Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli
Abstract
:Simple Summary
Abstract
1. Introduction
1.1. Multi-Drug Resistance (MDR)
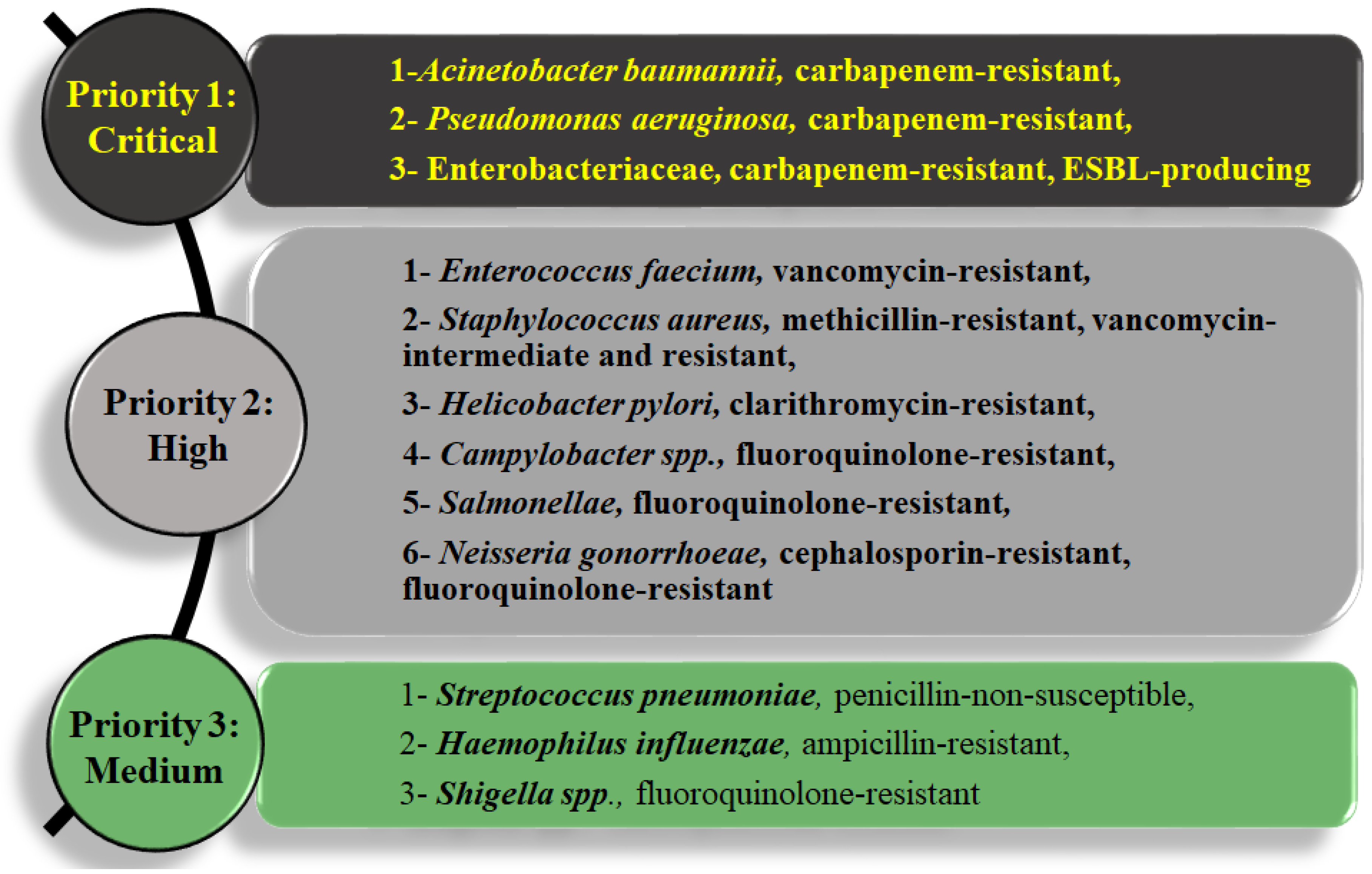
1.2. Gram-Negative Bacteria Are Difficult to Treat
1.3. Antibiotic Resistance Mechanisms and MDR
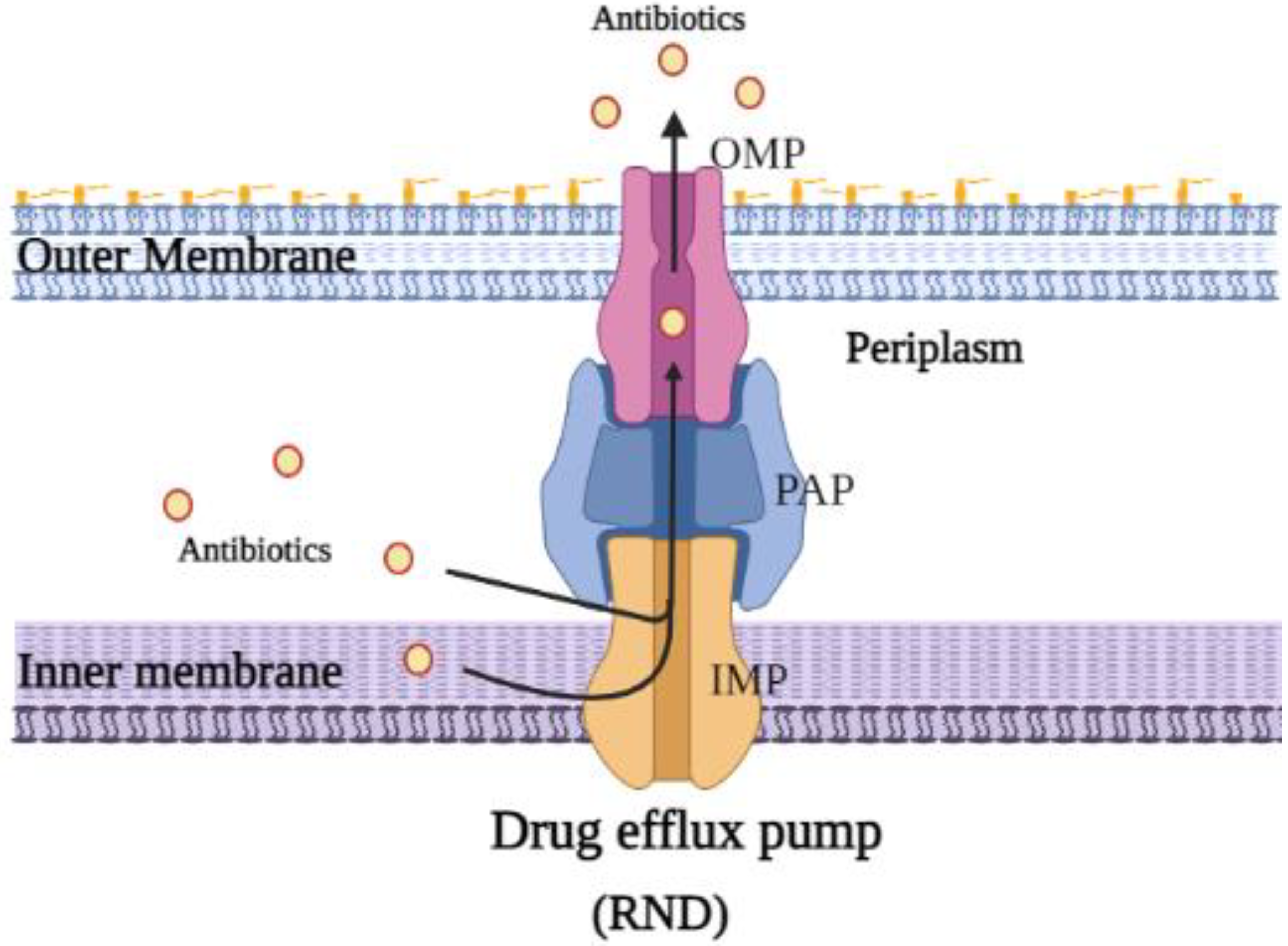
1.4. Drug Efflux Pumps and Antibiotic Resistance
2. Resistance–Nodulation–Division (RND) Superfamily Efflux Pump
2.1. Structure and Function of E. coli AcrAB-TolC Drug Efflux Pump Complex
2.2. Overview of the Structure of AcrB
2.3. Movement of Substrates through AcrB
2.4. Substrate Entry Sites into the AcrB Promoter
2.5. Substrate Binding Pockets
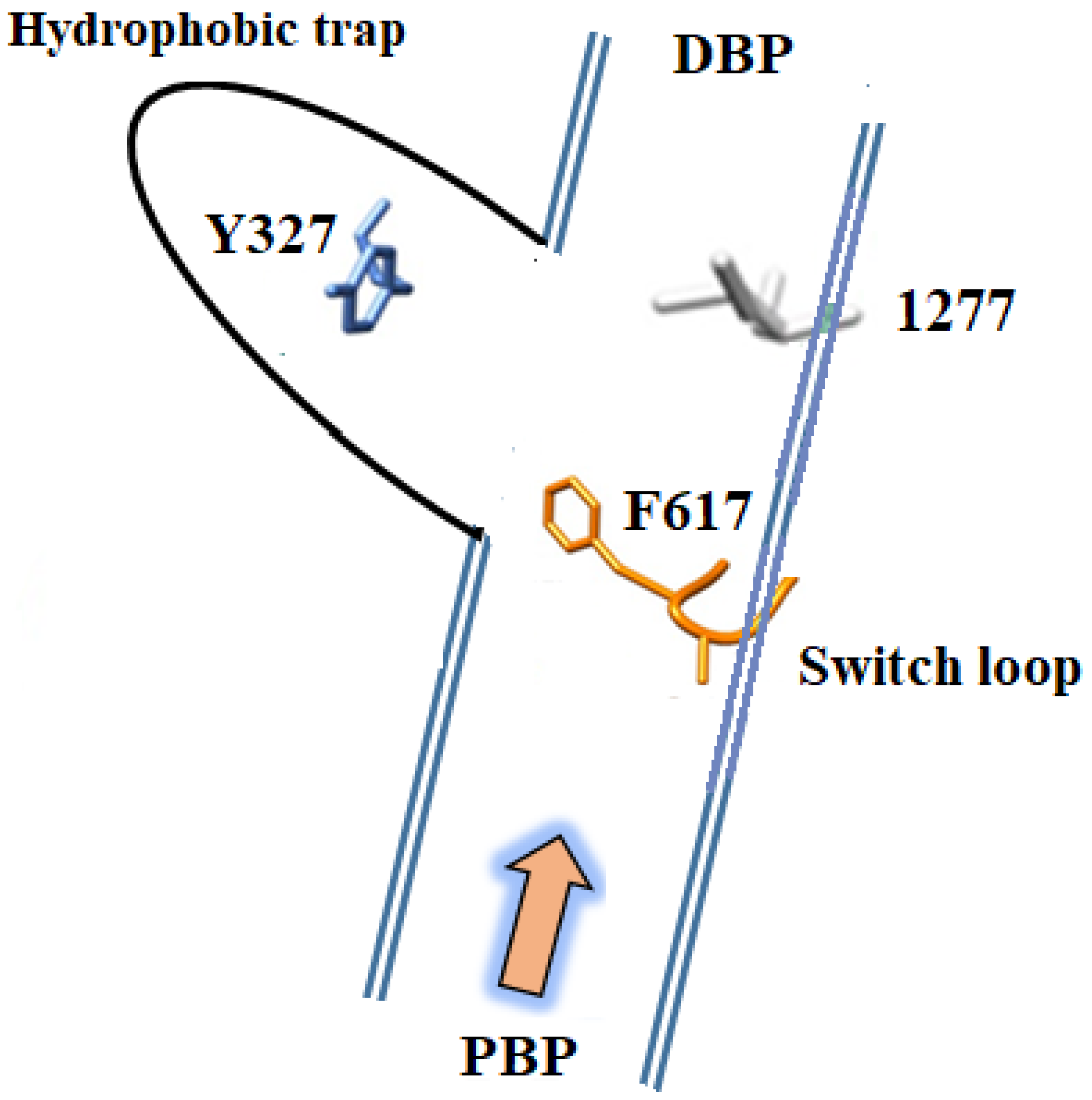
3. Structures of Inhibitors Bound to AcrB—The Hydrophobic Trap
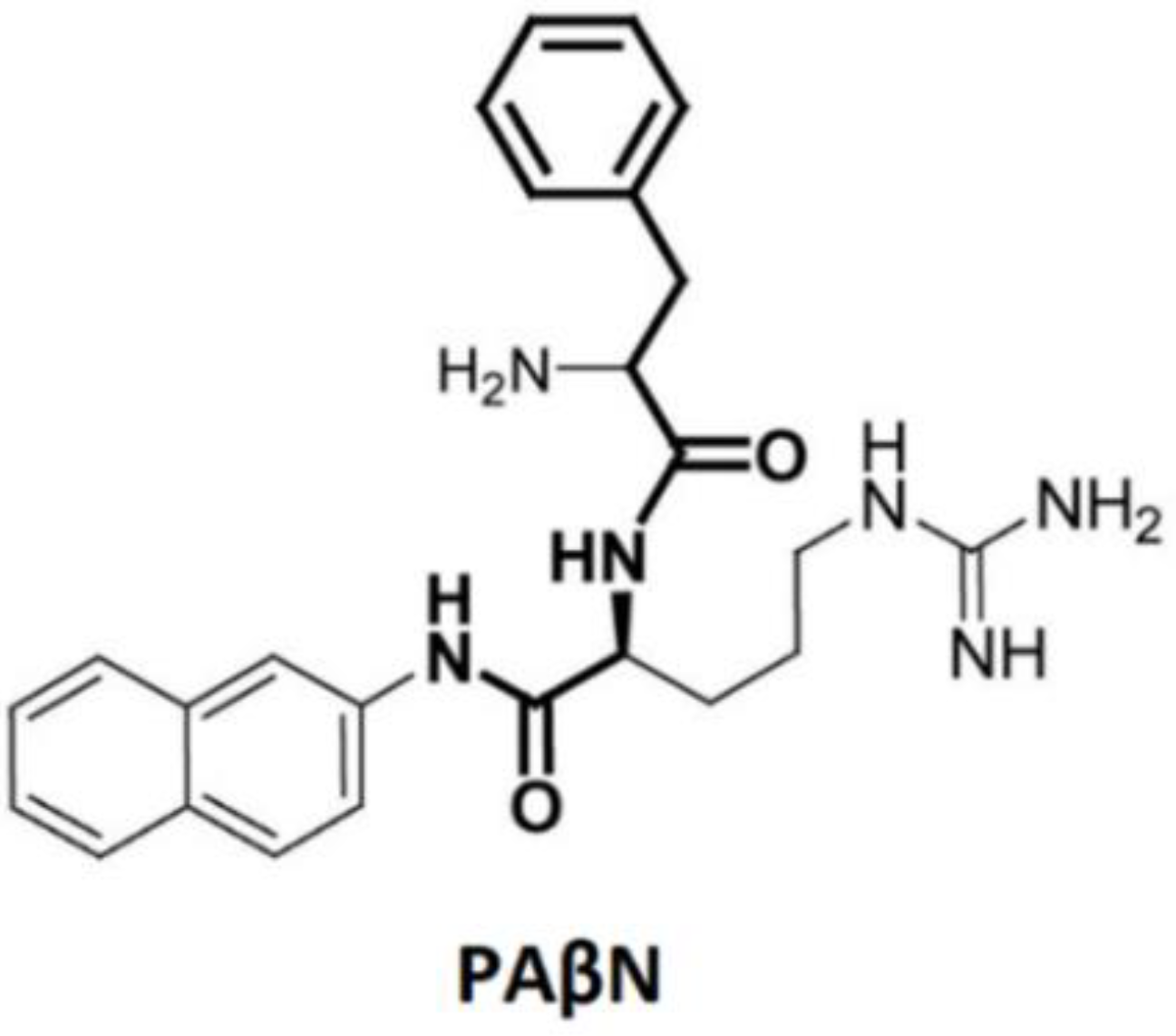
4. Small Molecules That Inhibit AcrB; Efflux Pump Inhibitors (EPI)
5. Conclusions
6. Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Cooper, M.A.; Shlaes, D. Fix the antibiotics pipeline. Nature 2011, 472, 32. [Google Scholar] [CrossRef]
- Blair, J.; Webber, M.A.; Baylay, A.J.; Ogbolu, D.O.; Piddock, L.J. Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 2014, 13, 42–51. [Google Scholar] [CrossRef]
- Venter, H. Reversing resistance to counter antimicrobial resistance in the World Health Organisation’s critical priority of most dangerous pathogens. Biosci. Rep. 2019, 39, 4. [Google Scholar] [CrossRef]
- O’neill, J.I.M. Antimicrobial resistance: Tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. 2014, 20, 1–16. [Google Scholar]
- Nikaido, H.; Pagès, J.M. Broad-specificity efflux pumps and their role in multidrug resistance of Gram-negative bacteria. FEMS Microbiol. Rev. 2012, 36, 340–363. [Google Scholar] [CrossRef]
- Venter, H.; Mowla, R.; Ohene-Agyei, T.; Ma, S. RND-type drug efflux pumps from Gram-negative bacteria: Molecular mechanism and inhibition. Front. Microbiol. 2015, 6, 377. [Google Scholar] [CrossRef]
- Piddock, L.J. Clinically relevant chromosomally encoded multidrug resistance efflux pumps in bacteria. Clin. Microbiol. Rev. 2006, 19, 382–402. [Google Scholar] [CrossRef]
- Webber, M.A.; Piddock, L.J.V. The importance of efflux pumps in bacterial antibiotic resistance. J. Antimicrob. Chemother. 2003, 51, 9–11. [Google Scholar] [CrossRef]
- Opperman, T.J.; Kwasny, S.M.; Kim, H.S.; Nguyen, S.T.; Houseweart, C.; D’Souza, S.; Walker, G.C.; Peet, N.P.; Nikaido, H.; Bowlin, T.L. Characterization of a novel pyranopyridine inhibitor of the AcrAB efflux pump of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 722–733. [Google Scholar] [CrossRef]
- Törnroth-Horsefield, S.; Gourdon, P.; Horsefield, R.; Brive, L.; Yamamoto, N.; Mori, H.; Snijder, A.; Neutze, R. Crystal structure of AcrB in complex with a single transmembrane subunit reveals another twist. Structure 2007, 15, 1663–1673. [Google Scholar] [CrossRef]
- Wang, Y.; Mowla, R.; Guo, L.; Ogunniyi, A.D.; Rahman, T.; Lopes, M.A.D.B.; Ma, S.; Venter, H. Evaluation of a series of 2-napthamide derivatives as inhibitors of the drug efflux pump AcrB for the reversal of antimicrobial resistance. Bioorganic Med. Chem. Lett. 2017, 27, 733–739. [Google Scholar] [CrossRef]
- Ohene-Agyei, T.; Mowla, R.; Rahman, T.; Venter, H. Phytochemicals increase the antibacterial activity of antibiotics by acting on a drug efflux pump. Microbiologyopen 2014, 3, 885–896. [Google Scholar] [CrossRef]
- Sulaiman, K.A.; Al Qahtani, N.; Al Muqrin, M.; Al Dossari, M.; Al Wabel, A.; Al Sulaiman, T.; Vishwakarma, R.; Alolayan, A.; Abudayah, F.; Korayem, G.B.; et al. The correlation between non-O blood group type and recurrent catheter-associated urinary tract infections in critically ill patients: A retrospective study. J. Int. Med. Res. 2022, 50, 7. [Google Scholar] [CrossRef]
- Roca, I.; Akova, M.; Baquero, F.; Carlet, J.; Cavaleri, M.; Coenen, S.; Cohen, J.; Findlay, D.; Gyssens, I.; Heure, O.E.; et al. The global threat of antimicrobial resistance: Science for intervention. New Microbes New Infect. 2015, 6, 22–29. [Google Scholar] [CrossRef]
- Exner, M.; Bhattacharya, S.; Christiansen, B.; Gebel, J.; Goroncy-Bermes, P.; Hartemann, P.; Heeg, P.; Ilschner, C.; Kramer, A.; Larson, E.; et al. Antibiotic resistance: What is so special about multidrug-resistant Gram-negative bacteria? GMS Hyg. Infect. Control 2017, 12. [Google Scholar]
- Du, D.; Wang-Kan, X.; Neuberger, A.; Van Veen, H.W.; Pos, K.M.; Piddock, L.J.; Luisi, B.F. Multidrug efflux pumps: Structure, function and regulation. Nat. Rev. Microbiol. 2018, 16, 523–539. [Google Scholar] [CrossRef]
- World Health Organization. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed. Available online: https://www.who.int/news-room/detail/27-02-2017-who-publishes-list-of-bacteria-for-which-new-antibiotics-are-urgently-needed (accessed on 9 March 2020).
- Nikaido, H. Structure and mechanism of RND-type multidrug efflux pumps. Adv. Enzymol. Relat. Areas Mol. Biol. 2011, 77, 1. [Google Scholar]
- Cag, Y.; Caskurlu, H.; Fan, Y.; Cao, B.; Vahaboglu, H. Resistance mechanisms. Ann. Transl. Med. 2016, 4, 17. [Google Scholar] [CrossRef]
- Ali, A.S.; Al-Qahtani, A.; Nabi, S.; Altaf, K.; Sonekhi, G.B.; Ahmed, J.; Solangi, F.A.; Ali, K.S.; Ajaz, A. A short review on antibiotics and ever-changing microbial resistance mechanisms. Br. J. Pharm. 2018, 3, 1–7. [Google Scholar] [CrossRef]
- Arzanlou, M.; Chai, W.C.; Venter, H. Intrinsic, adaptive and acquired antimicrobial resistance in Gram-negative bacteria. Essays Biochem. 2017, 61, 49–59. [Google Scholar]
- Sun, J.; Deng, Z.; Yan, A. Bacterial multidrug efflux pumps: Mechanisms, physiology and pharmacological exploitations. Biochem. Biophys. Res. Commun. 2014, 453, 254–267. [Google Scholar] [CrossRef]
- Lloyd, N.A.; Nazaret, S.; Barkay, T. Genome-facilitated discovery of RND efflux pump-mediated resistance to cephalosporins in Vibrio spp. isolated from the mummichog fish gut. J. Glob. Antimicrob. Resist. 2019, 19, 294–300. [Google Scholar] [CrossRef]
- Navidifar, T.; Amin, M.; Rashno, M. Effects of sub-inhibitory concentrations of meropenem and tigecycline on the expression of genes regulating pili, efflux pumps and virulence factors involved in biofilm formation by Acinetobacter baumannii. Infect. Drug Resist. 2019, 12, 1099. [Google Scholar] [CrossRef]
- Pérez-Varela, M.; Corral, J.; Aranda, J.; Barbé, J. Roles of efflux pumps from different superfamilies in the surface-associated motility and virulence of Acinetobacter baumannii ATCC 17978. Antimicrob. Agents Chemother. 2019, 63, e02190-18. [Google Scholar] [CrossRef] [Green Version]
- Subhadra, B.; Oh, M.H.; Choi, C.H. RND efflux pump systems in Acinetobacter, with special emphasis on their role in quorum sensing. J. Bacteriol. Virol. 2019, 49, 1–11. [Google Scholar] [CrossRef]
- Hassan, K.A.; Jackson, S.M.; Penesyan, A.; Patching, S.G.; Tetu, S.G.; Eijkelkamp, B.A.; Brown, M.H.; Henderson, P.J.; Paulsen, I.T. Transcriptomic and biochemical analyses identify a family of chlorhexidine efflux proteins. Proc. Natl. Acad. Sci. USA 2013, 110, 20254–20259. [Google Scholar] [CrossRef]
- Hassan, K.A.; Liu, Q.; Henderson, P.J.; Paulsen, I.T. Homologs of the Acinetobacter baumannii AceI transporter represent a new family of bacterial multidrug efflux systems. MBio 2015, 6, e01982-14. [Google Scholar] [CrossRef] [PubMed]
- Munita, J.M.; Arias, C.A. Mechanisms of antibiotic resistance. Microbiol. Spectrum 2016, 4. [Google Scholar] [CrossRef]
- Yamaguchi, A.; Nakashima, R.; Sakurai, K. Structural basis of RND-type multidrug exporters. Front. Microbiol. 2015, 6, 327. [Google Scholar] [CrossRef]
- Du, D.; van Veen, H.W.; Luisi, B.F. Assembly and operation of bacterial tripartite multidrug efflux pumps. Trends Microbiol. 2015, 23, 311–319. [Google Scholar] [CrossRef]
- Du, D.; Venter, H.; Pos, K.M.; Luisi, B. The machinery and mechanism of multidrug efflux in gram-negative bacteria. In Microbial Efflux Pumps: Current Research; Caister Academic Press: Wymondham, UK, 2013; pp. 35–48. [Google Scholar]
- Eicher, T.; Cha, H.J.; Seeger, M.A.; Brandstätter, L.; El-Delik, J.; Bohnert, J.A.; Kern, W.V.; Verrey, F.; Grütter, M.G.; Diederichs, K.; et al. Transport of drugs by the multidrug transporter AcrB involves an access and a deep binding pocket that are separated by a switch-loop. Proc. Natl. Acad. Sci. USA 2012, 109, 5687–5692. [Google Scholar] [CrossRef] [PubMed]
- Symmons, M.F.; Bokma, E.; Koronakis, E.; Hughes, C.; Koronakis, V. The assembled structure of a complete tripartite bacterial multidrug efflux pump. Proc. Natl. Acad. Sci. USA 2009, 106, 7173–7178. [Google Scholar] [CrossRef] [PubMed]
- Elkins, C.A.; Nikaido, H. Substrate specificity of the RND-type multidrug efflux pumps AcrB and AcrD of Escherichia coli is determined predominately by two large periplasmic loops. J. Bacteriol. 2002, 184, 6490–6498. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Lu, W.; Ye, C.; Wang, Z.; Zhong, M.; Chai, Q.; Sheetz, M.; Wei, Y. Role of a conserved residue R780 in Escherichia coli multidrug transporter AcrB. Biochemistry 2013, 52, 6790–6796. [Google Scholar] [CrossRef] [PubMed]
- Zgurskaya, H.I. Covalently linked AcrB giant offers a new powerful tool for mechanistic analysis of multidrug efflux in bacteria. J. Bacteriol. 2009, 191, 1727–1728. [Google Scholar] [CrossRef]
- Zwama, M.; Yamaguchi, A.; Nishino, K. Phylogenetic and functional characterisation of the Haemophilus influenzae multidrug efflux pump AcrB. Commun. Biol. 2019, 2, 340. [Google Scholar] [CrossRef]
- Eicher, T.; Seeger, M.A.; Anselmi, C.; Zhou, W.; Brandstätter, L.; Verrey, F.; Diederichs, K.; Faraldo-Gómez, J.D.; Pos, K.M. Coupling of remote alternating-access transport mechanisms for protons and substrates in the multidrug efflux pump AcrB. elife 2014, 3, e03145. [Google Scholar] [CrossRef]
- Hung, L.W.; Kim, H.B.; Murakami, S.; Gupta, G.; Kim, C.Y.; Terwilliger, T.C. Crystal structure of AcrB complexed with linezolid at 3.5 Å resolution. J. Struct. Funct. Genom. 2013, 14, 71–75. [Google Scholar] [CrossRef]
- Mousa, J.J.; Yang, Y.; Tomkovich, S.; Shima, A.; Newsome, R.C.; Tripathi, P.; Oswald, E.; Bruner, S.D.; Jobin, C. MATE transport of the E. coli-derived genotoxin colibactin. Nat. Microbiol. 2016, 1, 15009. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Matsumoto, T.; Yamaguchi, A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature 2006, 443, 173–179. [Google Scholar] [CrossRef]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Nishino, K.; Yamaguchi, A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature 2011, 480, 565. [Google Scholar] [CrossRef] [PubMed]
- Sjuts, H.; Vargiu, A.V.; Kwasny, S.M.; Nguyen, S.T.; Kim, H.S.; Ding, X.; Ornik, A.R.; Ruggerone, P.; Bowlin, T.L.; Nikaido, H.; et al. Molecular basis for inhibition of AcrB multidrug efflux pump by novel and powerful pyranopyridine derivatives. Proc. Natl. Acad. Sci. USA 2016, 113, 3509–3514. [Google Scholar] [CrossRef] [PubMed]
- Du, D.; Wang, Z.; James, N.R.; Voss, J.E.; Klimont, E.; Ohene-Agyei, T.; Venter, H.; Chiu, W.; Luisi, B.F. Structure of the AcrAB–TolC multidrug efflux pump. Nature 2014, 509, 512–515. [Google Scholar] [CrossRef] [PubMed]
- Spengler, G.; Kincses, A.; Gajdács, M.; Amaral, L. New roads leading to old destinations: Efflux pumps as targets to reverse multidrug resistance in bacteria. Molecules 2017, 22, 468. [Google Scholar] [CrossRef] [Green Version]
- Seeger, M.A.; Schiefner, A.; Eicher, T.; Verrey, F.; Diederichs, K.; Pos, K.M. Structural asymmetry of AcrB trimer suggests a peristaltic pump mechanism. Science 2006, 313, 1295–1298. [Google Scholar] [CrossRef]
- Murakami, S.; Nakashima, R.; Yamashita, E.; Yamaguchi, A. Crystal structure of bacterial multidrug efflux transporter AcrB. Nature 2002, 419, 587–593. [Google Scholar] [CrossRef]
- Su, C.C.; Li, M.; Gu, R.; Takatsuka, Y.; McDermott, G.; Nikaido, H.; Yu, E.W. Conformation of the AcrB multidrug efflux pump in mutants of the putative proton relay pathway. J. Bacteriol. 2006, 188, 7290–7296. [Google Scholar] [CrossRef]
- Seeger, M.A.; von Ballmoos, C.; Verrey, F.; Pos, K.M. Crucial role of Asp408 in the proton translocation pathway of multidrug transporter AcrB: Evidence from site-directed mutagenesis and carbodiimide labeling. Biochemistry 2009, 48, 5801–5812. [Google Scholar] [CrossRef]
- Pos, K.M. Drug transport mechanism of the AcrB efflux pump. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2009, 1794, 782–793. [Google Scholar] [CrossRef]
- Seeger, M.A.; Diederichs, K.; Eicher, T.; Brandstatter, L.; Schiefner, A.; Verrey, F.; Pos, K.M. The AcrB efflux pump: Conformational cycling and peristalsis lead to multidrug resistance. Curr. Drug Targets 2008, 9, 729–749. [Google Scholar] [CrossRef]
- Ababou, A.; Koronakis, V. Structures of gate loop variants of the AcrB drug efflux pump bound by erythromycin substrate. PLoS ONE 2016, 11, e0159154. [Google Scholar]
- Vargiu, A.V.; Ramaswamy, V.K.; Malloci, G.; Malvacio, I.; Atzori, A.; Ruggerone, P. Computer simulations of the activity of RND efflux pumps. Res. Microbiol. 2018, 169, 384–392. [Google Scholar] [CrossRef] [PubMed]
- Vargiu, A.V.; Nikaido, H. Multidrug binding properties of the AcrB efflux pump characterized by molecular dynamics simulations. Proc. Natl. Acad. Sci. USA 2012, 109, 20637–20642. [Google Scholar] [CrossRef] [PubMed]
- Jamshidi, S.; Sutton, J.M.; Rahman, K.M. Mapping the dynamic functions and structural features of acrb efflux pump transporter using accelerated molecular dynamics simulations. Sci. Rep. 2018, 8, 10470. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Fan, G.; Hryc, C.F.; Blaza, J.N.; Serysheva, I.I.; Schmid, M.F.; Chiu, W.; Luisi, B.F.; Du, D. An allosteric transport mechanism for the AcrAB-TolC multidrug efflux pump. elife 2017, 6, e24905. [Google Scholar] [CrossRef]
- Sakurai, K.; Yamasaki, S.; Nakao, K.; Nishino, K.; Yamaguchi, A.; Nakashima, R. Crystal structures of multidrug efflux pump MexB bound with high-molecular-mass compounds. Sci. Rep. 2019, 9, 7843. [Google Scholar] [CrossRef]
- Nakashima, R.; Sakurai, K.; Yamasaki, S.; Hayashi, K.; Nagata, C.; Hoshino, K.; Onodera, Y.; Nishino, K.; Yamaguchi, A. Structural basis for the inhibition of bacterial multidrug exporters. Nature 2013, 500, 102–106. [Google Scholar] [CrossRef]
- Sennhauser, G.; Bukowska, M.A.; Briand, C.; Grütter, M.G. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa. J. Mol. Biol. 2009, 389, 134–145. [Google Scholar] [CrossRef]
- Aron, Z.; Opperman, T.J. The hydrophobic trap—The Achilles heel of RND efflux pumps. Res. Microbiol. 2018, 169, 393–400. [Google Scholar] [CrossRef]
- Sharma, A.; Gupta, V.K.; Pathania, R. Efflux pump inhibitors for bacterial pathogens: From bench to bedside. Indian J. Med. Res. 2019, 149, 129. [Google Scholar]
- Bal, A.M.; Kumar, A.; Gould, I.M. Antibiotic heterogeneity: From concept to practice. Ann. N. Y. Acad. Sci. 2010, 1213, 81–91. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Van Bambeke, F.; Pagès, J.M.; Lee, V.J. Inhibitor of bacterial efflux pumps as adjuvants in antibacterial therapy and diagnostic tools for detection of resistance by efflux. Front. Anti-Infect. Drug Discov. 2010, 1, 157–175. [Google Scholar]
- Pagès, J.M.; Amaral, L. Mechanisms of drug efflux and strategies to combat them: Challenging the efflux pump of Gram-negative bacteria. Biochim. Et Biophys. Acta (BBA)-Proteins Proteom. 2009, 1794, 826–833. [Google Scholar] [CrossRef] [PubMed]
- Abdali, N.; Parks, J.M.; Haynes, K.M.; Chaney, J.L.; Green, A.T.; Wolloscheck, D.; Walker, J.K.; Rybenkov, V.V.; Baudry, J.; Smith, J.C.; et al. Reviving antibiotics: Efflux pump inhibitors that interact with AcrA, a membrane fusion protein of the AcrAB-TolC multidrug efflux pump. ACS Infect. Dis. 2017, 3, 89–98. [Google Scholar] [CrossRef]
- Opperman, T.J.; Nguyen, S.T. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015, 6, 421. [Google Scholar] [CrossRef]
- Lomovskaya, O.; Warren, M.S.; Lee, A.; Galazzo, J.; Fronko, R.; Lee, M.A.Y.; Blais, J.; Cho, D.; Chamberland, S.; Renau, T.; et al. Identification and characterization of inhibitors of multidrug resistance efflux pumps in Pseudomonas aeruginosa: Novel agents for combination therapy. Antimicrob. Agents Chemother. 2001, 45, 105–116. [Google Scholar] [CrossRef] [Green Version]
- Aron, Z.; Opperman, T.J. Optimization of a novel series of pyranopyridine RND efflux pump inhibitors. Curr. Opin. Microbiol. 2016, 33, 1–6. [Google Scholar] [CrossRef]
- Renau, T.E.; Léger, R.; Filonova, L.; Flamme, E.M.; Wang, M.; Yen, R.; Madsen, D.; Griffith, D.; Chamberland, S.; Dudley, M.N.; et al. Conformationally-restricted analogues of efflux pump inhibitors that potentiate the activity of levofloxacin in Pseudomonas aeruginosa. Bioorganic Med. Chem. Lett. 2003, 13, 2755–2758. [Google Scholar] [CrossRef]
- Wang, Y.; Venter, H.; Ma, S. Efflux pump inhibitors: A novel approach to combat efflux-mediated drug resistance in bacteria. Curr. Drug Targets 2016, 17, 702–719. [Google Scholar] [CrossRef]
- Cortez-Cordova, J.; Kumar, A. Activity of the efflux pump inhibitor phenylalanine-arginine β-naphthylamide against the AdeFGH pump of Acinetobacter baumannii. Int. J. Antimicrob. Agents 2011, 37, 420–424. [Google Scholar] [CrossRef] [PubMed]
- Kurinčič, M.; Klančnik, A.; Smole Možina, S. Effects of efflux pump inhibitors on erythromycin, ciprofloxacin, and tetracycline resistance in Campylobacter spp. isolates. Microb. Drug Resist. 2012, 18, 492–501. [Google Scholar] [CrossRef] [PubMed]
- Lomovskaya, O.; Bostian, K.A. Practical applications and feasibility of efflux pump inhibitors in the clinic—A vision for applied use. Biochem. Pharmacol. 2006, 71, 910–918. [Google Scholar] [CrossRef] [PubMed]
- Malléa, M.; Chevalier, J.; Eyraud, A.; Pagès, J.M. Inhibitors of antibiotic efflux pump in resistant Enterobacter aerogenes strains. Biochem. Biophys. Res. Commun. 2002, 293, 1370–1373. [Google Scholar] [CrossRef]
- Mamelli, L.; Amoros, J.P.; Pagès, J.M.; Bolla, J.M. A phenylalanine–arginine β-naphthylamide sensitive multidrug efflux pump involved in intrinsic and acquired resistance of Campylobacter to macrolides. Int. J. Antimicrob. Agents 2003, 22, 237–241. [Google Scholar] [CrossRef]
- Renau, T.E.; Léger, R.; Flamme, E.M.; Sangalang, J.; She, M.W.; Yen, R.; Gannon, C.L.; Griffith, D.; Chamberland, S.; Lomovskaya, O.; et al. Inhibitors of efflux pumps in Pseudomonas aeruginosa potentiate the activity of the fluoroquinolone antibacterial levofloxacin. J. Med. Chem. 1999, 42, 4928–4931. [Google Scholar] [CrossRef]
- Vera-Leiva, A.; Carrasco-Anabalón, S.; Lima, C.A.; Villagra, N.; Domínguez, M.; Bello-Toledo, H.; González-Rocha, G. The efflux pump inhibitor phenylalanine-arginine β-naphthylamide (PAβN) increases resistance to carbapenems in Chilean clinical isolates of KPC-producing Klebsiella pneumoniae. J. Glob. Antimicrob. Resist. 2018, 12, 73–76. [Google Scholar] [CrossRef]
- Bohnert, J.A.; Kern, W.V. Selected arylpiperazines are capable of reversing multidrug resistance in Escherichia coli overexpressing RND efflux pumps. Antimicrob. Agents Chemother. 2005, 49, 849–852. [Google Scholar] [CrossRef]
- Kern, W.V.; Steinke, P.; Schumacher, A.; Schuster, S.; Baum, H.V.; Bohnert, J.A. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 339–343. [Google Scholar] [CrossRef]
- Pannek, S.; Higgins, P.G.; Steinke, P.; Jonas, D.; Akova, M.; Bohnert, J.A.; Seifert, H.; Kern, W.V. Multidrug efflux inhibition in Acinetobacter baumannii: Comparison between 1-(1-naphthylmethyl)-piperazine and phenyl-arginine-β-naphthylamide. J. Antimicrob. Chemother. 2006, 57, 970–974. [Google Scholar] [CrossRef]
- Schumacher, A.; Steinke, P.; Bohnert, J.A.; Akova, M.; Jonas, D.; Kern, W.V. Effect of 1-(1-naphthylmethyl)-piperazine, a novel putative efflux pump inhibitor, on antimicrobial drug susceptibility in clinical isolates of Enterobacteriaceae other than Escherichia coli. J. Antimicrob. Chemother. 2006, 57, 344–348. [Google Scholar] [CrossRef] [PubMed]
- Sonnet, P.; Izard, D.; Mullié, C. Prevalence of efflux-mediated ciprofloxacin and levofloxacin resistance in recent clinical isolates of Pseudomonas aeruginosa and its reversal by the efflux pump inhibitors 1-(1-naphthylmethyl)-piperazine and phenylalanine-arginine-β-naphthylamide. Int. J. Antimicrob. Agents 2012, 39, 77–80. [Google Scholar] [CrossRef] [PubMed]
- Thorarensen, A.; Presley-Bodnar, A.L.; Marotti, K.R.; Boyle, T.P.; Heckaman, C.L.; Bohanon, M.J.; Tomich, P.K.; Zurenko, G.E.; Sweeney, M.T.; Yagi, B.H. 3-Arylpiperidines as potentiators of existing antibacterial agents. Bioorganic Med. Chem. Lett. 2001, 11, 1903–1906. [Google Scholar] [CrossRef]
- Chevalier, J.; Atifi, S.; Eyraud, A.; Mahamoud, A.; Barbe, J.; Pagès, J.M. New Pyridoquinoline Derivatives as Potential Inhibitors of the Fluoroquinolone Efflux Pump in Resistant Enterobacter aerogenes Strains. J. Med. Chem. 2001, 44, 4023–4026. [Google Scholar] [CrossRef] [PubMed]
- Gallo, S.; Chevalier, J.; Mahamoud, A.; Eyraud, A.; Pagès, J.M.; Barbe, J. 4-alkoxy and 4-thioalkoxyquinoline derivatives as chemosensitizers for the chloramphenicol-resistant clinical Enterobacter aerogenes 27 strain. Int. J. Antimicrob. Agents 2003, 22, 270–273. [Google Scholar] [CrossRef]
- Ghisalberti, D.; Mahamoud, A.; Chevalier, J.; Baitiche, M.; Martino, M.; Pagès, J.M.; Barbe, J. Chloroquinolines block antibiotic efflux pumps in antibiotic-resistant Enterobacter aerogenes isolates. Int. J. Antimicrob. Agents 2006, 27, 565–569. [Google Scholar] [CrossRef]
- Mahamoud, A.; Chevalier, J.; Davin-Regli, A.; Barbe, J. Quinoline derivatives as promising inhibitors of antibiotic efflux pump in multidrug resistant Enterobacter aerogenes isolates. Curr. Drug Targets 2006, 7, 843–847. [Google Scholar] [CrossRef]
- Malléa, M.; Mahamoud, A.; Chevalier, J.; Alibert-Franco, S.; Brouant, P.; Barbe, J.; Pagès, J.M. Alkylaminoquinolines inhibit the bacterial antibiotic efflux pump in multidrug-resistant clinical isolates. Biochem. J. 2003, 376, 801–805. [Google Scholar] [CrossRef]
- Chevalier, J.; Mahamoud, A.; Baitiche, M.; Adam, E.; Viveiros, M.; Smarandache, A.; Militaru, A.; Pascu, M.L.; Amaral, L.; Pagès, J.M. Quinazoline derivatives are efficient chemosensitizers of antibiotic activity in Enterobacter aerogenes, Klebsiella pneumoniae and Pseudomonas aeruginosa resistant strains. Int. J. Antimicrob. Agents 2010, 36, 164–168. [Google Scholar] [CrossRef]
- Bailey, A.M.; Paulsen, I.T.; Piddock, L.J. RamA confers multidrug resistance in Salmonella enterica via increased expression of acrB, which is inhibited by chlorpromazine. Antimicrob. Agents Chemother. 2008, 52, 3604–3611. [Google Scholar] [CrossRef]
- Chan, Y.Y.; Ong, Y.M.; Chua, K.L. Synergistic interaction between phenothiazines and antimicrobial agents against Burkholderia pseudomallei. Antimicrob. Agents Chemother. 2007, 51, 623–630. [Google Scholar] [CrossRef] [PubMed]
- Martins, M.; Dastidar, S.G.; Fanning, S.; Kristiansen, J.E.; Molnar, J.; Pages, J.M.; Schelz, Z.; Spengler, G.; Viveiros, M.; Amaral, L. Potential role of non-antibiotics (helper compounds) in the treatment of multidrug-resistant Gram-negative infections: Mechanisms for their direct and indirect activities. Int. J. Antimicrob. Agents 2008, 31, 198–208. [Google Scholar] [CrossRef]
- Handzlik, J.; Szymańska, E.; Chevalier, J.; Otrębska, E.; Kieć-Kononowicz, K.; Pagès, J.M.; Alibert, S. Amine–alkyl derivatives of hydantoin: New tool to combat resistant bacteria. Eur. J. Med. Chem. 2011, 46, 5807–5816. [Google Scholar] [CrossRef] [PubMed]
- Otręebska-Machaj, E.; Chevalier, J.; Handzlik, J.; Szymańska, E.; Schabikowski, J.; Boyer, G.; Bolla, J.M.; Kieć-Kononowicz, K.; Pagès, J.M.; Alibert, S. Efflux pump blockers in Gram-negative bacteria: The new generation of hydantoin based-modulators to improve antibiotic activity. Front. Microbiol. 2016, 7, 622. [Google Scholar] [CrossRef]
- Piddock, L.J.; Garvey, M.I.; Rahman, M.M.; Gibbons, S. Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65, 1215–1223. [Google Scholar] [CrossRef]
- Nelson, M.L.; Park, B.H.; Levy, S.B. Molecular requirements for the inhibition of the tetracycline antiport protein and the effect of potent inhibitors on the growth of tetracycline-resistant bacteria. J. Med. Chem. 1994, 37, 1355–1361. [Google Scholar] [CrossRef]
- Nelson, M.L.; Park, B.H.; Andrews, J.S.; Georgian, V.A.; Thomas, R.C.; Levy, S.B. Inhibition of the tetracycline efflux antiport protein by 13-thio-substituted 5-hydroxy-6-deoxytetracyclines. J. Med. Chem. 1993, 36, 370–377. [Google Scholar] [CrossRef]
- Van Bambeke, F.; Lee, V.J. Inhibitors of bacterial efflux pumps as adjuvants in antibiotic treatments and diagnostic tools for detection of resistance by efflux. Recent Pat. Anti-Infect. Drug Discov. 2006, 1, 157–175. [Google Scholar] [CrossRef]
- Zeng, B.; Wang, H.; Zou, L.; Zhang, A.; Yang, X.; Guan, Z. Evaluation and target validation of indole derivatives as inhibitors of the AcrAB-TolC efflux pump. Biosci. Biotechnol. Biochem. 2010, 74, 2237–2241. [Google Scholar] [CrossRef]
- Y Mahmood, H.; Jamshidi, S.; Mark Sutton, J.; M Rahman, K. Current advances in developing inhibitors of bacterial multidrug efflux pumps. Curr. Med. Chem. 2016, 23, 1062–1081. [Google Scholar] [CrossRef]
- Vargiu, A.V.; Ruggerone, P.; Opperman, T.J.; Nguyen, S.T.; Nikaido, H. Molecular mechanism of MBX2319 inhibition of Escherichia coli AcrB multidrug efflux pump and comparison with other inhibitors. Antimicrob. Agents Chemother. 2014, 58, 6224–6234. [Google Scholar] [CrossRef]
- Yoshida, K.I.; Nakayama, K.; Kuru, N.; Kobayashi, S.; Ohtsuka, M.; Takemura, M.; Hoshino, K.; Kanda, H.; Zhang, J.Z.; Lee, V.J.; et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 5: Carbon-substituted analogues at the C-2 position. Bioorganic Med. Chem. 2006, 14, 1993–2004. [Google Scholar] [CrossRef]
- Yoshida, K.I.; Nakayama, K.; Ohtsuka, M.; Kuru, N.; Yokomizo, Y.; Sakamoto, A.; Takemura, M.; Hoshino, K.; Kanda, H.; Nitanai, H.; et al. MexAB-OprM specific efflux pump inhibitors in Pseudomonas aeruginosa. Part 7: Highly soluble and in vivo active quaternary ammonium analogue D13-9001 2004, a potential preclinical candidate. Bioorganic Med. Chem. 2007, 15, 7087–7097. [Google Scholar] [CrossRef]
- Mu, Y.; Shen, Z.; Jeon, B.; Dai, L.; Zhang, Q. Synergistic effects of anti-CmeA and anti-CmeB peptide nucleic acids on sensitizing Campylobacter jejuni to antibiotics. Antimicrob. Agents Chemother. 2013, 57, 4575–4577. [Google Scholar] [CrossRef]
- Lee, M.D.; Galazzo, J.L.; Staley, A.L.; Lee, J.C.; Warren, M.S.; Fuernkranz, H.; Chamberland, S.; Lomovskaya, O.; Miller, G.H. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Il Farm. 2001, 56, 81–85. [Google Scholar] [CrossRef]
- Blanchard, C.; Barnett, P.; Perlmutter, J.; Dunman, P.M. Identification of Acinetobacter baumannii serum-associated antibiotic efflux pump inhibitors. Antimicrob. Agents Chemother. 2014, 58, 6360–6370. [Google Scholar] [CrossRef]
- Wang, Y.; Mowla, R.; Ji, S.; Guo, L.; Lopes, M.A.D.B.; Jin, C.; Song, D.; Ma, S.; Venter, H. Design, synthesis and biological activity evaluation of novel 4-subtituted 2-naphthamide derivatives as AcrB inhibitors. Eur. J. Med. Chem. 2018, 143, 699–709. [Google Scholar] [CrossRef]
- Stavri, M.; Piddock, L.J.; Gibbons, S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef]
- Chan, B.C.; Ip, M.; Lau, C.B.; Lui, S.L.; Jolivalt, C.; Ganem-Elbaz, C.; Litaudon, M.; Reiner, N.E.; Gong, H.; See, R.H.; et al. Synergistic effects of baicalein with ciprofloxacin against NorA over-expressed methicillin-resistant Staphylococcus aureus (MRSA) and inhibition of MRSA pyruvate kinase. J. Ethnopharmacol. 2011, 137, 767–773. [Google Scholar] [CrossRef]
- Fujita, M.; Shiota, S.; Kuroda, T.; Hatano, T.; Yoshida, T.; Mizushima, T.; Tsuchiya, T. Remarkable Synergies between Baicalein and Tetracycline, and Baicalein and β-Lactams against Methicillin-Resistant Staphylococcus aureus. Microbiol. Immunol. 2005, 49, 391–396. [Google Scholar] [CrossRef] [PubMed]
- Gibbons, S.; Moser, E.; Kaatz, G.W. Catechin gallates inhibit multidrug resistance (MDR) in Staphylococcus aureus. Planta Med. 2004, 70, 1240–1242. [Google Scholar] [CrossRef] [PubMed]
- Oluwatuyi, M.; Kaatz, G.W.; Gibbons, S. Antibacterial and resistance modifying activity of Rosmarinus officinalis. Phytochemistry 2004, 65, 3249–3254. [Google Scholar] [CrossRef] [PubMed]
- Bag, A.; Chattopadhyay, R.R. Efflux-pump inhibitory activity of a gallotannin from Terminalia chebula fruit against multidrug-resistant uropathogenic Escherichia coli. Nat. Prod. Res. 2014, 28, 1280–1283. [Google Scholar] [CrossRef]
- Dwivedi, G.R.; Tyagi, R.; Sanchita Tripathi, S.; Pati, S.; Srivastava, S.K.; Darokar, M.P.; Sharma, A. Antibiotics potentiating potential of catharanthine against superbug Pseudomonas aeruginosa. J. Biomol. Struct. Dyn. 2018, 36, 4270–4284. [Google Scholar] [CrossRef]
- Prasch, S.; Bucar, F. Plant derived inhibitors of bacterial efflux pumps: An update. Phytochem. Rev. 2015, 14, 961–974. [Google Scholar] [CrossRef]
- Maurya, A.; Dwivedi, G.R.; Darokar, M.P.; Srivastava, S.K. Antibacterial and Synergy of Clavine Alkaloid Lysergol and its Derivatives Against Nalidixic Acid-Resistant Escherichia coli. Chem. Biol. Drug Des. 2013, 81, 484–490. [Google Scholar] [CrossRef]
- Dwivedi, G.R.; Upadhyay, H.C.; Yadav, D.K.; Singh, V.; Srivastava, S.K.; Khan, F.; Darmwal, N.S.; Darokar, M.P. 4-Hydroxy-α-Tetralone and its Derivative as Drug Resistance Reversal Agents in Multi Drug Resistant Escherichia coli. Chem. Biol. Drug Des. 2014, 83, 482–492. [Google Scholar] [CrossRef]
- Chitemerere, T.A.; Mukanganyama, S. Evaluation of cell membrane integrity as a potential antimicrobial target for plant products. BMC Complement. Altern. Med. 2014, 14, 278. [Google Scholar] [CrossRef] [PubMed]
- Aparna, V.; Dineshkumar, K.; Mohanalakshmi, N.; Velmurugan, D.; Hopper, W. Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS ONE 2014, 9, e101840. [Google Scholar] [CrossRef]
- Dwivedi, G.R.; Maurya, A.; Yadav, D.K.; Khan, F.; Darokar, M.P.; Srivastava, S.K. Drug resistance reversal potential of ursolic acid derivatives against nalidixic acid-and multidrug-resistant Escherichia coli. Chem. Biol. Drug Des. 2015, 86, 272–283. [Google Scholar] [CrossRef]
- Maisuria, V.B.; Hosseinidoust, Z.; Tufenkji, N. Polyphenolic extract from maple syrup potentiates antibiotic susceptibility and reduces biofilm formation of pathogenic bacteria. Appl. Environ. Microbiol. 2015, 81, 3782–3792. [Google Scholar] [CrossRef] [PubMed]
- Kovač, J.; Šimunović, K.; Wu, Z.; Klančnik, A.; Bucar, F.; Zhang, Q.; Možina, S.S. Antibiotic resistance modulation and modes of action of (-)-α-pinene in Campylobacter jejuni. PLoS ONE 2015, 10, e0122871. [Google Scholar] [CrossRef] [PubMed]
- Aghayan, S.S.; Mogadam, H.K.; Fazli, M.; Darban-Sarokhalil, D.; Khoramrooz, S.S.; Jabalameli, F.; Yaslianifard, S.; Mirzaii, M. The effects of berberine and palmatine on efflux pumps inhibition with different gene patterns in Pseudomonas aeruginosa isolated from burn infections. Avicenna J. Med. Biotechnol. 2017, 9, 2. [Google Scholar]
- Siriyong, T.; Srimanote, P.; Chusri, S.; Yingyongnarongkul, B.E.; Suaisom, C.; Tipmanee, V.; Voravuthikunchai, S.P. Conessine as a novel inhibitor of multidrug efflux pump systems in Pseudomonas aeruginosa. BMC Complement. Altern. Med. 2017, 17, 405. [Google Scholar] [CrossRef]
- Willyard, C. Drug-resistant bacteria ranked. Nature 2017, 543, 15. [Google Scholar] [CrossRef]
- Lamut, A.; Peterlin Mašič, L.; Kikelj, D.; Tomašič, T. Efflux pump inhibitors of clinically relevant multidrug resistant bacteria. Med. Res. Rev. 2019, 39, 2460–2504. [Google Scholar] [CrossRef]
- Pouch, S.M.; Patel, G.; AST Infectious Diseases Community of Practice. Multidrug-resistant Gram-negative bacterial infections in solid organ transplant recipients—Guidelines from the American Society of Transplantation Infectious Diseases Community of Practice. Clin. Transplant. 2019, 33, e13594. [Google Scholar] [CrossRef]
- Tyers, M.; Wright, G.D. Drug combinations: A strategy to extend the life of antibiotics in the 21st century. Nat. Rev. Microbiol. 2019, 17, 141–155. [Google Scholar] [CrossRef]
- Slipski, C.J.; Zhanel, G.G.; Bay, D.C. Biocide selective TolC-independent efflux pumps in Enterobacteriaceae. J. Membr. Biol. 2018, 251, 15–33. [Google Scholar] [CrossRef]
- Abraham, V.C.; Towne, D.L.; Waring, J.F.; Warrior, U.; Burns, D.J. Application of a high-content multiparameter cytotoxicity assay to prioritize compounds based on toxicity potential in humans. SLAS Discov. 2008, 13, 527–537. [Google Scholar] [CrossRef]
- Garvey, M.I.; Rahman, M.M.; Gibbons, S.; Piddock, L.J. Medicinal plant extracts with efflux inhibitory activity against Gram-negative bacteria. Int. J. Antimicrob. Agents 2011, 37, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Handzlik, J.; Matys, A.; Kieć-Kononowicz, K. Recent advances in multi-drug resistance (MDR) efflux pump inhibitors of Gram-positive bacteria S. aureus. Antibiotics 2013, 2, 28–45. [Google Scholar] [CrossRef] [PubMed]
- Bettiol, E.; Wetherington, J.D.; Schmitt, N.; Harbarth, S. Challenges and solutions for clinical development of new antibacterial agents: Results of a survey among pharmaceutical industry professionals. Antimicrob. Agents Chemother. 2015, 59, 3695–3699. [Google Scholar] [CrossRef] [PubMed]
- González-Bello, C. Antibiotic adjuvants—A strategy to unlock bacterial resistance to antibiotics. Bioorganic Med. Chem. Lett. 2017, 27, 4221–4228. [Google Scholar] [CrossRef]





| EPI Type | Bacteria | Efflux Substrate | References |
|---|---|---|---|
| Peptidomimetics | E. coli, P. aeruginosa, K. pneumoniae, S. enterica, Campylobacter spp., E. aerogenes, A. baumanii | CAM, FQs, macrolides, CAR, TETs | [66,73,74,75,76,77,78,79] |
| Arylpiperazines | A. baumanii, P. aeruginosa, C. jejuni, Enterobacteriaceae (not Serratia) | CAM, TETs, FQs, macrolides, LZD | [74,80,81,82,83,84] |
| Arylpiperidines | E. coli | LZD | [85] |
| Quinoline derivatives | E. aerogenes, K. pneumoniae | CAM, TET, NOR | [86,87,88,89,90] |
| Quinazoline derivatives | E. aerogenes, P. aeruginosa, | CAM, NAL, SPX | [91] |
| Phenothiazines | E. coli, S. enterica, B. pseudomallei | CAM, TETs, NAL, LVX, triclosan, ERY, aminoglycosides | [92,93,94] |
| Hydantoins | E. coli, E. aerogenes | CAM, NAL, SPX, doxycycline, ERY | [95,96] |
| Antibiotics globomycin | E. aerogenes | CAM, NOR | [76] |
| Trimethoprim | Enterobacteriaceae, P. aeruginosa | CAM, TET, CIP, ERY | [97] |
| Antibiotic analogs, Tetracycline analogs | E. coli | TETs | [98,99] |
| Fluoroquinolone analogs | E. coli, P. aeruginosa | FQs, macrolides | [100] |
| Aminoglycoside analogs | H. influenza | TET, GEN | [100] |
| Indole derivatives | E. coli | CAM, ERY, CIP, TET | [101] |
| Substituted polyamines | H. influenza | – | [102] |
| Pyranopyridines | Enterobacteriaceae | FQs, PIP | [9,44,70,103] |
| Pyridopyrimidines | P. aeruginosa | FQs, β-lactams | [104,105] |
| sRNA and antisense oligonucleotides | E. coli, C. jejuni | CIP, ERY | [100,106] |
| Microbial EPIs | P. aeruginosa | LVX | [107] |
| Serum compounds | A. baumanii, P. aeruginosa | MIN, CIP | [108] |
| Epinephrine | Enterobacteriaceae, P. aeruginosa | CAM, TET, CIP, ERY | [97] |
| Naphthamides | E. coli | CAM, TPP, ERY | [11,109] |
| Extract Compound | Plant | Bacteria Affected | References |
|---|---|---|---|
| Plumbagin | Plumbago indica | E. coli | [12] |
| Nordihydroguaretic acid | Larrea tridentata | ||
| Shikonin | Lithospermum erythrorhizon | ||
| Lysergol | Ipomea muricata | [118] | |
| 4-Hydroxy-α-tetralone + semisynthetic derivatives | Ammannia spp. | [119] | |
| Ethanolic extract | Baccharoides adoensis, Callistemon citrinus | Pseudomonas aeruginosa | [120] |
| Lanatoside C | Digitalis lanata | Pseudomonas aeruginosa, E. coli | [121] |
| Ursolic acid | Eucalyptus tereticornis | E. coli | [122] |
| Daidzein | Glycine max | ||
| Phenolic-rich maple syrup extracts (PRMSE) | Acer saccharum | E. coli, Proteus mirabilis, Pseudomonas aeruginosa | [123] |
| (−)-α-Pinene | Alpinia katsumadai | Campylobacter jejuni | [124] |
| Berberine, palmatine | Berveris bulgaris | Pseudomonas aeruginosa | [125] |
| Conessine | Holarrhena antidysenterica | [126] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alenazy, R. Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology 2022, 11, 1328. https://doi.org/10.3390/biology11091328
Alenazy R. Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology. 2022; 11(9):1328. https://doi.org/10.3390/biology11091328
Chicago/Turabian StyleAlenazy, Rawaf. 2022. "Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli" Biology 11, no. 9: 1328. https://doi.org/10.3390/biology11091328
APA StyleAlenazy, R. (2022). Drug Efflux Pump Inhibitors: A Promising Approach to Counter Multidrug Resistance in Gram-Negative Pathogens by Targeting AcrB Protein from AcrAB-TolC Multidrug Efflux Pump from Escherichia coli. Biology, 11(9), 1328. https://doi.org/10.3390/biology11091328






