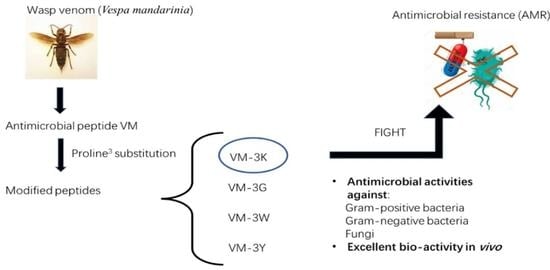
Evaluation of the Antimicrobial Properties of a Natural Peptide from Vespa mandarinia Venom and Its Synthetic Analogues as a Possible Route to Defeat Drug-Resistant Microbes
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Peptide Design and Synthesis
2.2. The Determination of Peptide Secondary Structures and Physicochemical Characteristics
2.3. Antimicrobial Assays
2.4. Antibiofilm Assays
2.5. Kinetic Time Killing Assay
2.6. Membrane Permeabilisation Assay
2.7. Haemolysis Assay
2.8. Efficacy Evaluation against S. aureus In Vivo
2.9. Resistance Induction by Serial Passage
3. Results
3.1. The Physicochemical Properties of VM and Modified Analogues
3.2. Conformational Study by CD
3.3. Anti-Microbial Activity of VM and Its Analogues
3.4. Anti-Biofilm Activity of VM and Its Analogues
3.5. Time-Killing Kinetics of VM and Its Analogues
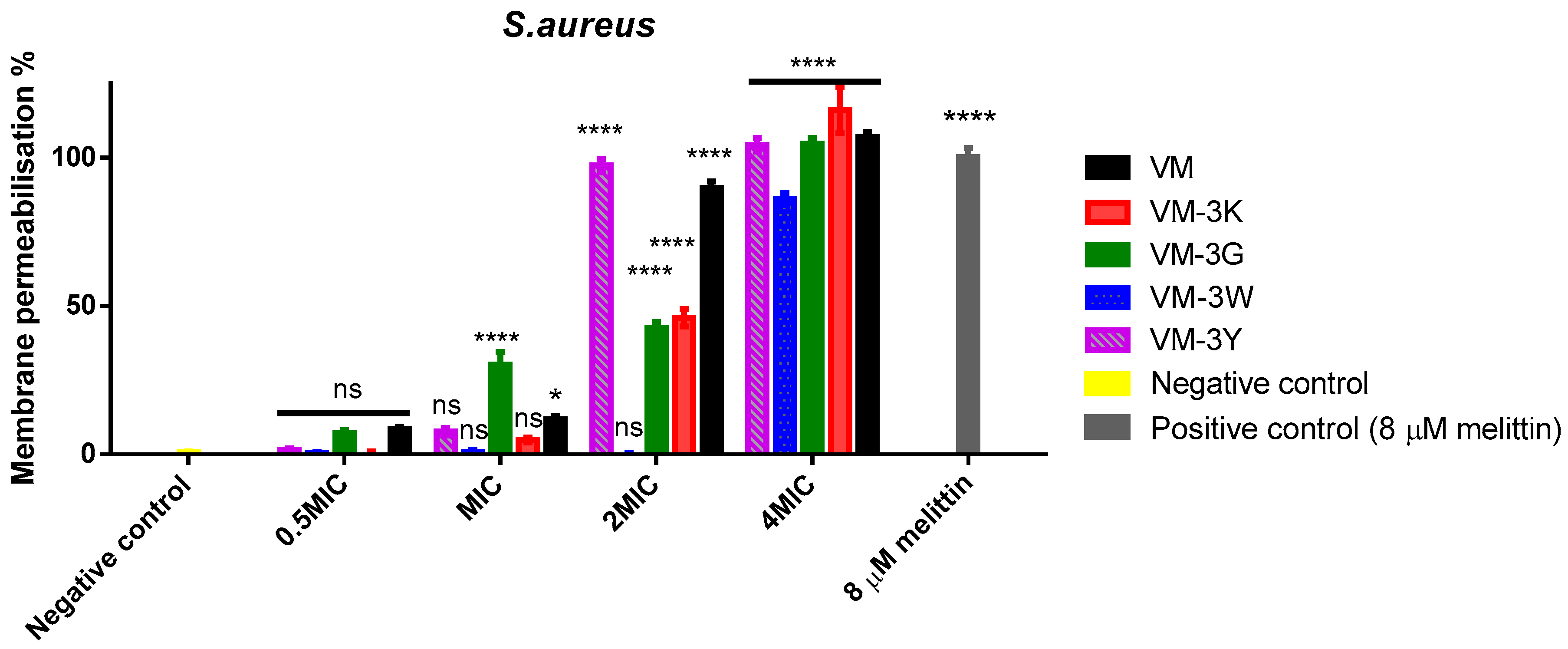
3.6. Bacterial Cell Membrane Permeabilisation of VM and Its Analogues
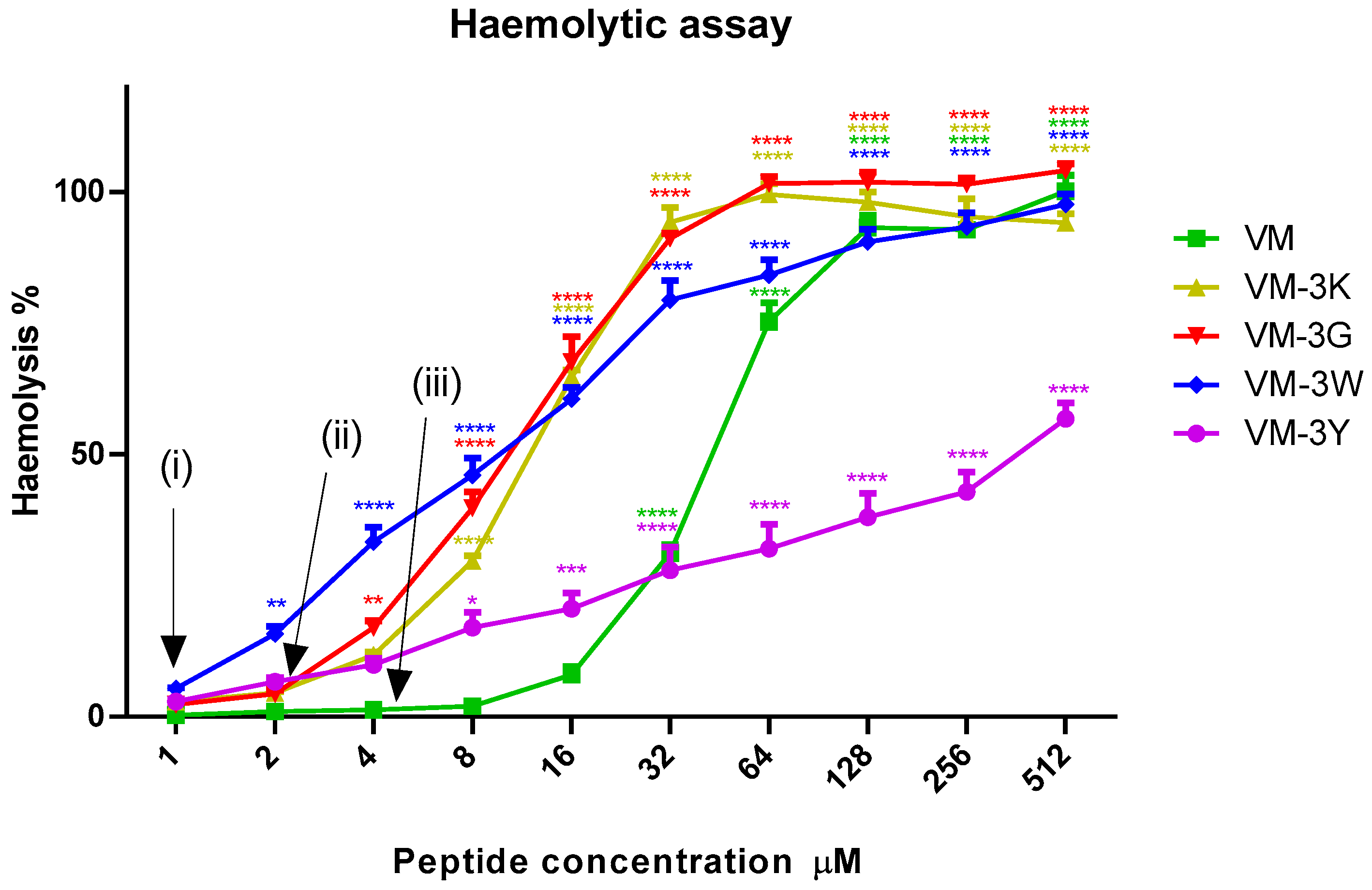
3.7. Haemolytic Activities of VM and Analogues
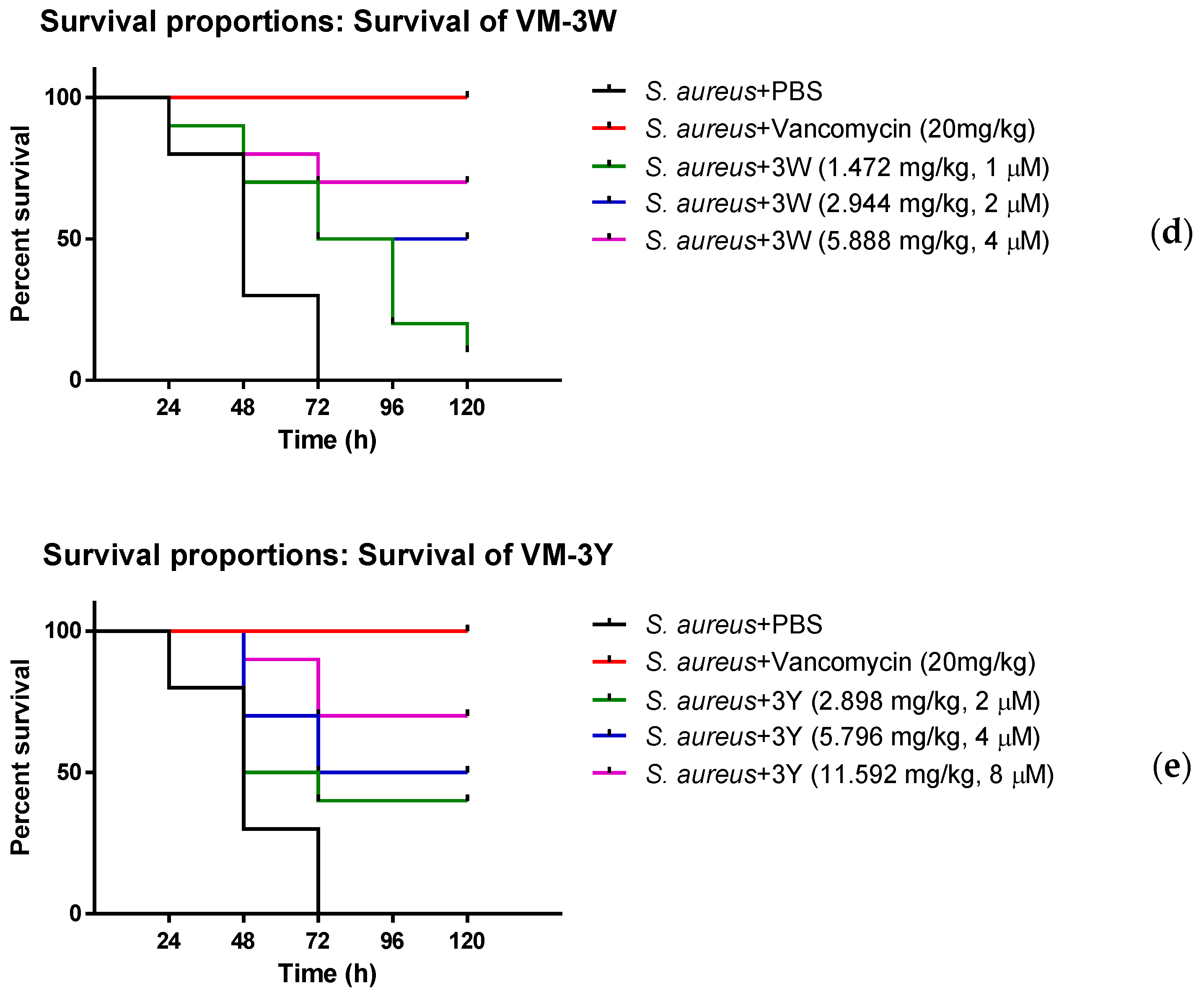
3.8. The Anti-Bacterial Efficiency of VM and Modified Peptides In Vivo
3.9. Resistance Induction by Serial Passage in S. aureus
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mobarki, N.; Almerabi, B.; Hattan, A. Antibiotic resistance crisis. IJMDC 2019, 40, 561–564. [Google Scholar] [CrossRef]
- Murray, C.J.; Ikuta, K.S.; Sharara, F.; Swetschinski, L.; Aguilar, G.R.; Gray, A.; Han, C.; Bisignano, C.; Rao, P.; Wool, E.; et al. Global burden of bacterial antimicrobial resistance in 2019: A systematic analysis. Lancet 2022, 399, 629–655. [Google Scholar] [CrossRef]
- Bhat, Z.; Kumar, S.; Bhat, H. Bioactive peptides of animal origin: A review. J. Food Sci. Technol. 2015, 52, 5377–5392. [Google Scholar] [CrossRef]
- Mokoena, M.P. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules 2017, 22, 1255. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Fang, Q.; Lu, Z.; Gao, Y.; Trembleau, L.; Ebel, R.; Andersen, J.H.; Philips, C.; Law, S.; Deng, H. Discovery and Biosynthetic Investigation of a New Antibacterial Dehydrated Non-Ribosomal Tripeptide. Angew. Chem. Int. Ed. 2021, 60, 3229–3237. [Google Scholar] [CrossRef] [PubMed]
- Lee, D.G.; Kim, H.K.; Kim, S.A.; Park, Y.; Park, S.C.; Jang, S.H.; Hahm, K.S. Fungicidal effect of indolicidin and its interaction with phospholipid membranes. Biochem. Biophys. Res. Commun. 2003, 305, 305–310. [Google Scholar] [CrossRef]
- Moerman, L.; Bosteels, S.; Noppe, W.; Willems, J.; Clynen, E.; Schoofs, L.; Thevissen, K.; Tytgat, J.; Van Eldere, J.; Van Der Walt, J.; et al. Antibacterial and antifungal properties of α-helical, cationic peptides in the venom of scorpions from southern Africa. Eur. J. Biochem. 2002, 269, 4799–4810. [Google Scholar] [CrossRef]
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 2559. [Google Scholar] [CrossRef]
- Powers, J.P.S.; Hancock, R.E.W. The relationship between peptide structure and antibacterial activity. Peptides 2003, 24, 1681–1691. [Google Scholar] [CrossRef]
- Xiao, Y.; Liu, C.; Lai, R. Antimicrobial peptides from amphibians. Biomol. Concepts 2011, 2, 27–38. [Google Scholar] [CrossRef]
- Gaspar, D.; Salomé, V.A.; Castanho, M.A.R.B. From antimicrobial to anticancer peptides. A review. Front. Microbiol. 2013, 4, 294. [Google Scholar] [CrossRef] [PubMed]
- Abd El-Wahed, A.; Yosri, N.; Sakr, H.; Du, M.; Algethami, A.F.M.; Zhao, C.; Abdelazeem, A.H.; Tahir, H.E.; Masry, S.H.D.; Abdel-Daim, M.M.; et al. Wasp Venom Biochemical Components and Their Potential in Biological Applications and Nanotechnological Interventions. Toxins 2021, 13, 206. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, E.; Siciliano, A.; Gesuele, R.; Di Onofrio, V.; Falanga, A.; Maione, A.; Liguori, R.; Libralato, G.; Guida, M. Melittin inhibition and eradication activity for resistant polymicrobial biofilm isolated from a dairy industry after disinfection. Int. J. Microbiol. 2019, 2019, 4012394. [Google Scholar] [CrossRef]
- Memariani, H.; Memariani, M. Anti-fungal properties and mechanisms of melittin. Appl. Microbiol. Biotechnol. 2020, 104, 6513–6526. [Google Scholar] [CrossRef]
- Moreno, M.; Giralt, E. Three valuable peptides from bee and wasp venoms for therapeutic and biotechnological use: Melittin, apamin and mastoparan. Toxins 2015, 7, 1126–1150. [Google Scholar] [CrossRef]
- Singulani, J.; Galeane, M.; Ramos, M.; Gomes, P.; Dos Santos, C.; de Souza, B.; Palma, M.; Fusco, A.A.; Mendes, G.M.J.S. Antifungal Activity, Toxicity, and Membranolytic Action of a Mastoparan Analog Peptide. Front. Cell Infect. Microbiol. 2019, 9, 419. [Google Scholar] [CrossRef]
- Yu, H.; Yang, H.; Ma, D.; Lv, Y.; Liu, T.; Lai, R.; Liu, J. Vespid chemotactic peptide precursor from the wasp, Vespa magnifica (Smith). Toxicon 2007, 50, 377–382. [Google Scholar] [CrossRef]
- Tsukamoto, Y.; Fukutani, S.; Morikawa, Y.; Fujimoto., H.; Tanaka, Y.; Mori, M.; Yasuhara, T.; Nakajima, T.; Kitada, C.; Fujino, M. Structure and Chemotactic Activity of Peptides from the Venom Sac of Vespinae. Jpn. J. Oral Biol. 1985, 27, 1254–1256. [Google Scholar] [CrossRef]
- Lu, Y.; Li, J.; Yu, H.; Xu, X.; Liang, J.; Tian, D.; Ma, D.; Lin, G.; Huang, G.; Lai, R. Two families of antimicrobial peptides with multiple functions from skin of rufous-spotted torrent frog, Amolops loloensis. Peptides 2006, 27, 3085–3091. [Google Scholar] [CrossRef]
- Misconai, A.; Bulyá, É.; Kardos, J. BeStSel: From Secondary Structure Analysis to Protein Fold Prediction by Circular Dichroism Spectroscopy. Methods Mol. Biol. 2021, 2199, 175–189. [Google Scholar]
- Gautier, R.; Douguet, D.; Antonny, B.; Drin, G. HELIQUEST: A web server to screen sequences with specific α-helical properties. Bioinformatics 2008, 24, 2101–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pollard, J.; Snarr, J.; Chaudhary, V.; Jennings, J.D.; Shaw, H.; Christiansen, B.; Wright, J.; Jia, W.; Bishop, R.E.; Savage, P.B. In vitro evaluation of the potential for resistance development to ceragenin CSA-13. J. Antimicrob. Chemother. 2012, 67, 2665–2672. [Google Scholar] [CrossRef] [PubMed]
- Dolzani, L.; Milan, A.; Scocchi, M.; Lagatolla, C.; Bressan, R.; Benincasa, M. Sub-MIC effects of a proline-rich antibacterial peptide on clinical isolates of Acinetobacter baumannii. J. Med. Microbiol. 2019, 68, 1253–1265. [Google Scholar] [CrossRef] [PubMed]
- Ilić, N.; Novković, M.; Guida, F.; Xhindoli, D.; Benincasa, M.; Tossi, A.; Juretić, D. Selective antimicrobial activity and mode of action of adepantins, glycine-rich peptide antibiotics based on anuran antimicrobial peptide sequences. Biochim. Biophys. Acta Biomembr. 2013, 1828, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.-X.; Pang, X. Electrostatic Interactions in Protein Structure, Folding, Binding, and Condensation. Physiol. Behav. 2017, 176, 139–148. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Sun, L.; Huang, S.; Zhu, C.; Li, P.; He, J.; Mackey, V.; Coy, D.H.; He, Q. The antimicrobial peptides and their potential clinical applications. Am. J. Transl. Res. 2019, 11, 3919–3931. [Google Scholar] [PubMed]
- Omardien, S.; Brul, S.; Zaat, S.A.J. Antimicrobial activity of cationic antimicrobial peptides against gram-positives: Current progress made in understanding the mode of action and the response of bacteria. Front. Cell Dev. Biol. 2016, 4, 111. [Google Scholar] [CrossRef]
- Kumar, P.; Kizhakkedathu, J.N.; Straus, S.K. Antimicrobial peptides: Diversity, mechanism of action and strategies to improve the activity and biocompatibility in vivo. Biomolecules 2018, 8, 4. [Google Scholar] [CrossRef]
- Doucette, K.A.; Chaiyasit, P.; Calkins, D.; Martinez, K.N.; Cleave, C.V.; Knebel, C.A.; Tongraar, A.; Crans, D.C. The interfacial interactions of glycine and short glycine peptides in model membrane systems. Int. J. Mol. Sci. 2021, 22, 162. [Google Scholar] [CrossRef]
- Lee, M.T.; Hung, W.; Chen, F.; Huang, H. Mechanism and kinetics of pore formation in membranes by water-soluble amphipathic peptides. Proc. Natl. Acad. Sci. USA 2008, 105, 5087–5092. [Google Scholar] [CrossRef] [PubMed]
- Rathinakumar, R.; Walkenhorst, W.F.; William, C.W. Broad-spectrum Antimicrobial Peptides by Rational Combinatorial Design and High-throughput Screening: The Importance of Interfacial Activity. J. Am. Chem. Soc. 2009, 131, 7609–7617. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedrich, C.L.; Moyles, D.; Beveridge, T.; Hancock, R.E.W. Antibacterial action of structurally diverse cationic peptides on gram-positive bacteria. AAC 2000, 44, 2086–2092. [Google Scholar] [CrossRef] [PubMed]
- Gottenbos, B.; Grijpma, D.; Van Der Mei, H.; Feijen, J.; Busscher, H.J. Antimicrobial effects of positively charged surfaces on adhering Gram-positive and Gram-negative bacteria. J. Antimicrob. Chemother. 2001, 48, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Ghosh, A.; Jayaraman, N.; Chatterji, D. Small-Molecule Inhibition of Bacterial Biofilm. ACS Omega 2020, 5, 3108–3115. [Google Scholar] [CrossRef] [PubMed]
- Olson, M.E.; Ceri, H.; Morck, D.; Buret, A.G.; Read, R.R. Biofilm bacteria: Formation and comparative susceptibility to antibiotics. Can. J. Vet. Res. 2002, 66, 86–92. [Google Scholar]
- Lerch, T.Z.; Chenu, C.; Dignac, M.; Barriuso, E.; Mariotti, A. Biofilm vs. Planktonic Lifestyle: Consequences for Pesticide 2,4-D Metabolism by Cupriavidus necator JMP134. Front. Microbiol. 2017, 8, 1–11. [Google Scholar] [CrossRef]
- Roy, R.; Tiwari, M.; Donelli, G.; Tiwari, V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence 2018, 9, 522–554. [Google Scholar] [CrossRef]
- Idiong, G.; Won, A.; Ruscito, A.; Leung, B.O.; Hitchcock, A.P.; Ianoul, A. Investigating the effect of a single glycine to alanine substitution on interactions of antimicrobial peptide latarcin 2a with a lipid membrane. Eur. Biophys. 2011, 40, 1087–1100. [Google Scholar] [CrossRef]
- Nam, H.; Choi, J.; Kumar, S.; Nielson, J.E.; Kyeong, M.; Wang, S.; Kang, D.; Lee, Y.; Lee, J.; Yoon, M.H.; et al. Helicity Modulation Improves the Selectivity of Antimicrobial Peptoids. ACS Infect. Dis. 2020, 6, 2732–2744. [Google Scholar] [CrossRef]
- Mondal, A.; Verma, P.; Sengupta, N.; Dutta, S.; Bhushan, P.S.; Chattopadhyay, K. Tyrosine in the hinge region of the pore-forming motif regulates oligomeric β-barrel pore formation by Vibrio cholerae cytolysin. Mol. Microbiol. 2021, 115, 508–525. [Google Scholar] [CrossRef]
- Tsai, C.; Loh, J.M.S.; Proft, T. Galleria mellonella infection models for the study of bacterial diseases and for antimicrobial drug testing. Virulence 2016, 7, 214–229. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, N.; Zhang, W.; Cheng, X.; Yan, Z.; Shao, G.; Wang, X.; Wang, R.; Fu, C. Therapeutic peptides: Current applications and future directions. Signal Transduct. Target. Ther. 2022, 7, 48. [Google Scholar] [CrossRef]
- Blower, R.; Popov, S.; Van Hoek, M. Cathelicidin peptide rescues G. mellonella infected with B. anthracis. Virulence 2018, 9, 287–293. [Google Scholar] [CrossRef] [PubMed]
- Greco, I.; Molchanova, N.; Holmedal, E.; Jenssen, H.; Hummel, B.D.; Watts, J.L.; Håkansson, J.; Hansen, P.R.; Svenson, J. Correlation between hemolytic activity, cytotoxicity and systemic in vivo toxicity of synthetic antimicrobial peptides. Sci. Rep. 2020, 10, 13206. [Google Scholar] [CrossRef]
- Garima, K.; Saurabh, S.; Elongavan, A. Action and resistance mechanisms of antibiotics: A guide for clinicians. JOACP 2019, 33, 300–305. [Google Scholar] [CrossRef]
- Pontes, D.S.; de Araujo, R.S.A.; Dantas, N.; Scotti, L.; Scotti, M.T.; de Moura, R.O.; Mendonca-Junior, F.J.B. Genetic Mechanisms of Antibiotic Resistance and the Role of Antibiotic Adjuvants. Curr. Top. Med. Chem. 2018, 18, 42–74. [Google Scholar] [CrossRef]
- Lewies, A.; Du Plessis, L.H.; Wentzel, J.F. Antimicrobial Peptides: The Achilles’ Heel of Antibiotic Resistance? Probiotics Antimicrob. Proteins 2019, 11, 370–381. [Google Scholar] [CrossRef]





| Peptide Name | Peptide Sequence |
|---|---|
| VM | FLPIIGKLLSGLL-NH2 |
| VM-3K | FLKIIGKLLSGLL-NH2 |
| VM-3G | FLGIIGKLLSGLL-NH2 |
| VM-3W | FLWIIGKLLSGLL-NH2 |
| VM-3Y | FLYIIGKLLSGLL-NH2 |
| Peptide | Hydrophobicity (<H>) | Hydrophobic Moment <μM> | Net Charge |
|---|---|---|---|
| VM | 1.045 | 0.618 | +1 |
| VM-3K | 0.913 | 0.744 | +2 |
| VM-3G | 0.989 | 0.671 | +1 |
| VM-3W | 1.162 | 0.508 | +1 |
| VM-3Y | 1.063 | 0.601 | +1 |
| % of α-Helix in 10 mM NH4Ac | % of α-Helix in 50% TFE/NH4Ac | |
|---|---|---|
| VM | 0.3 | 64.3 |
| VM-3K | 2.5 | 60 |
| VM-3G | 11.7 | 75.8 |
| VM-3W | 24 | 63.1 |
| VM-3Y | 38.0 | 75.1 |
| S. aureus | MRSA | E. faecalis | E. coli | K. pneumoniae | P. aeruginosa | C. albicans | |
|---|---|---|---|---|---|---|---|
| VM | 4/4 | 4/4 | 16/16 | 256/256 | >512 | >512 | 32/32 |
| VM-3K | 1/1 | 1/2 | 2/2 | 8/8 | 32/32 | 16/16 | 32/32 |
| VM-3G | 1/1 | 2/2 | 4/4 | 64/128 | >512 | 128/128 | >512 |
| VM-3W | 1/1 | 2/2 | 4/4 | >512 | >512 | >512 | >512 |
| VM-3Y | 2/2 | 2/2 | 4/4 | >512 | >512 | >512 | >512 |
| S. aureus | MRSA | E. faecalis | E. coli | K. pneumoniae | P. aeruginosa | |
|---|---|---|---|---|---|---|
| VM | 8/64 | 8/64 | 16>512 | >512/>512 | >512/>512 | >512/>512 |
| VM-3K | 2/32 | 2/64 | 4/>512 | 16/>512 | 64/>512 | 32/>512 |
| VM-3G | 2/32 | 2/32 | 8/>512 | 256/>512 | >512/>512 | >512/>512 |
| VM-3W | 4/128 | 4/128 | 16/>512 | >512/>512 | >512/>512 | >512/>512 |
| VM-3Y | 2/128 | 2/64 | 64/>512 | >512/>512 | >512/>512 | >512/>512 |
| Melittin | 4/ND | 8/ND | 8/ND | 32/ND | 128/ND | 64/ND |
| Peptide Name | HC50 (µM) | SI 1 |
|---|---|---|
| VM | 44.43 | 11.11 |
| VM-3K | 11.20 | 11.20 |
| VM-3G | 9.686 | 9.686 |
| VM-3W | 9.682 | 9.682 |
| VM-3Y | 211.2 | 105.6 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.; Sun, R.; Chen, Z.; Zhou, C.; Ma, C.; Zhou, M.; Chen, X.; Chen, T.; Shaw, C.; Wang, L. Evaluation of the Antimicrobial Properties of a Natural Peptide from Vespa mandarinia Venom and Its Synthetic Analogues as a Possible Route to Defeat Drug-Resistant Microbes. Biology 2022, 11, 1263. https://doi.org/10.3390/biology11091263
Zhang J, Sun R, Chen Z, Zhou C, Ma C, Zhou M, Chen X, Chen T, Shaw C, Wang L. Evaluation of the Antimicrobial Properties of a Natural Peptide from Vespa mandarinia Venom and Its Synthetic Analogues as a Possible Route to Defeat Drug-Resistant Microbes. Biology. 2022; 11(9):1263. https://doi.org/10.3390/biology11091263
Chicago/Turabian StyleZhang, Jin, Ruize Sun, Zhiwei Chen, Chunyuan Zhou, Chengbang Ma, Mei Zhou, Xiaoling Chen, Tianbao Chen, Chris Shaw, and Lei Wang. 2022. "Evaluation of the Antimicrobial Properties of a Natural Peptide from Vespa mandarinia Venom and Its Synthetic Analogues as a Possible Route to Defeat Drug-Resistant Microbes" Biology 11, no. 9: 1263. https://doi.org/10.3390/biology11091263
APA StyleZhang, J., Sun, R., Chen, Z., Zhou, C., Ma, C., Zhou, M., Chen, X., Chen, T., Shaw, C., & Wang, L. (2022). Evaluation of the Antimicrobial Properties of a Natural Peptide from Vespa mandarinia Venom and Its Synthetic Analogues as a Possible Route to Defeat Drug-Resistant Microbes. Biology, 11(9), 1263. https://doi.org/10.3390/biology11091263