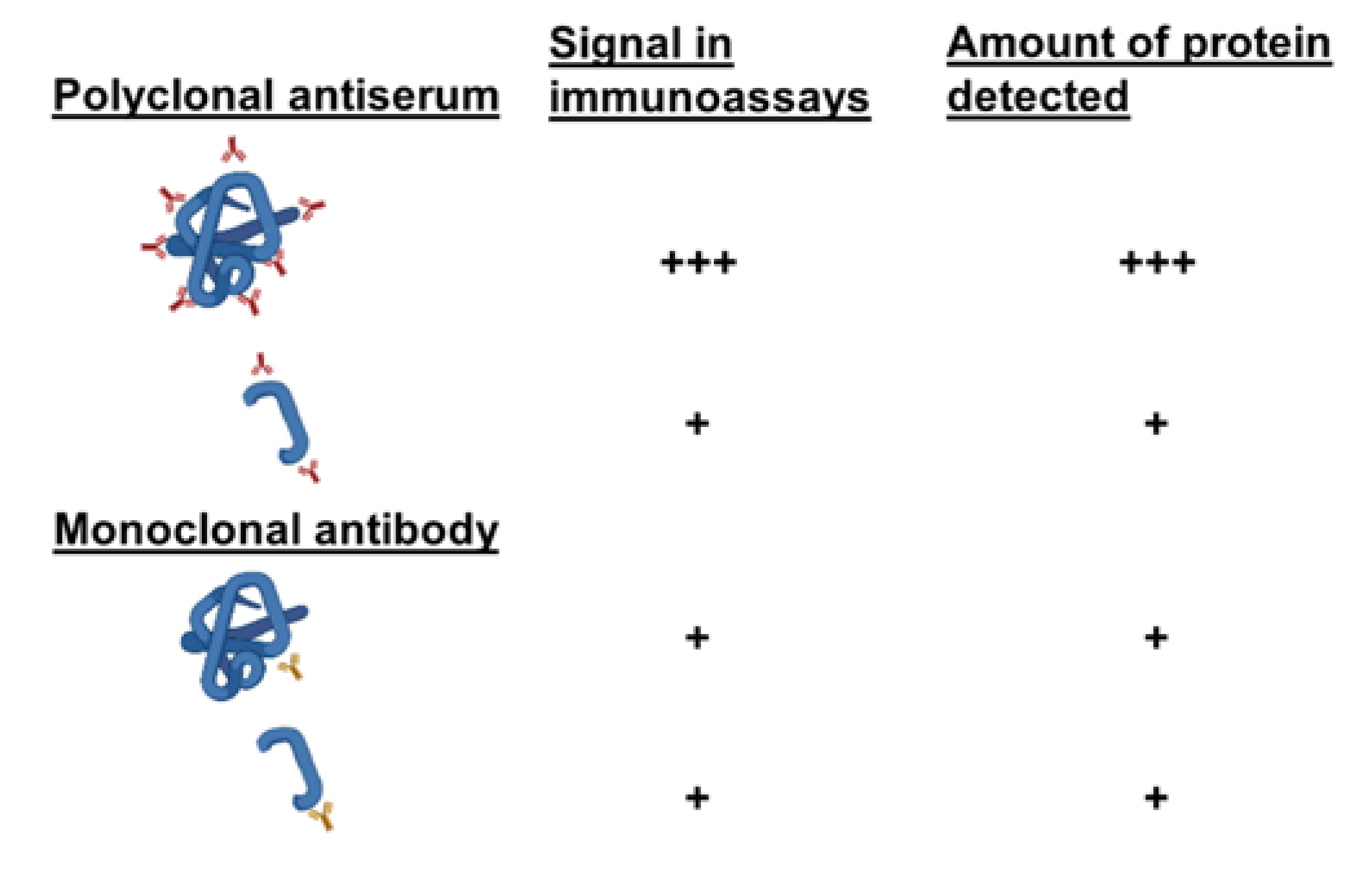
4.5. Monoclonal Antibodies to Study the Integrity of Fossil Proteins
4.6. Proteins Other than Collagen in Fossils
4.7. Twenty-First-Century Paleoproteomics and the Venta Micena Fossils
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| ELISA | Enzyme-linked immunosorbent assay |
| IgG | Immunoglobulin G |
| MS | Mass spectrometry |
| RIA | Radioimmunoassay |
References
- Wilson, A.C.; Cann, R.L. The recent African genesis of humans. Sci. Am. 1992, 266, 68–73. [Google Scholar] [CrossRef] [PubMed]
- Abelson, P. Organic Constituents of Fossils; Carnegie Institute of Washington Yearbook: Washington, DC, USA, 1954; Volume 53, p. 5. [Google Scholar]
- Wykoff, R.W.G. The Biochemistry of Animal Fossils; Scientechnica: Bristol, UK, 1972. [Google Scholar]
- Higuchi, R.; Bowman, B.; Freiberger, M.; Ryder, O.A.; Wilson, A.C. DNA sequences from the quagga, an extinct member of the horse family. Nature 1984, 312, 282–284. [Google Scholar] [CrossRef] [PubMed]
- Curry, G.B.; Eglinton, G. Molecules through time—Fossil molecules and biochemical systematics—Proceedings of a Royal-Society discussion meeting on Biomolecular Paleontology held on 20 and 21 march, 1991—Preface. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1991, 333, 311–312. [Google Scholar] [CrossRef]
- Hare, P.E.; Hoering, T.C.; King, K. (Eds.) Biogeochemistry of Amino Acids; John Wiley & Sons Inc: New York, NY, USA, 1980. [Google Scholar]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef]
- Lindahl, T. Facts and artifacts of ancient DNA. Cell 1997, 90, 1–3. [Google Scholar] [CrossRef]
- Prufer, K.; Racimo, F.; Patterson, N.; Jay, F.; Sankararaman, S.; Sawyer, S.; Heinze, A.; Renaud, G.; Sudmant, P.H.; de Filippo, C.; et al. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature 2014, 505, 43–49. [Google Scholar] [CrossRef]
- Meyer, M.; Fu, Q.; Aximu-Petri, A.; Glocke, I.; Nickel, B.; Arsuaga, J.L.; Martinez, I.; Gracia, A.; de Castro, J.M.; Carbonell, E.; et al. A mitochondrial genome sequence of a hominin from Sima de los Huesos. Nature 2014, 505, 403–406. [Google Scholar] [CrossRef]
- Orlando, L.; Ginolhac, A.; Zhang, G.; Froese, D.; Albrechtsen, A.; Stiller, M.; Schubert, M.; Cappellini, E.; Petersen, B.; Moltke, I.; et al. Recalibrating Equus evolution using the genome sequence of an early Middle Pleistocene horse. Nature 2013, 499, 74–78. [Google Scholar] [CrossRef]
- van der Valk, T.; Pecnerova, P.; Diez-del-Molino, D.; Bergstrom, A.; Oppenheimer, J.; Hartmann, S.; Xenikoudakis, G.; Thomas, J.A.; Dehasque, M.; Saglican, E.; et al. Million-year-old DNA sheds light on the genomic history of mammoths. Nature 2021, 591, 265–269. [Google Scholar] [CrossRef]
- Woodward, S.R.; Weyand, N.J.; Bunnell, M. DNA sequence from Cretaceous period bone fragments. Science 1994, 266, 1229–1232. [Google Scholar] [CrossRef] [PubMed]
- Zischler, H.; Hoss, M.; Handt, O.; von Haeseler, A.; van der Kuyl, A.C.; Goudsmit, J.; Pääbo, S. Detecting dinosaur DNA. Science 1995, 268, 1192–1193. [Google Scholar] [CrossRef] [PubMed]
- Herman, A.; Addadi, L.; Weiner, S. Interactions of sea-urchin skeleton macromolecules with growing calcite crystals—A study of intracrystalline proteins. Nature 1988, 331, 546–548. [Google Scholar] [CrossRef]
- Smith, A.J.; Matthews, J.B.; Wilson, C.; Sewell, H.F. Plasma proteins in human cortical bone: In vitro binding studies. Calcif. Tissue Int. 1985, 37, 208–210. [Google Scholar] [CrossRef]
- Buus, S.; Rockberg, J.; Forsstrom, B.; Nilsson, P.; Uhlen, M.; Schafer-Nielsen, C. High-resolution mapping of linear antibody epitopes using ultrahigh-density peptide microarrays. Mol. Cell. Proteom. 2012, 11, 1790–1800. [Google Scholar] [CrossRef]
- Graves, P.R.; Haystead, T.A. Molecular biologist’s guide to proteomics. Microbiol. Mol. Biol. Rev. 2002, 66, 39–63. [Google Scholar] [CrossRef]
- Olivares, E.G. Not a first: Identifying hominin fossils from their proteins. Nature 2019, 573, 196. [Google Scholar] [CrossRef]
- Lowenstein, J.M. Immunological reactions from fossil material. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1981, 292, 143–149. [Google Scholar] [CrossRef] [PubMed]
- Lowenstein, J.M.; Scheuenstuhl, G. Immunological methods in molecular palaeontology. Philos. Trans. R. Soc. Lond. B Biol. Sci. 1991, 333, 375–380. [Google Scholar] [CrossRef]
- Rowley, M.J.; Rich, P.V.; Rich, T.H.; Mackay, I.R. Immunoreactive collagen in avian and mammalian fossils. Naturwissenschaften 1986, 73, 620–623. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, M.M.; Perizonius, W.R.; Spoor, C.F.; Sandberg, P.; Vermeer, C. Extraction of osteocalcin from fossil bones and teeth. Biochem. Biophys. Res. Commun. 1987, 149, 712–719. [Google Scholar] [CrossRef]
- Muyzer, G.; Sandberg, P.; Knapen, M.H.J.; Vermeer, C.; Collins, M.; Westbroek, P. Preservation of the bone protein osteocalcin in dinosaurs. Geology 1992, 20, 871–874. [Google Scholar] [CrossRef]
- Sarich, V.M.; Wilson, A.C. Immunological time scale for hominid evolution. Science 1967, 158, 1200–1203. [Google Scholar] [CrossRef]
- Lowenstein, J.M. Fossil proteins and evolutionary time. Pontif. Acad. Sci. Scr. Var. 1983, 50, 151–162. [Google Scholar]
- Lewin, R. Bones of Contention: Controversies in the Search for Human Origins, 2nd ed.; The University of Chicago Press: Chicago, IL, USA, 1987. [Google Scholar]
- Lowenstein, J.M.; Molleson, T.; Washburn, S.L. Piltdown jaw confirmed as orang. Nature 1982, 299, 294. [Google Scholar] [CrossRef]
- Gibert, J.; Ribot, F.; Ferrandez, C.; Martinez, B.; Caporicci, R.; Campillo, D. Anatomical study: Comparison of the cranial fragment from Venta Micena, (Orce; Spain) with fossil and extant mammals. Hum. Evol. 1989, 4, 283–305. [Google Scholar] [CrossRef]
- Scott, G.R.; Gibert, L.; Gibert, J. Magnetostratigraphy of the Orce Region (Baza Basin), SE Spain: New chronologies for Early Pleistocene faunas and hominid occupation sites. Quat. Sci. Rev. 2007, 26, 415–435. [Google Scholar] [CrossRef]
- Gibert, J.; Gibert Beotas, L.; Ferràndez-Cañadell, C.; Iglesias; González, F. Venta Micena, Barranco León-5 and Fuentenueva-3: Three archaeological sites in the Early Pleistocene deposits of Orce, south-east Spain. In The Human Evolution Source Book; Ciochon, R.L., Fleagle, J.G., Eds.; Pearson Prentice Hall: Englewood Cliffs, NJ, USA, 2006; pp. 327–335. [Google Scholar]
- Gibert, J.; Gibert, L.; Albadalejo, S.; Ribot, F.; Sánchez, F.; Gibert, P. Molar tooth fragment BL5-0: The oldest human remain found in the Plio-Pleistocene of Orce (Granada province, Spain). Hum. Evol. 1999, 14, 3–19. [Google Scholar] [CrossRef]
- Ribot, F.; Gibert, L.; Ferrandez-Canadell, C.; Garcia Olivares, E.; Sanchez, F.; Leria, M. Two Deciduous Human Molars from the Early Pleistocene Deposits of Barranco Leon (Orce, Spain). Curr. Anthropol. 2015, 56, 134–142. [Google Scholar] [CrossRef]
- Toro-Moyano, I.; Martinez-Navarro, B.; Agusti, J.; Souday, C.; Bermudez de Castro, J.M.; Martinon-Torres, M.; Fajardo, B.; Duval, M.; Falgueres, C.; Oms, O.; et al. The oldest human fossil in Europe, from Orce (Spain). J. Hum. Evol. 2013, 65, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Barsky, D.; Titton, S.; Sala-Ramos, R.; Bargalló, A.; Grégoire, S.; Saos, T.; Serrano-Ramos, A.; Oms, O.; Solano García, J.-A.; Toro-Moyano, I.; et al. The Significance of Subtlety: Contrasting Lithic Raw Materials Procurement and Use Patterns at the Oldowan Sites of Barranco León and Fuente Nueva 3 (Orce, Andalusia, Spain). Front. Earth Sci. 2022, 10, 893776. [Google Scholar] [CrossRef]
- Gibert, J.; Gibert, L.; Iglesias, A.; Maestro, E. Two ‘Oldowan’ assemblages in the Plio-Pleistocene deposits of the Orce region, southeast Spain. Antiquity 1998, 72, 17–25. [Google Scholar] [CrossRef]
- Alba, D.M. A fistful of fossils: The rise and fall of the Orce Man and the politics of paleoanthropological science. J. Hum. Evol. 2022, 165, 103166. [Google Scholar] [CrossRef]
- Gibert, J.; Campillo, D.; Arques, J.M.; Garcia-Olivares, E.; Borja, C.; Lowenstein, J.M. Hominid status of the Orce cranial fragment reasserted. J. Hum. Evol. 1998, 34, 203–217. [Google Scholar] [CrossRef]
- Agusti, J.; Moya-Sola, S. On the identity of the cranial fragment attributed to homo-sp in Venta Micena, Granada, Spain. Estud. Geol.-Madr. 1987, 43, 535–538. [Google Scholar]
- Martinez-Navarro, B. The skull of Orce: Parietal bones or frontal bones? J. Hum. Evol. 2002, 43, 265–270. [Google Scholar] [CrossRef]
- Tobias, P.V. Some comments on the case for Early Pleistocene hominids in South-Eastern Spain. Hum. Evol. 2006, 13, 91–96. [Google Scholar] [CrossRef]
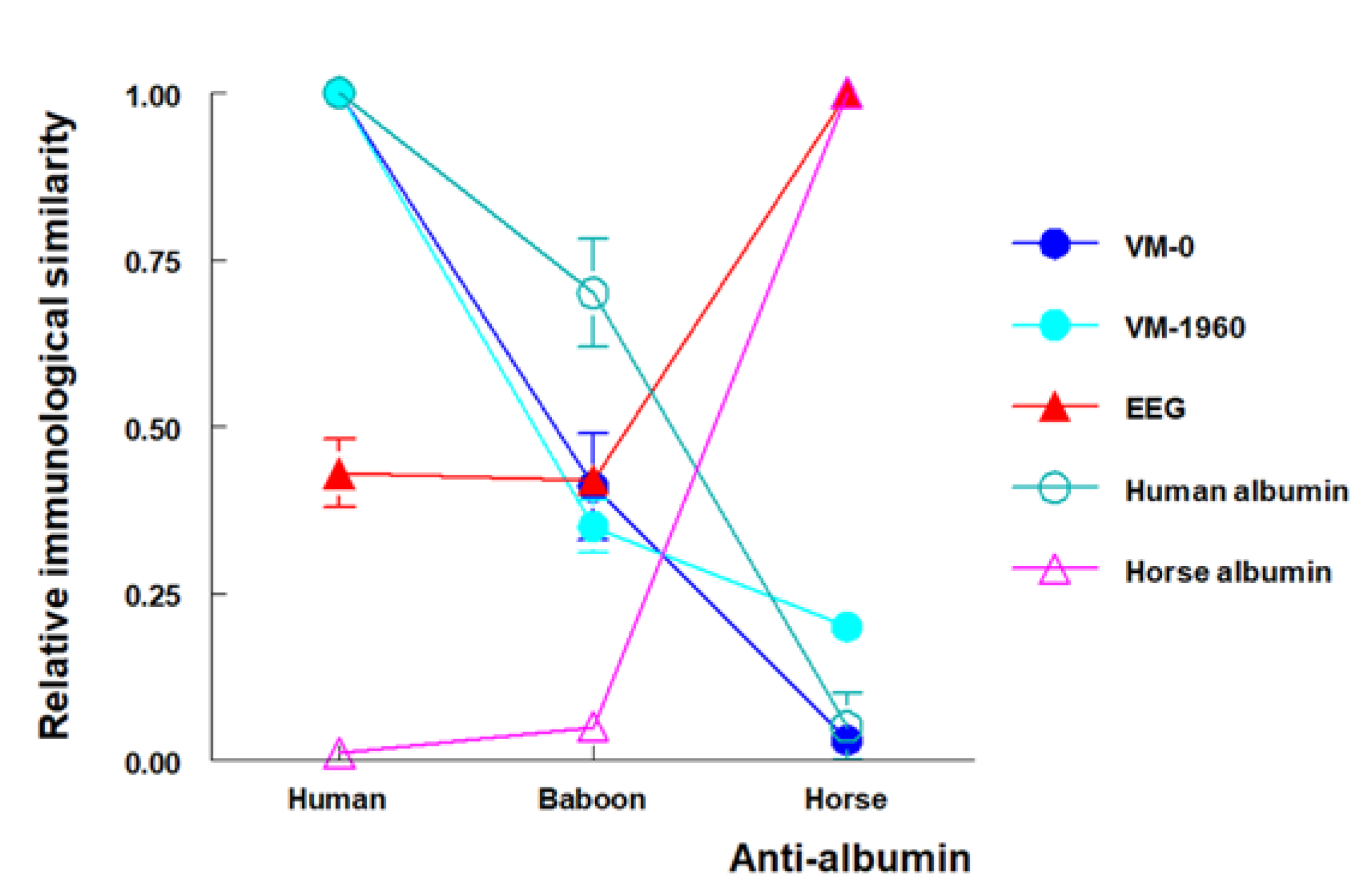
- Borja, C.; Garcia-Pacheco, M.; Olivares, E.G.; Scheuenstuhl, G.; Lowenstein, J.M. Immunospecificity of albumin detected in 1.6 million-year-old fossils from Venta Micena in Orce, Granada, Spain. Am. J. Phys. Anthropol. 1997, 103, 433–441. [Google Scholar] [CrossRef]
- Gibert, L.; Scott, G.R.; Scholz, D.; Budsky, A.; Ferrandez, C.; Ribot, F.; Martin, R.A.; Leria, M. Chronology for the Cueva Victoria fossil site (SE Spain): Evidence for Early Pleistocene Afro-Iberian dispersals. J. Hum. Evol. 2016, 90, 183–197. [Google Scholar] [CrossRef]
- Kooyman, B.; Newman, M.E.; Ceri, H. Verifying the reliability of blood residue analysis on archaeological tools. J. Archaeol. Sci. 1992, 19, 265–269. [Google Scholar] [CrossRef]
- Lowenstein, J.M.; Borja, C.; García-Olivares, E. Species-specific albumin in fossil bones from Orce, Granada, Spain. Hum. Evol. 1999, 14, 21–28. [Google Scholar] [CrossRef]
- Cattaneo, C.; Gelsthorpe, K.; Phillips, P.; Sokol, R.J. Blood residues on stone tools: Indoor and outdoor experiments. World Archaeol. 1993, 25, 29–43. [Google Scholar] [CrossRef]
- Palmqvist, P. A critical re-evaluation of the evidence for the presence of hominids in Lower Pleistocene times at Venta Micena, Southern Spain. J. Hum. Evol. 1997, 33, 83–89. [Google Scholar] [CrossRef]
- Poinar, G.O. Recovery of antediluvian DNA. Nature 1993, 365, 700. [Google Scholar] [CrossRef]
- Gibert, L.; Orti, F.; Rosell, L. Plio-Pleistocene lacustrine evaporites of the Baza Basin (Betic Chain, SE Spain). Sediment. Geol. 2007, 200, 89–116. [Google Scholar] [CrossRef]
- Singh, S.; Kumar, N.K.; Dwiwedi, P.; Charan, J.; Kaur, R.; Sidhu, P.; Chugh, V.K. Monoclonal Antibodies: A Review. Curr. Clin. Pharmacol. 2018, 13, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Torres, J.M.; Borja, C.; Olivares, E.G. Immunoglobulin G in 1.6 million-year-old fossil bones from Venta Micena (Granada, Spain). J. Archaeol. Sci. 2002, 29, 167–175. [Google Scholar] [CrossRef][Green Version]
- Tuross, N. Albumin preservation in the Taima-taima mastodon skeleton. Appl. Geochem. 1989, 4, 255–259. [Google Scholar] [CrossRef]
- Tuross, N.; Behrensmeyer, A.K.; Eanes, E.D.; Fisher, L.W.; Hare, P.E. Molecular preservation and crystallographic alterations in a weathering sequence of wildebeest bones. Appl. Geochem. 1989, 4, 261–270. [Google Scholar] [CrossRef]
- Wilson, A.C.; Carlson, S.S.; White, T.J. Biochemical evolution. Ann. Rev. Biochem. 1977, 46, 573–639. [Google Scholar] [CrossRef]
- Lowenstein, J.M.; Ryder, O.A. Immunological systematics of the extinct quagga (equidae). Experientia 1985, 41, 1192–1193. [Google Scholar] [CrossRef]
- Lowenstein, J.M.; Sarich, V.M.; Richardson, B.J. Albumin systematics of the extinct mammoth and tasmanian wolf. Nature 1981, 291, 409–411. [Google Scholar] [CrossRef] [PubMed]
- Rainey, W.E.; Lowenstein, J.M.; Sarich, V.M.; Magor, D.M. Sirenian molecular systematics including the extinct stellers sea cow (Hydrodamalis-gigas). Naturwissenschaften 1984, 71, 586–588. [Google Scholar] [CrossRef] [PubMed]
- Shoshani, J.; Lowenstein, J.M.; Walz, D.A.; Goodman, M. Proboscidean origins of mastodon and woolly mammoth demonstrated immunologically. Paleobiology 1985, 11, 429–437. [Google Scholar] [CrossRef]
- Thomas, R.H.; Schaffner, W.; Wilson, A.C.; Paabo, S. DNA phylogeny of the extinct marsupial wolf. Nature 1989, 340, 465–467. [Google Scholar] [CrossRef] [PubMed]
- Pawlicki, R.; Nowogrodzka-Zagorska, M. Blood vessels and red blood cells preserved in dinosaur bones. Ann. Anat. -Anat. Anz. 1998, 180, 73–77. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Johnson, C.; Zocco, T.G.; Horner, J.R.; Starkey, J.R. Preservation of biomolecules in cancellous bone of Tyrannosaurus rex. J. Vertebr. Paleontol. 1997, 17, 349–359. [Google Scholar] [CrossRef]
- Montgelard, C. Albumin preservation in fossil bones and systematics of Malpaisomys insularis (Muridae, Rodentia), an extinct rodent of the Canary Islands. Hist. Biol. 1992, 6, 293–302. [Google Scholar] [CrossRef]
- Welker, F.; Ramos-Madrigal, J.; Gutenbrunner, P.; Mackie, M.; Tiwary, S.; Rakownikow Jersie-Christensen, R.; Chiva, C.; Dickinson, M.R.; Kuhlwilm, M.; de Manuel, M.; et al. The dental proteome of Homo antecessor. Nature 2020, 580, 235–238. [Google Scholar] [CrossRef]
- Cappellini, E.; Jensen, L.J.; Szklarczyk, D.; Ginolhac, A.; da Fonseca, R.A.; Stafford, T.W.; Holen, S.R.; Collins, M.J.; Orlando, L.; Willerslev, E.; et al. Proteomic analysis of a pleistocene mammoth femur reveals more than one hundred ancient bone proteins. J. Proteome Res. 2012, 11, 917–926. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Marshall, M.; Carron, K.; Bohle, D.S.; Arnold, E.V.; Barnard, D.; Horner, J.R.; Starkey, J.R. Heme compounds in dinosaur trabecular bone. Proc. Natl. Acad. Sci. USA 1997, 94, 6291–6296. [Google Scholar] [CrossRef]
- Schweitzer, M.H.; Chiappe, L.; Garrido, A.C.; Lowenstein, J.M.; Pincus, S.H. Molecular preservation in Late Cretaceous sauropod dinosaur eggshells. Proc. Biol. Sci. 2005, 272, 775–784. [Google Scholar] [CrossRef] [PubMed]
- Borja, C. Detección y Caracterización de Proteínas Fósiles Mediante Técnicas Inmunes. Ph.D. Thesis, Universidad de Granada, Granada, Spain, 1995. [Google Scholar]
- Poinar, H.N.; Hofreiter, M.; Spaulding, W.G.; Martin, P.S.; Stankiewicz, B.A.; Bland, H.; Evershed, R.P.; Possnert, G.; Paabo, S. Molecular coproscopy: Dung and diet of the extinct ground sloth Nothrotheriops shastensis. Science 1998, 281, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Buckley, M. Paleoproteomics: An Introduction to the Analysis of Ancient Proteins by Soft Ionisation Mass Spectrometry. In Paleogenomics: Genome-Scale Analysis of Ancient DNA; Lindqvist, C., Rajora, O.P., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 31–52. [Google Scholar]
- Cleland, T.P.; Schroeter, E.R. A Comparison of Common Mass Spectrometry Approaches for Paleoproteomics. J. Proteome Res. 2018, 17, 936–945. [Google Scholar] [CrossRef] [PubMed]
- Asara, J.M.; Schweitzer, M.H.; Freimark, L.M.; Phillips, M.; Cantley, L.C. Protein sequences from mastodon and Tyrannosaurus rex revealed by mass spectrometry. Science 2007, 316, 280–285. [Google Scholar] [CrossRef] [PubMed]
- Schroeter, E.R.; Cleland, T.P.; Schweitzer, M.H. Deep Time Paleoproteomics: Looking Forward. J. Proteome Res. 2022, 21, 9–19. [Google Scholar] [CrossRef]
- Low, T.Y.; Syafruddin, S.E.; Mohtar, M.A.; Vellaichamy, A.; ARahman, N.S.; Pung, Y.F.; Tan, C.S.H. Recent progress in mass spectrometry-based strategies for elucidating protein-protein interactions. Cell. Mol. Life Sci. 2021, 78, 5325–5339. [Google Scholar] [CrossRef]
- ten Have, S.; Boulon, S.; Ahmad, Y.; Lamond, A.I. Mass spectrometry-based immuno-precipitation proteomics—the user’s guide. Proteomics 2011, 11, 1153–1159. [Google Scholar] [CrossRef] [PubMed]
- Brown, S.; Higham, T.; Slon, V.; Pääbo, S.; Meyer, M.; Douka, K.; Brock, F.; Comeskey, D.; Procopio, N.; Shunkov, M.; et al. Identification of a new hominin bone from Denisova Cave, Siberia using collagen fingerprinting and mitochondrial DNA analysis. Sci. Rep. 2016, 6, 23559. [Google Scholar] [CrossRef]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Torres, J.M.; Borja, C.; Gibert, L.; Ribot, F.; Olivares, E.G. Twentieth-Century Paleoproteomics: Lessons from Venta Micena Fossils. Biology 2022, 11, 1184. https://doi.org/10.3390/biology11081184
Torres JM, Borja C, Gibert L, Ribot F, Olivares EG. Twentieth-Century Paleoproteomics: Lessons from Venta Micena Fossils. Biology. 2022; 11(8):1184. https://doi.org/10.3390/biology11081184
Chicago/Turabian StyleTorres, Jesús M., Concepción Borja, Luis Gibert, Francesc Ribot, and Enrique G. Olivares. 2022. "Twentieth-Century Paleoproteomics: Lessons from Venta Micena Fossils" Biology 11, no. 8: 1184. https://doi.org/10.3390/biology11081184
APA StyleTorres, J. M., Borja, C., Gibert, L., Ribot, F., & Olivares, E. G. (2022). Twentieth-Century Paleoproteomics: Lessons from Venta Micena Fossils. Biology, 11(8), 1184. https://doi.org/10.3390/biology11081184






