The Divergent and Conserved Expression Profile of Turtle Nanog Gene Comparing with Fish and Mammals
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals and Tissue Collection
2.2. Cloning PsNanog cDNA Sequence and Its Promoter
2.3. Bioinformatic Analysis of PsNanog Gene and Its Promoter
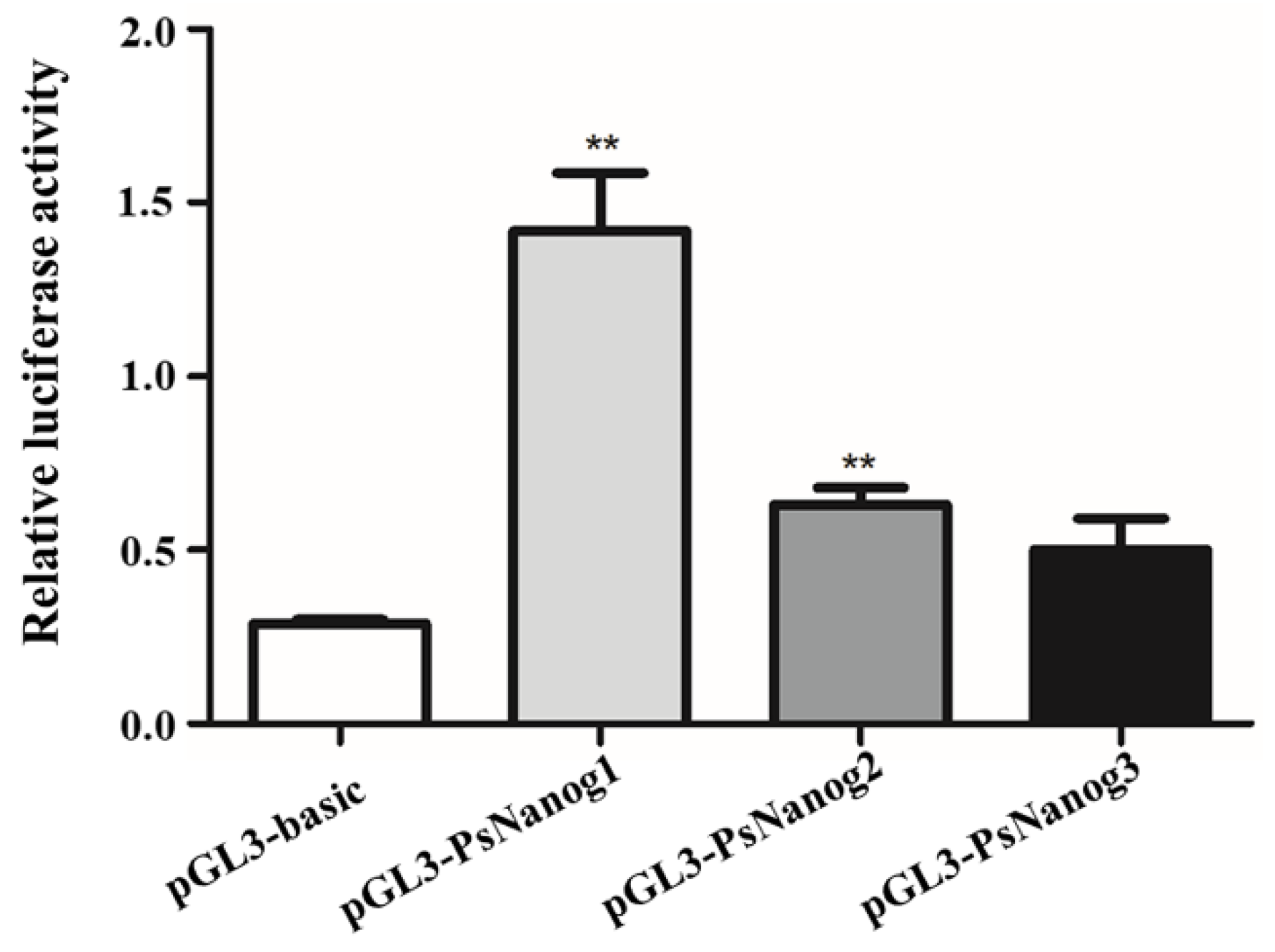
2.4. Luciferase Assay
2.5. Expression Analysis of Nanog mRNA by RT-PCR and RT-qPCR
2.6. Cryostat Sections of Testis and Ovary
2.7. In Situ Hybridization
2.8. Preparation of Anti-Nanog Antibody
2.9. Immunofluorescence Staining of Ovary and Testis Sections
3. Results
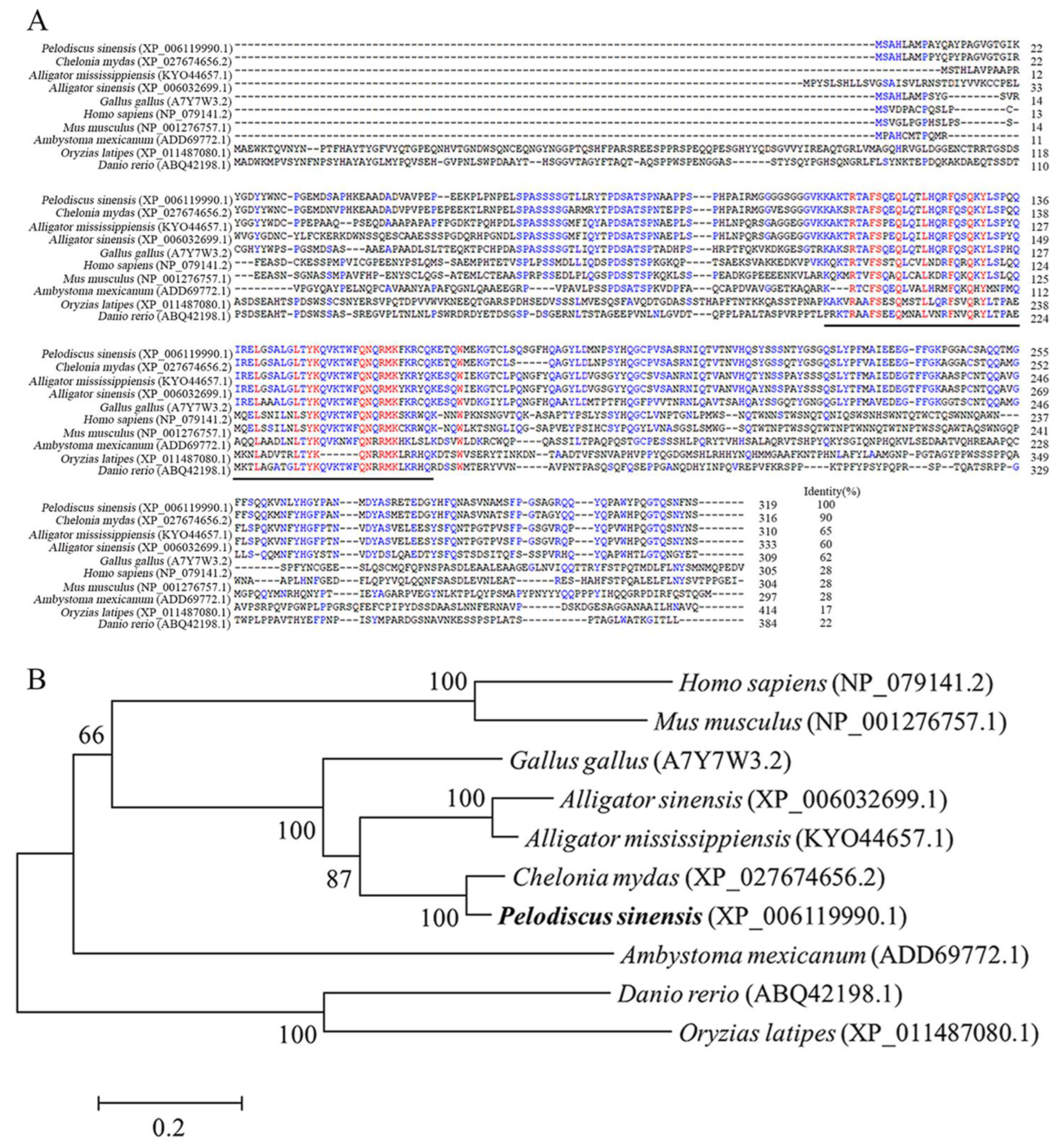
3.1. Molecular Characterization of PsNanog and Its Promoter
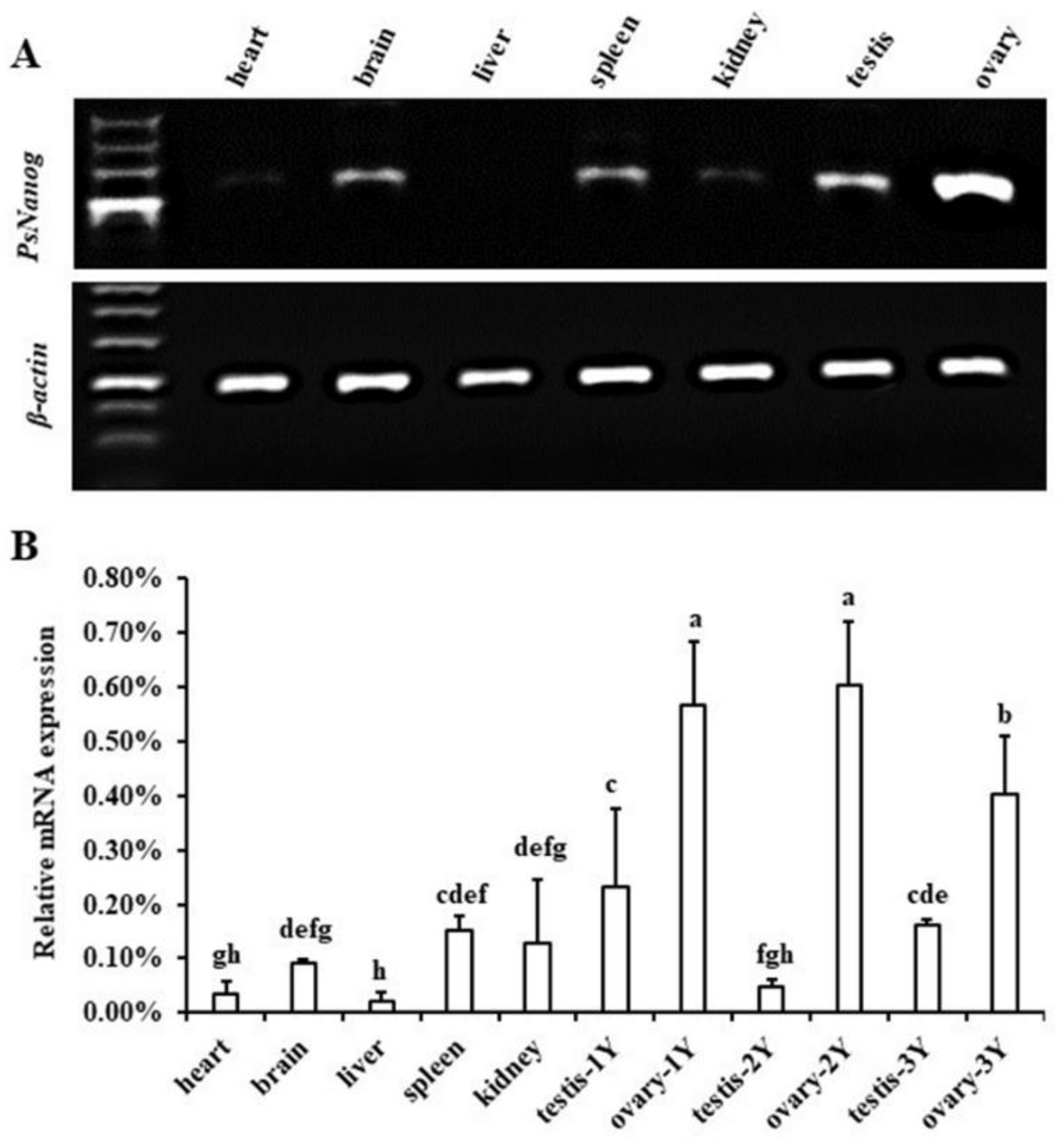
3.2. Transcription Expression of PsNanog in Different Tissues
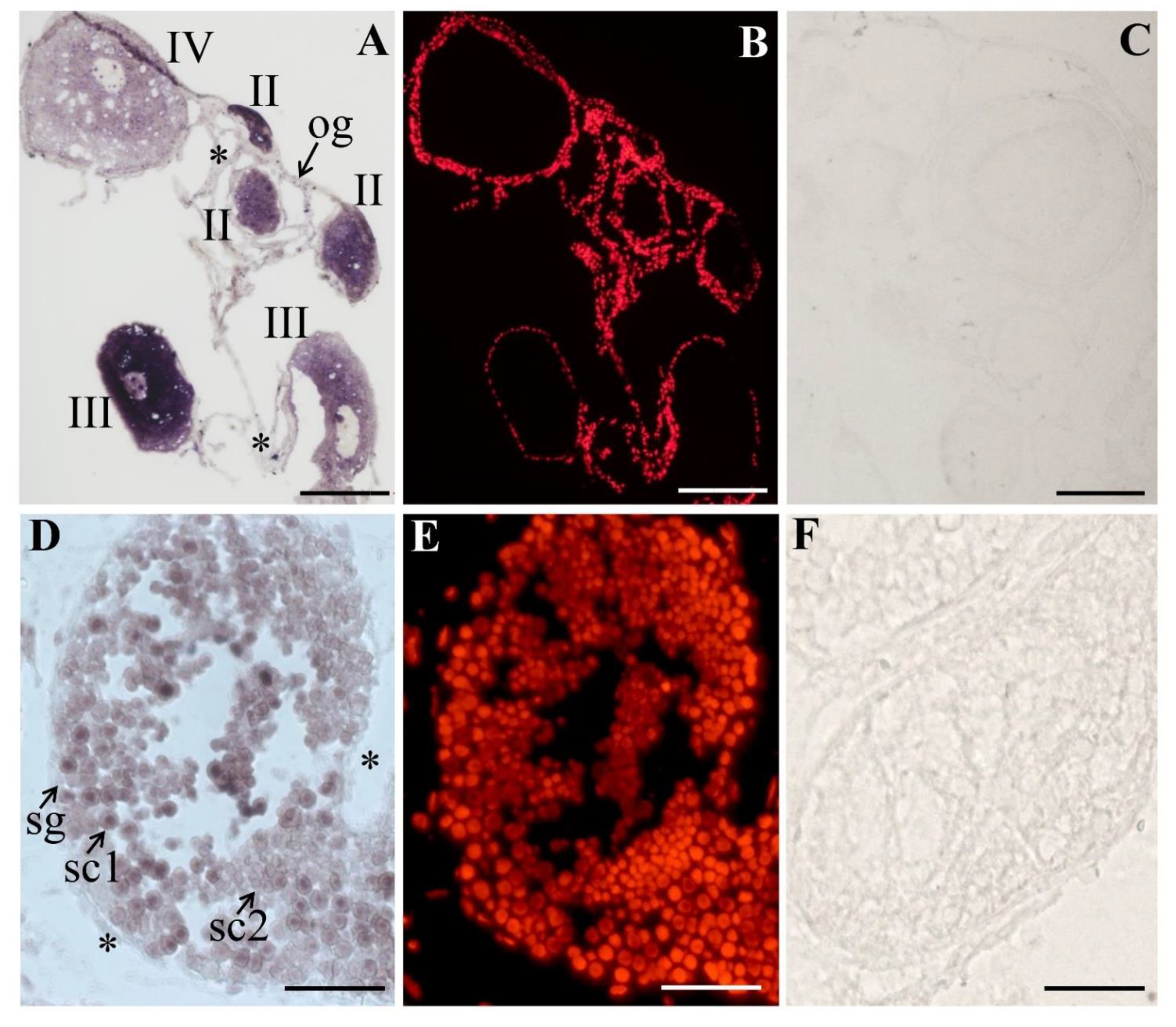
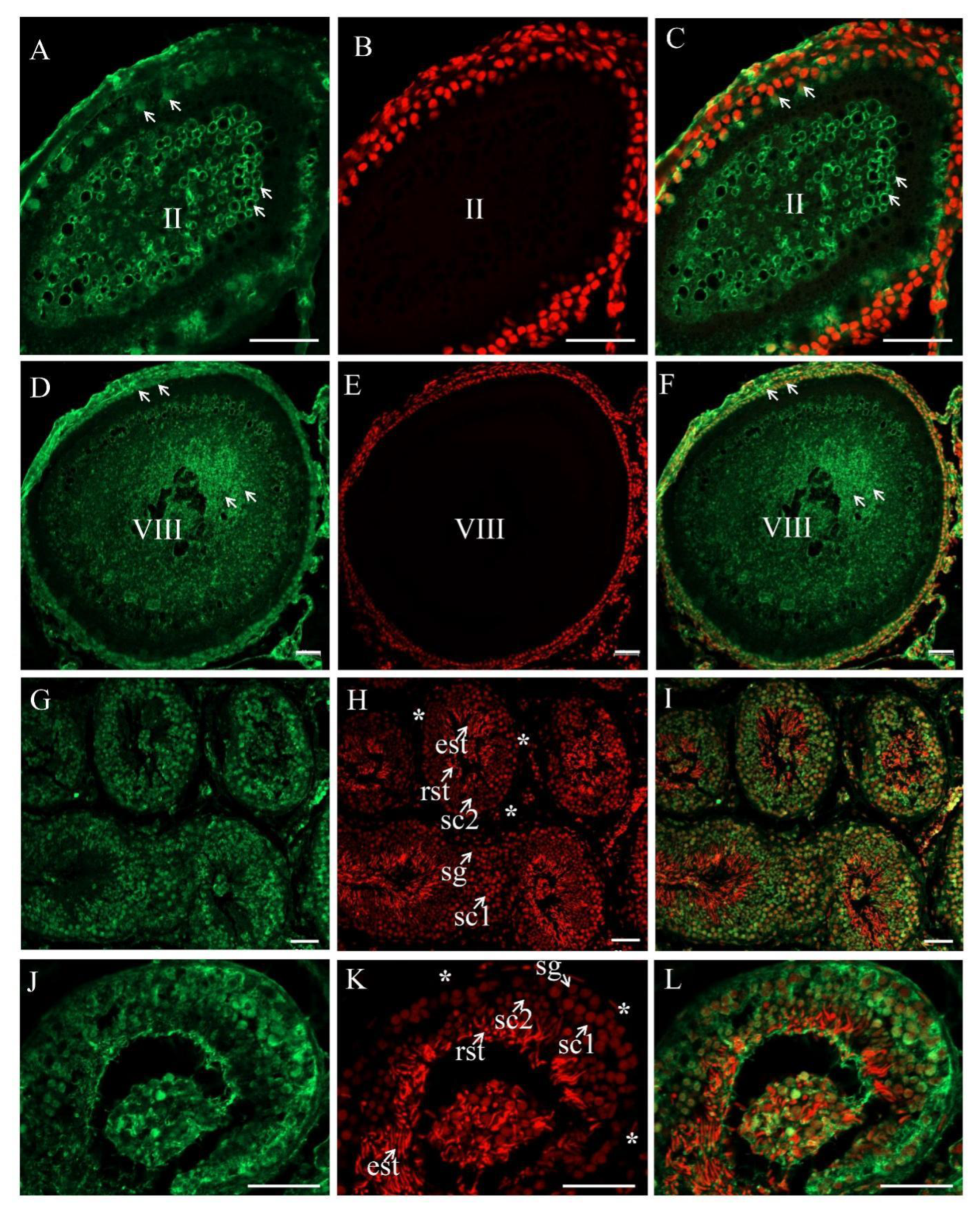
3.3. The Expression and Cellular Localization of PsNanog mRNA in Chinese Soft-Shell Turtle Ovary during Oogenesis and Testis during Spermatogenesis
3.4. The Expression and Cellular Localization of PsNanog Protein in Chinese Soft-Shell Turtle Ovary during Oogenesis and Testis during Spermatogenesis
4. Discussion
4.1. Identification of PsNanog Gene
4.2. PsNanog Expression in Different Tissues
4.3. PsNanog mRNA Expression in Chinese Soft-Shell Turtle Germ Cells
4.4. PsNanog Protein Expression in Chinese Soft-Shell Turtle Germ Cells
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Chambers, I.; Colby, D.; Robertson, M.; Nichols, J.; Lee, S.; Tweedie, S.; Smith, A. Functional expression cloning of Nanog, a pluripotency sustaining factor in embryonic stem cells. Cell 2003, 113, 643–655. [Google Scholar] [CrossRef]
- Mitsui, K.; Tokuzawa, Y.; Itoh, H.; Segawa, K.; Murakami, M.; Takahashi, K.; Maruyama, M.; Maeda, M.; Yamanaka, S. The homeoprotein Nanog is required for maintenance of pluripotency in mouse epiblast and ES cells. Cell 2003, 113, 631–642. [Google Scholar] [CrossRef]
- Wang, J.; Rao, S.; Chu, J.; Shen, X.; Levasseur, D.N.; Theunissen, T.W.; Orkin, S.H. A protein interaction network for pluripotency of embryonic stem cells. Nature 2006, 444, 364–368. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, T.W.; Costa, Y.; Radzisheuskaya, A.; van Oosten, A.L.; Lavial, F.; Pain, B.; Castro, L.F.; Silva, J.C. Reprogramming capacity of Nanog is functionally conserved in vertebrates and resides in a unique homeodomain. Development 2011, 138, 4853–4865. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.-H.; Tsai, M.-S.; Chiang, M.-F.; Li, H. A novel NK-type homeobox gene, ENK (early embryo specific NK), preferentially expressed in embryonic stem cells. Gene Expr. Patterns 2003, 3, 99–103. [Google Scholar] [CrossRef]
- Hart, A.H.; Hartley, L.; Ibrahim, M.; Robb, L. Identification, cloning and expression analysis of the pluripotency promoting Nanog genes in mouse and human. Dev. Dyn. 2004, 230, 187–198. [Google Scholar] [CrossRef]
- Cañón, S.; Herranz, C.; Manzanares, M. Germ cell restricted expression of chick Nanog. Dev. Dyn. 2006, 235, 2889–2894. [Google Scholar] [CrossRef]
- Lavial, F.; Acloque, H.; Bertocchini, F.; Macleod, D.J.; Boast, S.; Bachelard, E.; Montillet, G.; Thenot, S.; Sang, H.M.; Stern, C.D.; et al. The Oct4 homologue PouV and Nanog regulate pluripotency in chicken embryonic stem cells. Development 2007, 134, 3549–3563. [Google Scholar] [CrossRef] [PubMed]
- Dixon, J.E.; Allegrucci, C.; Redwood, C.; Kump, K.; Bian, Y.; Chatfield, J.; Chen, Y.H.; Sottile, V.; Voss, S.R.; Alberio, R.; et al. Axolotl Nanog activity in mouse embryonic stem cells demonstrates that ground state pluripotency is conserved from urodele amphibians to mammals. Development 2010, 137, 2973–2980. [Google Scholar] [CrossRef]
- Camp, E.; Sánchez-Sánchez, A.V.; Garcia-Espana, A.; Desalle, R.; Odqvist, L.; Enrique O’Connor, J.; Mullor, J.L. Nanog regulates proliferation during early fish development. Stem Cells 2009, 27, 2081–2091. [Google Scholar] [CrossRef]
- Wang, D.; Manali, D.; Wang, T.; Bhat, N.; Hong, N.; Li, Z.; Wang, L.; Yan, Y.; Liu, R.; Hong, Y. Identification of pluripotency genes in the fish medaka. Int. J. Biol. Sci. 2011, 7, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Wang, J.; Jiang, J.; Fan, L.; Wang, W.; Liu, J.; Zhang, Q.; Wang, X. Identification and characterization of a nanog homolog in Japanese flounder (Paralichthys olivaceus). Gene 2013, 531, 411–421. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Liu, Y.; Ye, D.; Li, J.; Liu, J.; Deng, F. Knockdown of zebrafish Nanog increases primordial germ cells during early embryonic development. Dev. Growth Differ. 2016, 58, 355–366. [Google Scholar] [CrossRef]
- Patra, S.K.; Vemulawada, C.; Soren, M.M.; Sundaray, J.K.; Panda, M.K.; Barman, H.K. Molecular characterization and expression patterns of Nanog gene validating its involvement in the embryonic development and maintenance of spermatogonial stem cells of farmed carp, Labeo rohita. J. Anim. Sci. Biotechnol. 2018, 9, 45. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Kimura, H.; Tada, M.; Nakatsuji, N.; Tada, T. Nanog expression in mouse germ cell development. Gene Expr. Patterns 2005, 5, 639–646. [Google Scholar] [CrossRef] [PubMed]
- Yamaguchi, S.; Kurimoto, K.; Yabuta, Y.; Sasaki, H.; Nakatsuji, N.; Saitou, M.; Tada, T. Conditional knockdown of Nanog induces apoptotic cell death in mouse migrating primordial germ cells. Development 2009, 136, 4011–4020. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Li, M.; Gui, J.; Hong, Y. Fish germ cells. Sci. China Life Sci. 2010, 53, 435–446. [Google Scholar] [CrossRef]
- Sánchez-Sánchez, A.V.; Camp, E.; Leal-Tassias, A.; Atkinson, S.P.; Armstrong, L.; Diaz-Llopis, M.; Mullor, J.L. Nanog regulates primordial germ cell migration through Cxcr4b. Stem Cells 2010, 28, 1457–1464. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, W.; Zhu, L.; Ye, D.; Zhu, X.; Wang, H.; Sun, Y.; Deng, F. Nanog suppresses the expression of vasa by directly regulating nlk1 in the early zebrafish embryo. Biochimie 2017, 142, 93–101. [Google Scholar] [CrossRef]
- Schuff, M.; Siegel, D.; Philipp, M.; Bundschu, K.; Heymann, N.; Donow, C.; Knochel, W. Characterization of Danio rerio Nanog and functional comparison to Xenopus Vents. Stem Cells Dev. 2012, 21, 1225–1238. [Google Scholar] [CrossRef] [Green Version]
- Han, X.K.; Zhang, L.; Hei, N.N.; Chen, Q.S. Sperm storage in male and female soft-shelled turtles, Trionyx sinensis in hibernation. Chin. J. Fish. Sci. 2007, 14, 706–711. [Google Scholar]
- Tang, Z.; Zhang, P.; Chu, Z.; Zhu, X.; Li, W.; Wu, X.; Xu, H. The cloning and expression analysis of dazl in germ cells in the Chinese soft-shelled turtle (Pelodiscus sinensis). J. Fish. China 2019, 43, 400–409. [Google Scholar]
- Li, W.; Zhang, P.; Wu, X.; Zhu, X.; Xu, H. A novel dynamic expression of vasa in male germ cells during spermatogenesis in the Chinese soft-shell turtle (Pelidiscus sinensis). J. Exp. Zool. Mol. Dev. Evol. 2017, 328, 1–10. [Google Scholar] [CrossRef]
- Liu, X.; Tang, Z.; Zhang, P.; Zhu, X.; Chu, Z.; Li, W.; Xu, H. Identification and characterization of DAZ family genes in Chinese soft-shell turtle (Pelodiscus sinensis). J. Exp. Zool. Mol. Dev. Evol. 2019, 332, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Han, X.K.; Qi, Y.Y.; Liu, Y.; Chen, Q.S. Seasonal effects on apoptosis and proliferation of germ cells in the testes of the Chinese soft-shelled turtle, Pelodiscus sinensis. Theriogenology 2008, 69, 1148–1158. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Gui, J.; Hong, Y. Differential expression of vasa RNA and protein during spermatogenesis and oogenesis in the gibel carp (Carassius auratus gibelio), a bisexually and gynogenetically reproducing vertebrate. Dev. Dyn. 2005, 233, 872–882. [Google Scholar] [CrossRef]
- Xu, H.; Zhu, X.; Li, W.; Tang, Z.; Zhao, Y.; Wu, X. Isolation and in vitro culture of ovarian stem cells in Chinese soft-shell turtle (Pelodiscus sinensis). J. Cell. Biochem. 2018, 119, 7667–7677. [Google Scholar] [CrossRef]
- Hei, N.; Yang, P.; Yang, Y.; Liu, J.; Bao, H.; Liu, H.; Zhang, H.; Chen, Q. Fine structural observation on the oogenesis and vitellogenesis of the Chinese soft-shelled turtle (Pelodiseus sinensis). Zygote 2009, 18, 109–120. [Google Scholar]
- Zhang, L.; Han, X.K.; Li, M.Y.; Bao, H.J.; Chen, Q.S. Spermiogenesis in soft-shelled turtle, Pelodiscus sinensis. Anat. Rec. 2007, 290, 1213–1222. [Google Scholar] [CrossRef] [PubMed]
- Pan, G.; Pei, D. The stem cell pluripotency factor NANOG activates transcription with two unusually potent subdomains at its C terminus. J. Biol. Chem. 2005, 280, 1401–1407. [Google Scholar] [CrossRef]
- Xu, C.; Xu, M.; Tan, L.; Yang, H.; Permuth-Wey, J.; Kruk, P.A.; Wenham, R.M.; Nicosia, S.V.; Lancaster, J.M.; Sellers, T.A.; et al. Microrna mir-214 regulates ovarian cancer cell stemness by targeting p53/nanog. J. Biol. Chem. 2012, 287, 34970–34978. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Ma, T.; Chi, X.; Pei, D. Aromatic residues in the c-terminal domain 2 are required for nanog to mediate lif-independent self-renewal of mouse embryonic stem cells. J. Biol. Chem. 2008, 283, 4480–4489. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.Y.; Yao, Z. Isolation and characterization of the murine Nanog gene promoter. Cell Res. 2005, 15, 317–324. [Google Scholar] [CrossRef] [PubMed]
- Saunders, A.; Faiola, F.; Wang, J. Concise review: Pursuing self-renewal and pluripotency with the stem cell factor Nanog. Stem Cells 2013, 31, 1227–1236. [Google Scholar] [CrossRef]
- Pereira, L.; Yi, F.; Merrill, B.J. Repression of Nanog gene transcription by Tcf3 limits embryonic stem cell self-renewal. Mol. Cell. Biol. 2006, 26, 7479–7491. [Google Scholar] [CrossRef]
- Lin, T.; Chao, C.; Saito, S.; Mazur, S.J.; Murphy, M.E.; Appella, E.; Xu, Y. p53 induces differentiation of mouse embryonic stem cells by suppressing Nanog expression. Nat. Cell Biol. 2005, 7, 165–171. [Google Scholar] [CrossRef]
- Nettersheim, D.; Biermann, K.; Gillis, A.J.; Steger, K.; Looijenga, L.H.; Schorle, H. NANOG promoter methylation and expression correlation during normal and malignant human germ cell development. Epigenetics 2011, 6, 114–122. [Google Scholar] [CrossRef]
- Wu, D.Y.; Yao, Z. Functional analysis of two Sp1/Sp3 binding sites in murine Nanog gene promoter. Cell Res. 2006, 16, 319–322. [Google Scholar] [CrossRef]
- Wang, J.; Levasseur, D.N.; Orkin, S.H. Requirement of Nanog dimerization for stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 2008, 105, 6326–6331. [Google Scholar] [CrossRef]
- Marandel, L.; Labbe, C.; Bobe, J.; Le Bail, P.Y. nanog 5′-upstream sequence, DNA methylation, and expression in gametes and early embryo reveal striking differences between teleosts and mammals. Gene 2012, 492, 130–137. [Google Scholar] [CrossRef]
- Yu, M.; Xue, T.; Chen, T.; Fang, J.; Pan, Q.; Deng, Y.; Li, L.; Chen, K.; Wang, Y. Maternal inheritance of Nanog ortholog in blunt-snout bream. J. Exp. Zool. Mol. Dev. Evol. 2017, 328, 749–759. [Google Scholar] [CrossRef] [PubMed]
- Grubelnik, G.; Bostjancic, E.; Pavlic, A.; Kos, M.; Zidar, N. NANOG expression in human development and cancerogenesis. Exp. Biol. Med. 2020, 245, 456–464. [Google Scholar] [CrossRef] [PubMed]
- Piazzolla, D.; Palla, A.R.; Pantoja, C.; Cañamero, M.; de Castro, I.P.; Ortega, S.; Gómez-López, G.; Dominguez, O.; Megías, D.; Roncador, G.; et al. Lineage-restricted function of the pluripotency factor NANOG in stratified epithelia. Nat. Commun. 2014, 5, 4226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Jahagirdar, B.N.; Reinhardt, R.L.; Schwartz, R.E.; Keene, C.D.; Ortiz-Gonzalez, X.R.; Reyes, M.; Lenvik, T.; Lund, T.; Blackstad, M.; et al. Pluripotency of mesenchymal stem cells derived from adult marrow. Nature 2002, 418, 41–49. [Google Scholar] [CrossRef] [PubMed]
- Nakanoh, S.; Fuse, N.; Takahashi, Y.; Agata, K. Verification of chicken Nanog as an epiblast marker and identification of chicken PouV as Pou5f3 by newly raised antibodies. Dev. Growth Differ. 2015, 57, 251–263. [Google Scholar] [CrossRef]
- Shinomiya, A.; Tanaka, M.; Kobayashi, T.; Nagahama, Y.; Hamaguchi, S. The vasa-like gene, olvas, identifies the migration path of primordial germ cells during embryonic body formation stage in the medaka, Oryzias latipes. Dev. Growth Differ. 2000, 42, 317–326. [Google Scholar] [CrossRef]
- Brons, I.G.; Smithers, L.E.; Trotter, M.W.; Rugg-Gunn, P.; Sun, B.; Chuva de Sousa Lopes, S.M.; Howlett, S.K.; Clarkson, A.; Ahrlund-Richter, L.; Pedersen, R.A.; et al. Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 2007, 448, 191–195. [Google Scholar] [CrossRef]
- Ortega, M.S.; Kelleher, A.M.; O’Neil, E.; Benne, J.; Cecil, R.; Spencer, T.E. NANOG is required to form the epiblast and maintain pluripotency in the bovine embryo. Mol. Reprod. Dev. 2020, 87, 152–160. [Google Scholar] [CrossRef]
- Verma, R.; Liu, J.; Holland, M.K.; Temple-Smith, P.; Williamson, M.; Verma, P.J. Nanog is an essential factor for induction of pluripotency in somatic cells from endangered felids. BioResearch Open Access 2013, 2, 72–76. [Google Scholar] [CrossRef] [Green Version]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, K.; Xu, J.; Ban, W.; Tian, J.; Tan, Z.; Tang, Z.; Lei, L.; Li, W.; Zhu, X.; Xu, H. The Divergent and Conserved Expression Profile of Turtle Nanog Gene Comparing with Fish and Mammals. Biology 2022, 11, 1342. https://doi.org/10.3390/biology11091342
Chen K, Xu J, Ban W, Tian J, Tan Z, Tang Z, Lei L, Li W, Zhu X, Xu H. The Divergent and Conserved Expression Profile of Turtle Nanog Gene Comparing with Fish and Mammals. Biology. 2022; 11(9):1342. https://doi.org/10.3390/biology11091342
Chicago/Turabian StyleChen, Kaili, Jianfei Xu, Wenzhuo Ban, Jiaming Tian, Zhiming Tan, Zhoukai Tang, Luo Lei, Wei Li, Xinping Zhu, and Hongyan Xu. 2022. "The Divergent and Conserved Expression Profile of Turtle Nanog Gene Comparing with Fish and Mammals" Biology 11, no. 9: 1342. https://doi.org/10.3390/biology11091342



