Comprehensive Transcriptome Analysis of Different Skin Colors to Evaluate Genes Related to the Production of Pigment in Celestial Goldfish
Abstract
:Simple Summary
Abstract
1. Introduction
2. Material and Method
2.1. Animal Sample Collection
2.2. RNA Extraction and Sequencing
2.3. Bioinformatic Analysis
2.4. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Enrichment Analysis
2.5. Validation of Differentially Expressed Transcripts
2.6. Analytical Statistics
3. Results
3.1. Blast Analysis of Transcriptome Sequencing
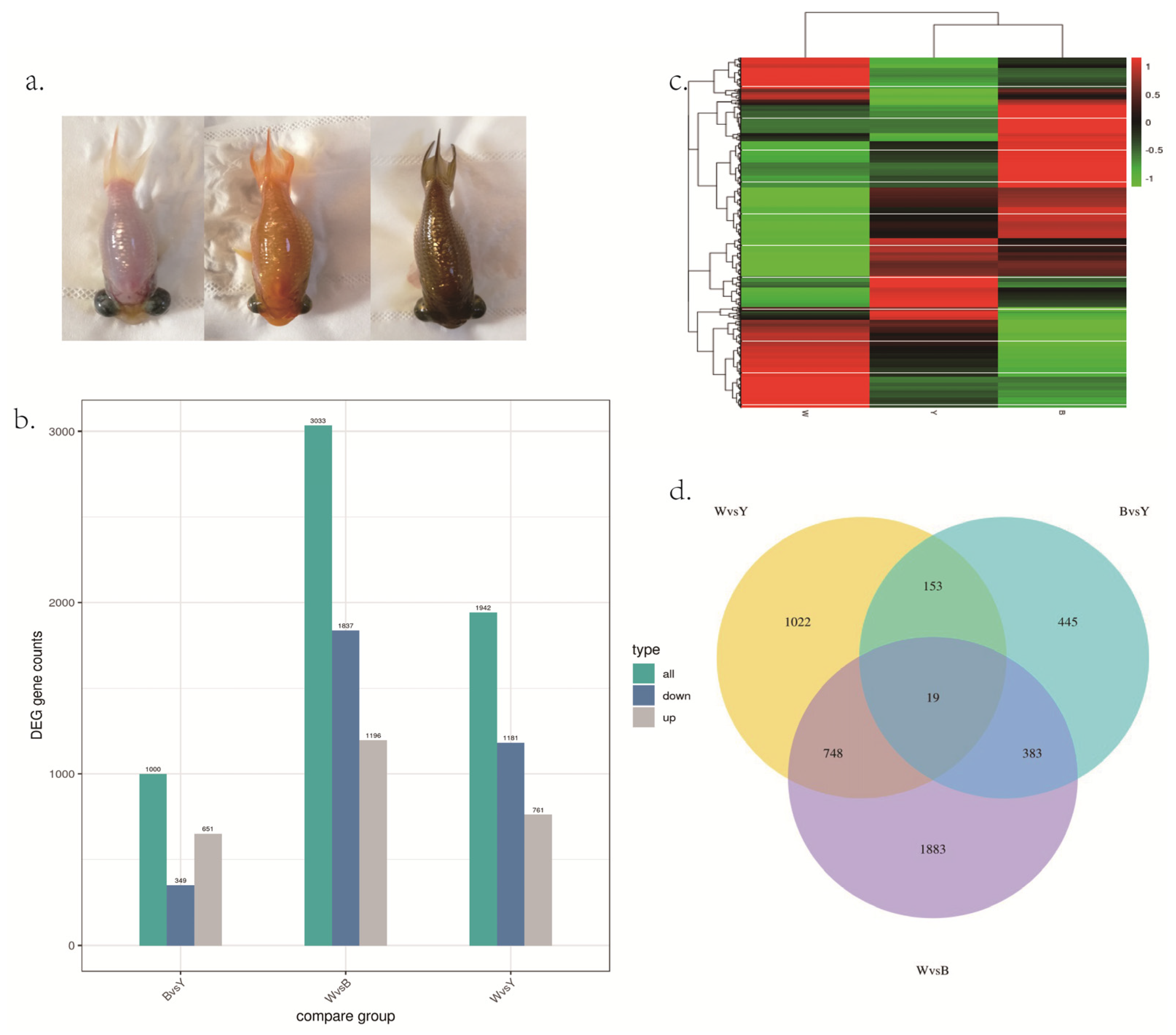
3.2. Differentially Expressed Genes in Goldfish with Different Skin Colors
3.3. Functional Analysis of Differentially Expressed Genes
3.4. Common Expression Gene Analysis
3.5. Validation of Differentially Expressed Genes Expression
4. Discussion
4.1. Composition Analysis of Pigment Cells in Different Skin Colors
4.2. Regulated Genes in Brown Skin Indicated the Molecular Mechanism of the Melanogenesis Pathway
4.3. Identification of Candidate Genes Related to the Production of Pigment
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Conflicts of Interest
References
- Kelsh, R.N. Genetics and Evolution of Pigment Patterns in Fish. Pigment Cell Res. 2010, 17, 326–336. [Google Scholar] [CrossRef] [PubMed]
- Barsh, G.S. The genetics of pigmentation: From fancy genes to complex traits. Trends Genet. 1996, 12, 299–305. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.A.; Mccue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Jiang, B.; Wang, L.; Luo, M.; Fu, J.; Dong, Z. Transcriptome Analysis of Skin Color Variation During and after Overwintering of Malaysian Red Tilapia. Fish Physiol. Biochem. 2022, 48, 669–682. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Chung, D.; Yu, W.; Chen, Z.; Song, S.; Ren, J. Global tissue transcriptomic analysis to improve genome annotation and unravel skin pigmentation in goldfish. Sci. Rep. 2021, 11, 1815. [Google Scholar] [CrossRef]
- Luo, M.; Wang, L.; Yin, H.; Zhu, W.; Dong, Z. Integrated analysis of long non-coding RNA and mRNA expression in different colored skin of koi carp. BMC Genom. 2019, 20, 515. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Gong, Q.; Lai, J.; Song, M.; Long, Z. Transcriptome analysis identifies candidate genes associated with skin color variation in Triplophysa siluroides. Comp. Biochem. Physiol. Part D Genom. Proteom. 2020, 35, 100682. [Google Scholar] [CrossRef]
- Hearing, V.J.; Jiménez, M. Mammalian tyrosinase—The critical regulatory control point in melanocyte pigmentation. Int. J. Biochem. 1987, 19, 1141–1147. [Google Scholar] [CrossRef]
- Ferrini, U.; Mileo, A.M.; Hearing, V.J. Microheterogeneity of melanosome-bound tyrosinase from the harding-passey murine melanoma. Int. J. Biochem. 1987, 19, 227–234. [Google Scholar] [CrossRef]
- Kowichi, J.; Hiroyuki, H.; Thuraiayah, V.; Dong, L.; Jamal, D. Molecular Control of Melanogenesis in Malignant Melanoma: Functional Assessment of Tyrosinase and Lamp Gene Families by UV Exposure and Gene Co-Transfection, and Cloning of a cDNA Encoding Calnexin, A Possible Melanogenesis “Chaperone”. J. Dermatol. 1994, 21, 894–906. [Google Scholar]
- Ginger, R.S.; Askew, S.E.; Ogborne, R.M.; Wilson, S.; Green, M.R. SLC24A5 Encodes a trans-Golgi Network Protein with Potassium-dependent Sodium-Calcium Exchange Activity That Regulates Human Epidermal Melanogenesis. J. Biol. Chem. 2008, 283, 5486. [Google Scholar] [CrossRef] [PubMed]
- Lamason, R.L.; Mohideen, M.A.P.; Mest, J.R.; Wong, A.C.; Norton, H.L.; Aros, M.C.; Jurynec, M.J.; Mao, X.; Humphreville, V.R.; Humbert, J.E.; et al. SLC24A5, a Putative Cation Exchanger, Affects Pigmentation in Zebrafish and Humans. Science 2005, 16, 5755. [Google Scholar]
- Braasch, I.; Schartl, M.; Volff, J. Evolution of pigment synthesis pathways by gene and genome duplication in fish. BMC Evol. Biol. 2007, 7, 74. [Google Scholar]
- Pelletier, I.; Boyle, P.; Mcdonald, T.; Hesslinger, C.; Ziegler, I. The Pteridine Pathway in the Zebrafish, Danio Rerio: Development in Neural Crest-Derived Cells and Its Control by GTP Cyclohydrolase I. In Chemistry and Biology of Pteridines and Folates; Springer: Boston, MA, USA, 2002. [Google Scholar]
- Wu, L.C.; Lin, Y.Y.; Yang, S.Y.; Weng, Y.T.; Tsai, Y.T. Antimelanogenic effect of c-phycocyanin through modulation of tyrosinase expression by upregulation of ERK and downregulation of p38 MAPK signaling pathways. J. Biomed. Sci. 2011, 18, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Liang, S.; Haitao, L.; Dechao, B.; Guoguang, Z.; Kuntao, Y.; Changhai, Z.; Yuanning, L.; Runsheng, C.; Yi, Z. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Res. 2013, 41, e166. [Google Scholar]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [Green Version]
- Kanehisa, M.; Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000, 28, 27–30. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. featureCounts: An efficient general-purpose read summarization program. Bioinformatics 2013, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Likun, W.; Zhixing, F.; Xi, W.; Xiaowo, W.; Xuegong, Z. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics (Oxf. Engl.) 2010, 26, 136–138. [Google Scholar]
- Granneman, J.G.; Kimler, V.A.; Zhang, H.; Ye, X.; Luo, X.; Postlethwait, J.H.; Thummel, R. Lipid droplet biology and evolution illuminated by the characterization of a novel perilipin in teleost fish. eLife 2017, 6, e21771. [Google Scholar] [CrossRef] [PubMed]
- Kimura, T.; Nagao, Y.; Hashimoto, H.; Yamamoto-Shiraishi, Y.I.; Yamamoto, S.; Yabe, T.; Takada, S.; Kinoshita, M.; Kuroiwa, A.; Naruse, K. Leucophores are similar to xanthophores in their specification and differentiation processes in medaka. Proc. Natl. Acad. Sci. USA 2014, 111, 7343–7348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teulings, H.E.; Willemsen, K.J.; Glykofridis, I.; Krebbers, G.; Komen, L.; Kemp, E.H.; Wolkerstorfer, A.; van der Veen, J.P.W.; Luiten, R.M.; Tjin, E.P. The antibody response against MART-1 differs in patients with melanoma-associated leucoderma and vitiligo. Pigment Cell Melanoma Res. 2015, 27, 1086–1096. [Google Scholar] [CrossRef] [PubMed]
- Heimsath, E.G.; Yim, Y.I.; Mustapha, M.; Hammer, J.A.; Cheney, R.E. Myosin-X knockout is semi-lethal and demonstrates that myosin-X functions in neural tube closure, pigmentation, hyaloid vasculature regression, and filopodia formation. Sci. Rep. 2017, 7, 17354. [Google Scholar] [CrossRef]
- Funkenstein, B.; Rebhan, Y.; Balas, V.; Pliatner, A.; Skopal, T.; Nadjar-Boger, E.; Du, S.J. New insights into the regulation of expression and biological activity of myostatin in fish: Lessons from sea bream. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2008, 151, S11–S12. [Google Scholar] [CrossRef]
- Wallat, G.K.; Lazur, A.M.; Chapman, F.A. Carotenoids of Different Types and Concentrations in Commercial Formulated Fish Diets Affect Color and Its Development in the Skin of the Red Oranda Variety of Goldfish. N. Am. J. Aquac. 2005, 67, 42–51. [Google Scholar] [CrossRef]
- Cunha, L.D.; Besen, K.P.; Ha, N.; Uczay, J.; Fabregat, T. Biofloc technology (BFT) improves skin pigmentation of goldfish (Carassius auratus). Aquaculture 2020, 522, 735132. [Google Scholar] [CrossRef]
- Kwon, B.S.; Haq, A.K.; Pomerantz, S.H.; Halaban, R. Isolation and sequence of a cDNA clone for human tyrosinase that maps at the mouse c-albino locus. Proc. Natl. Acad. Sci. USA 1987, 84, 7473–7477. [Google Scholar] [CrossRef] [Green Version]
- Giebel, L.B.; Strunk, K.M.; King, R.A.; Hanifin, J.M.; Spritz, R.A. A frequent tyrosinase gene mutation in classic, tyrosinase-negative (type IA) oculocutaneous albinism. Proc. Natl. Acad. Sci. USA 1990, 87, 3255–3258. [Google Scholar] [CrossRef] [Green Version]
- Goto, M.; Sato-Matsumura, K.C.; Sawamura, D.; Yokota, K.; Nakamura, H.; Shimizu, H. Tyrosinase gene analysis in Japanese patients with oculocutaneous albinism. J. Dermatol. Sci. 2004, 35, 215–220. [Google Scholar] [CrossRef]
- Jiang, Y.; Zhang, S.; Xu, J.; Feng, J.; Mahboob, S.; Al-Ghanim, K.A.; Sun, X.; Xu, P. Comparative Transcriptome Analysis Reveals the Genetic Basis of Skin Color Variation in Common Carp. PLoS ONE 2014, 9, e108200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hari, L.; Brault, V.; Kleber, M.; Lee, H.Y.; Ille, F.; Leimeroth, R.; Paratore, C.; Suter, U.; Kemler, R.; Sommer, L. Lineage-specific requirements of beta-catenin in neural crest development. J. Cell Biol. 2002, 159, 867–880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, E.J.; Erickson, C.A.; Takada, S.; Burrus, L.W. Wnt and BMP Signaling Govern Lineage Segregation of Melanocytes in the Avian Embryo. Dev. Biol. 2001, 233, 22–37. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dorsky, R.I.; Randall, T.; Raible, D. Control of neural crest cell fate by the Wnt signalling pathway. Nature 1998, 396, 370–373. [Google Scholar] [CrossRef] [PubMed]
- Del Bino, S.; Duval, C.; Bernerd, F. Clinical and Biological Characterization of Skin Pigmentation Diversity and Its Consequences on UV Impact. Int. J. Mol. Sci. 2018, 19, 2668. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.M.; Lee, E.C.; Lim, H.M.; Seo, Y.K. Rice Bran Ash Mineral Extract Increases Pigmentation through the p-ERK Pathway in Zebrafish (Danio rerio). Int. J. Mol. Sci. 2019, 20, 2172. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Béjar, J.; Hong, Y.; Schartl, M. Mitf expression is sufficient to direct differentiation of medaka blastula derived stem cells to melanocytes. Development 2003, 130, 6545–6553. [Google Scholar] [CrossRef] [Green Version]
- Chae, K.J.; Subedi, L.; Jeong, M.; Park, Y.U.; Kim, Y.C. Gomisin N Inhibits Melanogenesis through Regulating the PI3K/Akt and MAPK/ERK Signaling Pathways in Melanocytes. Int. J. Mol. Sci. 2017, 18, 471. [Google Scholar] [CrossRef]



| Gene ID | FPKM | Description | ||
|---|---|---|---|---|
| White | Yellow | Brown | ||
| 113116617 | 0.053316 | 14.85341 | 4.939858 | uncharacterized |
| 113059134 | 0.101642 | 11.21777 | 3.472121 | perilipin-3-like |
| novel.91 | 0.024732 | 41.3923 | 11.47289 | -- |
| 113094292 | 0 | 22.96619 | 9.53373 | putative defense protein 3 |
| 113054403 | 0 | 7.702924 | 1.333329 | diacylglycerol O-acyltransferase 2 |
| 113060382 | 0.021323 | 4.63168 | 1.063391 | retinol dehydrogenase 7-like |
| 113072589 | 0 | 3.77947 | 0.555086 | solute carrier family facilitated glucose transporter member 11-like |
| 113081155 | 0.065602 | 12.08961 | 3.294872 | melanoma antigen recognized by T-cells 1-like |
| novel.919 | 0.052276 | 2.696407 | 9.566884 | -- |
| 113056131 | 0.017281 | 4.126237 | 0.910635 | GTP cyclohydrolase 1-like |
| 113079820 | 0.023412 | 8.795228 | 2.98262 | melanoma antigen recognized by T-cells 1-like |
| 113116699 | 18.29243 | 526.6035 | 80.16275 | translation initiation factor IF-2-like |
| 113044679 | 0.872271 | 0.010594 | 0.200235 | uncharacterized |
| 113107421 | 0.154702 | 2.39033 | 0.707736 | RAS and EF-hand domain containing |
| 113059190 | 1.056176 | 0.085419 | 5.333497 | transcription elongation factor A protein 3-like |
| 113068772 | 0.148587 | 0.020619 | 1.319104 | myosin-6-like |
| 113119606 | 3.652859 | 1.230851 | 0.193719 | translocation protein SEC62-like |
| 113106535 | 0.017673 | 0.195103 | 1.180688 | C6-dihydroxyindole-2-carboxylic acid oxidase-like |
| novel.58 | 0.154461 | 0.976559 | 4.569909 | -- |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Sun, Y.; Cui, R.; Zhang, X. Comprehensive Transcriptome Analysis of Different Skin Colors to Evaluate Genes Related to the Production of Pigment in Celestial Goldfish. Biology 2023, 12, 7. https://doi.org/10.3390/biology12010007
Li R, Sun Y, Cui R, Zhang X. Comprehensive Transcriptome Analysis of Different Skin Colors to Evaluate Genes Related to the Production of Pigment in Celestial Goldfish. Biology. 2023; 12(1):7. https://doi.org/10.3390/biology12010007
Chicago/Turabian StyleLi, Rongni, Yansheng Sun, Ran Cui, and Xin Zhang. 2023. "Comprehensive Transcriptome Analysis of Different Skin Colors to Evaluate Genes Related to the Production of Pigment in Celestial Goldfish" Biology 12, no. 1: 7. https://doi.org/10.3390/biology12010007




