Using DeepLabCut as a Real-Time and Markerless Tool for Cardiac Physiology Assessment in Zebrafish
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Zebrafish Maintenance
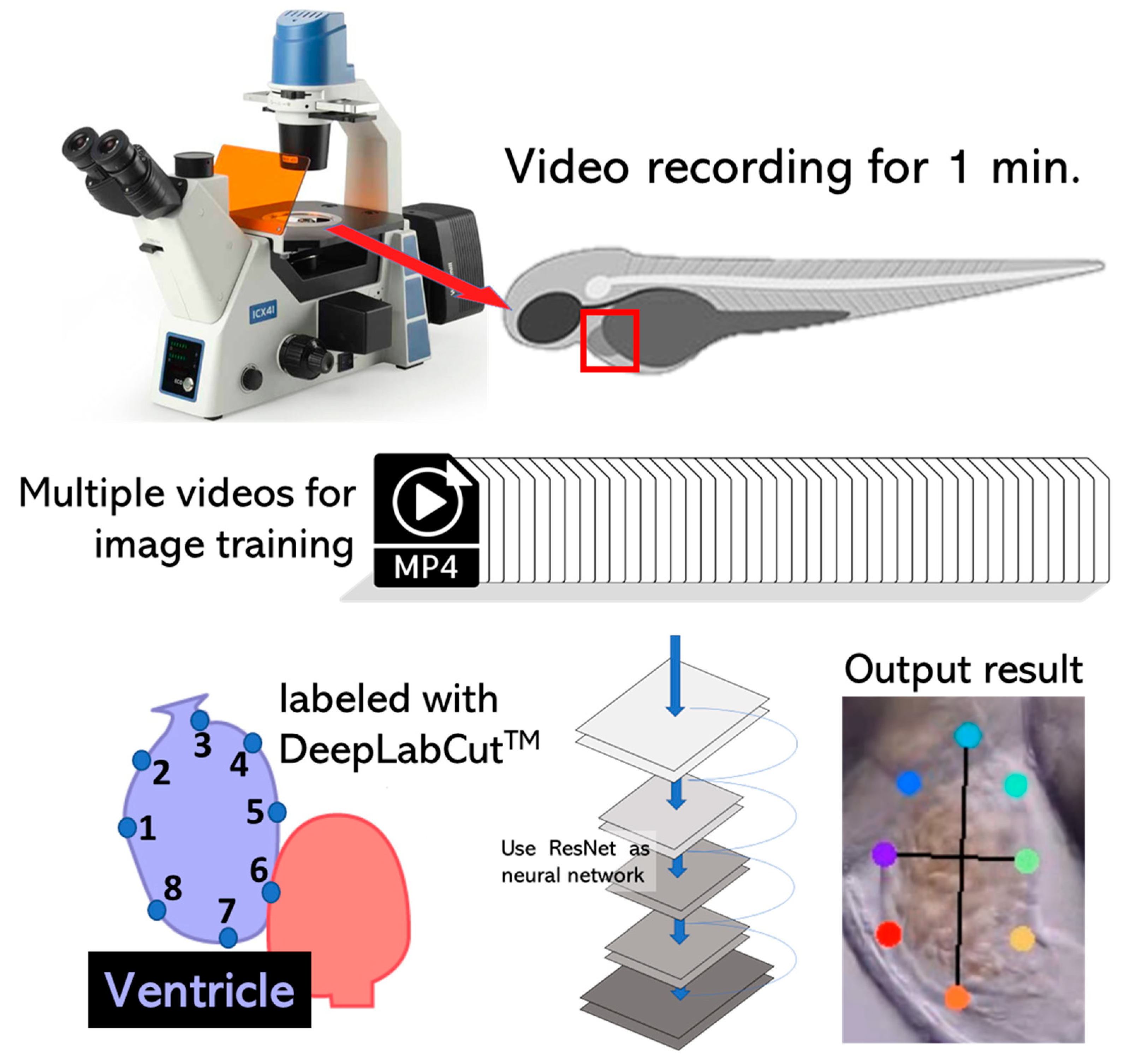
2.2. High-Definition Videography
2.3. DeepLabCut Training
2.4. Cardiac Parameter Calculation
2.5. Data Validation with ImageJ and Kymograph
2.6. Chemical Exposure to Induce Cardiac Abnormality
2.7. Heart Rate Variability Measurement by Poincaré Plot
2.8. Statistics
3. Results
3.1. Overview of Experimental Design
3.2. DeepLabCut Training for Zebrafish
3.3. Cardiac Physiology Comparison between DLC and ImageJ in Control Animals
3.4. Cardiac Physiology Assessment in Zebrafish Embryos after Chemical Treatment
4. Discussion
4.1. Advantages and Limitations
4.2. Cardiac Physiology Comparison between DLC and ImageJ Methods
4.3. Comparison of the Cardiac Parameters between Control and Chemical-Treated Zebrafish
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A



References
- Kanungo, J.; Cuevas, E.; Ali, S.F.; Paule, M.G. Zebrafish model in drug safety assessment. Curr. Pharm. Des. 2014, 20, 5416–5429. [Google Scholar] [CrossRef] [PubMed]
- Hussain, A.; Audira, G.; Malhotra, N.; Uapipatanakul, B.; Chen, J.-R.; Lai, Y.-H.; Huang, J.-C.; Chen, K.H.-C.; Lai, H.-T.; Hsiao, C.-D. Multiple screening of pesticides toxicity in zebrafish and daphnia based on locomotor activity alterations. Biomolecules 2020, 10, 1224. [Google Scholar] [CrossRef] [PubMed]
- Lessman, C.A. The developing zebrafish (Danio rerio): A vertebrate model for high-throughput screening of chemical libraries. Birth Defects Res. Part C Embryo Today Rev. 2011, 93, 268–280. [Google Scholar] [CrossRef]
- Kurnia, K.A.; Saputra, F.; Roldan, M.J.M.; Castillo, A.L.; Huang, J.-C.; Chen, K.H.-C.; Lai, H.-T.; Hsiao, C.-D. Measurement of Multiple Cardiac Performance Endpoints in Daphnia and Zebrafish by Kymograph. Inventions 2021, 6, 8. [Google Scholar] [CrossRef]
- Santoso, F.; Farhan, A.; Castillo, A.L.; Malhotra, N.; Saputra, F.; Kurnia, K.A.; Chen, K.H.-C.; Huang, J.-C.; Chen, J.-R.; Hsiao, C.-D. An overview of methods for cardiac rhythm detection in Zebrafish. Biomedicines 2020, 8, 329. [Google Scholar]
- De Luca, E.; Zaccaria, G.M.; Hadhoud, M.; Rizzo, G.; Ponzini, R.; Morbiducci, U.; Santoro, M.M. ZebraBeat: A flexible platform for the analysis of the cardiac rate in zebrafish embryos. Sci. Rep. 2014, 4, 4898. [Google Scholar] [CrossRef] [Green Version]
- Pylatiuk, C.; Sanchez, D.; Mikut, R.; Alshut, R.; Reischl, M.; Hirth, S.; Rottbauer, W.; Just, S. Automatic zebrafish heartbeat detection and analysis for zebrafish embryos. Zebrafish 2014, 11, 379–383. [Google Scholar] [CrossRef] [Green Version]
- Burns, C.G.; Milan, D.J.; Grande, E.J.; Rottbauer, W.; MacRae, C.A.; Fishman, M.C. High-throughput assay for small molecules that modulate zebrafish embryonic heart rate. Nat. Chem. Biol. 2005, 1, 263–264. [Google Scholar] [CrossRef]
- Zhong, K.; Meng, Y.; Wu, J.; Wei, Y.; Huang, Y.; Ma, J.; Lu, H. Effect of flupyradifurone on zebrafish embryonic development. Environ. Pollut. 2021, 285, 117323. [Google Scholar] [CrossRef]
- Santoso, F.; Sampurna, B.P.; Lai, Y.-H.; Liang, S.-T.; Hao, E.; Chen, J.-R.; Hsiao, C.-D. Development of a simple imagej-based method for dynamic blood flow tracking in zebrafish embryos and its application in drug toxicity evaluation. Inventions 2019, 4, 65. [Google Scholar] [CrossRef] [Green Version]
- Langlotz, C.P.; Allen, B.; Erickson, B.J.; Kalpathy-Cramer, J.; Bigelow, K.; Cook, T.S.; Flanders, A.E.; Lungren, M.P.; Mendelson, D.S.; Rudie, J.D. A roadmap for foundational research on artificial intelligence in medical imaging: From the 2018 NIH/RSNA/ACR/The Academy Workshop. Radiology 2019, 291, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Rogers, M.A.; Aikawa, E. Cardiovascular calcification: Artificial intelligence and big data accelerate mechanistic discovery. Nat. Rev. Cardiol. 2019, 16, 261–274. [Google Scholar] [CrossRef]
- Teixidó, E.; Kießling, T.R.; Krupp, E.; Quevedo, C.; Muriana, A.; Scholz, S. Automated morphological feature assessment for zebrafish embryo developmental toxicity screens. Toxicol. Sci. 2019, 167, 438–449. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Li, G.; Wu, D.; Liu, Y.; Greaves, J.R.; Hao, W.; Krakauer, N.J.; Krudy, L.; Perez, J.; Sreenivasan, V. Multi defect detection and analysis of electron microscopy images with deep learning. Comput. Mater. Sci. 2021, 199, 110576. [Google Scholar] [CrossRef]
- Naderi, A.M.; Bu, H.; Su, J.; Huang, M.-H.; Vo, K.; Torres, R.S.T.; Chiao, J.-C.; Lee, J.; Lau, M.P.; Xu, X. Deep learning-based framework for cardiac function assessment in embryonic zebrafish from heart beating videos. Comput. Biol. Med. 2021, 135, 104565. [Google Scholar] [CrossRef]
- Nasrat, S.; Marcato, D.; Hirth, S.; Reischl, M.; Pylatiuk, C. Semi-automated detection of fractional shortening in zebrafish embryo heart videos. Curr. Dir. Biomed. Eng. 2016, 2, 233–236. [Google Scholar] [CrossRef]
- Spomer, W.; Pfriem, A.; Alshut, R.; Just, S.; Pylatiuk, C. High-throughput screening of zebrafish embryos using automated heart detection and imaging. J. Lab. Autom. 2012, 17, 435–442. [Google Scholar] [CrossRef] [Green Version]
- Farhan, A.; Kurnia, K.A.; Saputra, F.; Chen, K.H.-C.; Huang, J.-C.; Roldan, M.J.M.; Lai, Y.-H.; Hsiao, C.-D. An OpenCV-Based Approach for Automated Cardiac Rhythm Measurement in Zebrafish from Video Datasets. Biomolecules 2021, 11, 1476. [Google Scholar] [CrossRef]
- Akerberg, A.A.; Burns, C.E.; Burns, C.G.; Nguyen, C. Deep learning enables automated volumetric assessments of cardiac function in zebrafish. Dis. Models Mech. 2019, 12, dmm040188. [Google Scholar] [CrossRef] [Green Version]
- Xing, Q.; Huynh, V.; Parolari, T.G.; Maurer-Morelli, C.V.; Peixoto, N.; Wei, Q. Zebrafish larvae heartbeat detection from body deformation in low resolution and low frequency video. Med. Biol. Eng. Comput. 2018, 56, 2353–2365. [Google Scholar] [CrossRef]
- Labuguen, R.; Bardeloza, D.K.; Negrete, S.B.; Matsumoto, J.; Inoue, K.; Shibata, T. Primate markerless pose estimation and movement analysis using DeepLabCut. In Proceedings of the 2019 Joint 8th International Conference on Informatics, Electronics & Vision (ICIEV) and 2019 3rd International Conference on Imaging, Vision & Pattern Recognition (icIVPR), Spokane, WA, USA, 30 May–2 June 2019; pp. 297–300. [Google Scholar]
- Mathis, A.; Mamidanna, P.; Cury, K.M.; Abe, T.; Murthy, V.N.; Mathis, M.W.; Bethge, M. DeepLabCut: Markerless pose estimation of user-defined body parts with deep learning. Nat. Neurosci. 2018, 21, 1281–1289. [Google Scholar] [CrossRef] [PubMed]
- Forys, B.J.; Xiao, D.; Gupta, P.; Murphy, T.H. Real-time selective markerless tracking of forepaws of head fixed mice using deep neural networks. Eneuro 2020, 7, 1–14. [Google Scholar] [CrossRef]
- Wu, S.; Tan, K.J.; Govindarajan, L.N.; Stewart, J.C.; Gu, L.; Ho, J.W.H.; Katarya, M.; Wong, B.H.; Tan, E.-K.; Li, D. Fully automated leg tracking of Drosophila neurodegeneration models reveals distinct conserved movement signatures. PLoS Biol. 2019, 17, e3000346. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fujimori, S.; Ishikawa, T.; Watanabe, H. Animal Behavior Classification Using DeepLabCut. In Proceedings of the 2020 IEEE 9th Global Conference on Consumer Electronics (GCCE), Kobe, Japan, 13–16 October 2020; pp. 254–257. [Google Scholar]
- Nath, T.; Mathis, A.; Chen, A.C.; Patel, A.; Bethge, M.; Mathis, M.W. Using DeepLabCut for 3D markerless pose estimation across species and behaviors. Nat. Protoc. 2019, 14, 2152–2176. [Google Scholar] [CrossRef] [PubMed]
- Avdesh, A.; Chen, M.; Martin-Iverson, M.T.; Mondal, A.; Ong, D.; Rainey-Smith, S.; Taddei, K.; Lardelli, M.; Groth, D.M.; Verdile, G. Regular care and maintenance of a zebrafish (Danio rerio) laboratory: An introduction. JoVE (J. Vis. Exp.) 2012, 69, e4196. [Google Scholar] [CrossRef] [Green Version]
- Jardim, S.; António, J.; Mora, C. Graphical Image Region Extraction with K-Means Clustering and Watershed. J. Imaging 2022, 8, 163. [Google Scholar] [CrossRef]
- Insafutdinov, E.; Pishchulin, L.; Andres, B.; Andriluka, M.; Schiele, B. Deepercut: A deeper, stronger, and faster multi-person pose estimation model. In Proceedings of the European Conference on Computer Vision, Amsterdam, The Netherlands, 11–14 October 2016; pp. 34–50. [Google Scholar]
- Santoso, F.; Krylov, V.V.; Castillo, A.L.; Saputra, F.; Chen, H.-M.; Lai, H.-T.; Hsiao, C.-D. Cardiovascular performance measurement in water fleas by utilizing high-speed videography and ImageJ software and its application for pesticide toxicity assessment. Animals 2020, 10, 1587. [Google Scholar] [CrossRef]
- Sampurna, B.P.; Audira, G.; Juniardi, S.; Lai, Y.-H.; Hsiao, C.-D. A simple imagej-based method to measure cardiac rhythm in zebrafish embryos. Inventions 2018, 3, 21. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Gao, A.; Wang, Y.; Chen, M.; Peng, J.; Yan, H.; Zhao, X.; Feng, X.; Chen, D. Alcohol exposure leads to unrecoverable cardiovascular defects along with edema and motor function changes in developing zebrafish larvae. Biol. Open 2016, 5, 1128–1133. [Google Scholar] [CrossRef] [Green Version]
- Cheng, S.; Jin, P.; Li, H.; Pei, D.; Shu, X. Evaluation of CML TKI Induced cardiovascular toxicity and development of potential rescue strategies in a zebrafish model. Front. Pharmacol. 2021, 12, 2866. [Google Scholar] [CrossRef]
- Hoshi, R.A.; Pastre, C.M.; Vanderlei, L.C.M.; Godoy, M.F. Poincaré plot indexes of heart rate variability: Relationships with other nonlinear variables. Auton. Neurosci. 2013, 177, 271–274. [Google Scholar] [CrossRef] [PubMed]
- Piskorski, J.; Guzik, P. Filtering poincare plots. Comput. Methods Sci. Technol. 2005, 11, 39–48. [Google Scholar] [CrossRef]
- Kosourikhina, V.; Kavanagh, D.; Richardson, M.J.; Kaplan, D.M. Validation of DeepLabCut as a tool for markerless 3D pose estimation. bioRxiv 2022. [Google Scholar] [CrossRef]
- Chen, Z.; Xie, Z.; Zhang, W.; Xu, X. ResNet and Model Fusion for Automatic Spoofing Detection. In Proceedings of the Interspeech, Stockholm, Sweden, 20–24 August 2017; pp. 102–106. [Google Scholar]
- Han, S.S.; Park, G.H.; Lim, W.; Kim, M.S.; Na, J.I.; Park, I.; Chang, S.E. Deep neural networks show an equivalent and often superior performance to dermatologists in onychomycosis diagnosis: Automatic construction of onychomycosis datasets by region-based convolutional deep neural network. PLoS ONE 2018, 13, e0191493. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, R.U.; Zhang, X.; Kumar, R.; Aboagye, E.O. Evaluating the performance of resnet model based on image recognition. In Proceedings of the 2018 International Conference on Computing and Artificial Intelligence, New York, NY, USA, 21–23 December 2018; pp. 86–90. [Google Scholar]
- Lee, S.; Waugh, B.; O’Dell, G.; Zhao, X.; Yoo, W.-S.; Kim, D.H. Predicting Fruit Fly Behaviour using TOLC device and DeepLabCut. In Proceedings of the 2021 IEEE 21st International Conference on Bioinformatics and Bioengineering (BIBE), Kragujevac, Serbia, 25–27 October 2021; pp. 1–6. [Google Scholar]
- Miura, G.I.; Yelon, D. A guide to analysis of cardiac phenotypes in the zebrafish embryo. Methods Cell Biol. 2011, 101, 161–180. [Google Scholar] [PubMed] [Green Version]
- Singleman, C.; Holtzman, N.G. Analysis of postembryonic heart development and maturation in the zebrafish, Danio rerio. Dev. Dyn. 2012, 241, 1993–2004. [Google Scholar] [CrossRef] [Green Version]
- Claireaux, G.; McKenzie, D.J.; Genge, A.G.; Chatelier, A.; Aubin, J.; Farrell, A.P. Linking swimming performance, cardiac pumping ability and cardiac anatomy in rainbow trout. J. Exp. Biol. 2005, 208, 1775–1784. [Google Scholar] [CrossRef] [Green Version]
- Echeazarra, L.; Hortigón-Vinagre, M.P.; Casis, O.; Gallego, M. Adult and developing zebrafish as suitable models for cardiac electrophysiology and pathology in research and industry. Front. Physiol. 2021, 11, 1692. [Google Scholar] [CrossRef]
- Yalcin, H.C.; Amindari, A.; Butcher, J.T.; Althani, A.; Yacoub, M. Heart function and hemodynamic analysis for zebrafish embryos. Dev. Dyn. 2017, 246, 868–880. [Google Scholar] [CrossRef] [Green Version]
- Shin, J.T.; Pomerantsev, E.V.; Mably, J.D.; MacRae, C.A. High-resolution cardiovascular function confirms functional orthology of myocardial contractility pathways in zebrafish. Physiol. Genom. 2010, 42, 300–309. [Google Scholar] [CrossRef] [Green Version]
- Pereira, T.D.; Aldarondo, D.E.; Willmore, L.; Kislin, M.; Wang, S.S.-H.; Murthy, M.; Shaevitz, J.W. Fast animal pose estimation using deep neural networks. Nat. Methods 2019, 16, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Lauer, J.; Zhou, M.; Ye, S.; Menegas, W.; Nath, T.; Rahman, M.M.; Di Santo, V.; Soberanes, D.; Feng, G.; Murthy, V.N. Multi-animal pose estimation and tracking with DeepLabCut. bioRxiv 2021. [Google Scholar] [CrossRef] [PubMed]
- Lucas, B.D.; Kanade, T. An Iterative Image Registration Technique with an Application to Stereo Vision; Canegie Mellon University: Pittsburgh, PA, USA, 1981. [Google Scholar]
- Bradski, G.; Kaehler, A. Learning OpenCV: Computer Vision with the OpenCV Library; O’Reilly Media, Inc.: Sebastopol, CA, USA, 2008. [Google Scholar]
- Mathis, M.W.; Mathis, A. Deep learning tools for the measurement of animal behavior in neuroscience. Curr. Opin. Neurobiol. 2020, 60, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mah, K.M.; Torres-Espín, A.; Hallworth, B.W.; Bixby, J.L.; Lemmon, V.P.; Fouad, K.; Fenrich, K.K. Automation of training and testing motor and related tasks in pre-clinical behavioural and rehabilitative neuroscience. Exp. Neurol. 2021, 340, 113647. [Google Scholar] [CrossRef]
- Arent, I.; Schmidt, F.P.; Botsch, M.; Dürr, V. Marker-Less Motion Capture of Insect Locomotion With Deep Neural Networks Pre-trained on Synthetic Videos. Front. Behav. Neurosci. 2021, 15, 637806. [Google Scholar] [CrossRef] [PubMed]
- Sarmah, S.; Marrs, J.A. Complex cardiac defects after ethanol exposure during discrete cardiogenic events in zebrafish: Prevention with folic acid. Dev. Dyn. 2013, 242, 1184–1201. [Google Scholar] [CrossRef] [Green Version]
- Dejana, E.; Tournier-Lasserve, E.; Weinstein, B.M. The control of vascular integrity by endothelial cell junctions: Molecular basis and pathological implications. Dev. Cell. 2009, 16, 209–221. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.P.; Glennon, M.S.; Umbarkar, P.; Gupte, M.; Galindo, C.L.; Zhang, Q.; Force, T.; Becker, J.R.; Lal, H. Ponatinib-induced cardiotoxicity: Delineating the signalling mechanisms and potential rescue strategies. Cardiovasc. Res. 2019, 115, 966–977. [Google Scholar] [CrossRef]
- Buza, V.; Rajagopalan, B.; Curtis, A.B. Cancer treatment–induced arrhythmias: Focus on chemotherapy and targeted therapies. Circul. Arrhythmia Electrophysiol. 2017, 10, e005443. [Google Scholar] [CrossRef]





| Species | Detection Method | Detection Endpoints | Literatures |
|---|---|---|---|
| Danio rerio (36–120 hpf) | Detection algorithms written in Matlab based on changes in pixel intensity and color segmentation | Heartbeats and heart rate irregularity | Pylatiuk et al., 2014 [7] |
| Danio rerio (3 dpf) | Using green fluorescent protein-expressing zebrafish Tg(cmlc2: GFP) to automate the myocardial phenotype screening | Number of heart contraction times based on subsite pixel intensities changes | Burns et al., 2005 [8] |
| Danio rerio (72 hpf) | Heart rate was calculated by the software “DanioScope” using a Noldus DanioVision system | Heart rate in beats per minute (BPM) based on video assessment of inter-beat intervals | Zhong et al., 2021 [9] |
| Danio rerio | Kymograph plugin in ImageJ | Heartbeat regularity, stroke volume, ejection fraction, shortening fraction, and cardiac output | Kurnia et al., 2021 [4] |
| Danio rerio | ImageJ based on the dynamic pixel changes method | Atrium rhythm and heartbeat frequency | Santoso et al., 2019 [10] |
| Species | Deep Learning Method | Detection Endpoints | Literatures |
|---|---|---|---|
| Danio rerio (3 dpf) | Automatic assessment of cardiovascular function based on a U-net deep learning model | Shortening fraction and ejection fraction of masked ventricles | Naderi et al., 2021 [15] |
| Danio rerio (embryonic) | A stand-alone software that uses C# language with the.NET Framework 4.5.2 | Shortening fraction based on two pairs of marking points from the diastolic and systolic heart edges of the ventricles | Nasrat et al., 2016 [16] |
| Danio rerio (48 to 96 hpf) | Automatic detection of the heart region by using an intelligent robotic microscope | Heart-region detection based on the intensity and difference images which can be used to distinguish the heart dysfunction | Spomer et al., 2012 [17] |
| Danio rerio | OpenCV-based approach | Heart rate and heartbeat regularity | Farhan et al., 2021 [18] |
| Danio rerio | Cardiac Functional Imaging Network (CFIN) | Shorteing fraction, ejection fraction, heart rate, and cardiac output | Akerberg et al., 2019 [19] |
| Danio rerio | Zebrafish Heart Rate Automatic Method (Z-HRAM) | Heartbeat detection based on zebrafish body expansion and contraction movements | Xing et al., 2018 [20] |
| Danio rerio (2–3 dpf) | DeepLabCutTM (DLC) using ResNet-152 | Calculation of volume change, heart rate, stroke volume, ejection fraction, shortening fraction, cardiac output, and heartbeat regularity based on 8-point labeling of heart edges for short and long axis lengths | In this study |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suryanto, M.E.; Saputra, F.; Kurnia, K.A.; Vasquez, R.D.; Roldan, M.J.M.; Chen, K.H.-C.; Huang, J.-C.; Hsiao, C.-D. Using DeepLabCut as a Real-Time and Markerless Tool for Cardiac Physiology Assessment in Zebrafish. Biology 2022, 11, 1243. https://doi.org/10.3390/biology11081243
Suryanto ME, Saputra F, Kurnia KA, Vasquez RD, Roldan MJM, Chen KH-C, Huang J-C, Hsiao C-D. Using DeepLabCut as a Real-Time and Markerless Tool for Cardiac Physiology Assessment in Zebrafish. Biology. 2022; 11(8):1243. https://doi.org/10.3390/biology11081243
Chicago/Turabian StyleSuryanto, Michael Edbert, Ferry Saputra, Kevin Adi Kurnia, Ross D. Vasquez, Marri Jmelou M. Roldan, Kelvin H.-C. Chen, Jong-Chin Huang, and Chung-Der Hsiao. 2022. "Using DeepLabCut as a Real-Time and Markerless Tool for Cardiac Physiology Assessment in Zebrafish" Biology 11, no. 8: 1243. https://doi.org/10.3390/biology11081243
APA StyleSuryanto, M. E., Saputra, F., Kurnia, K. A., Vasquez, R. D., Roldan, M. J. M., Chen, K. H.-C., Huang, J.-C., & Hsiao, C.-D. (2022). Using DeepLabCut as a Real-Time and Markerless Tool for Cardiac Physiology Assessment in Zebrafish. Biology, 11(8), 1243. https://doi.org/10.3390/biology11081243











