Shifts in the Microbial Populations of Bioleach Reactors Are Determined by Carbon Sources and Temperature
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Concentrate
2.2. Experimental Setup and Bio-Oxidaton
2.3. Cyanidation
2.4. Biomass Sampling and DNA Extraction
2.5. 16S rRNA Gene Amplification, Sequencing, and Analysis
2.6. Metagenomic Analysis
2.7. Deposition of Nucleotide Sequences
3. Results
3.1. Concentrate Bio-Oxidaton and Gold Recovery
3.2. A Taxonomic Analysis of the Composition of the Microbial Community
3.3. Metagenomic Analysis
3.3.1. Metagenome Sequencing and MAG Assembly
3.3.2. The Genome of Archaea of the A-Plasma Group of the Order Thermoplasmatales
4. Discussion
- –
- The experiments performed in continuous mode under conditions similar to those used at an industrial scale demonstrated that the proposed approach (the application of carbon sources) is promising from a practical point of view for increasing bio-oxidation performance;
- –
- A metagenomic analysis made it possible to characterize a novel group of uncultivated archaea which were detected in different habitats (including bioleach reactors) but not studied.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Johnson, D.B. Biomining—biotechnologies for extracting and recovering metals from ores and waste materials. Curr. Opin. Biotechnol. 2014, 30, 24–31. [Google Scholar] [CrossRef]
- Johnson, D.B. The Evolution, Current Status, and Future Prospects of Using Biotechnologies in the Mineral Extraction and Metal Recovery Sectors. Minerals 2018, 8, 343. [Google Scholar] [CrossRef]
- Mahmoud, A.; Cezac, P.; Hoadley, A.F.A.; Contaminea, F.; d’Hugues, P. A review of sulfide minerals microbially assisted leaching in stirred tank reactors. Int. Biodeterior. Biodegrad. 2017, 119, 118–146. [Google Scholar] [CrossRef]
- van Niekerk, J.A.; van Buuren, C.B.; Olivier, J.W. Bioprocessing of Refractory Gold Ores: The BIOX, MesoTHERM, and ASTER Processes. In Biomining Technologies: Extracting and Recovering Metals from Ores and Wastes; Johnson, D.B., Bryan, C.G., Schlömann, M., Roberto, F.F., Eds.; Springer: Cham, Switzerland, 2023; pp. 67–88. [Google Scholar] [CrossRef]
- Marsden, J.O.; House, C.I. The Chemistry of Gold Extraction, 2nd ed.; Society for Mining, Metallurgy, and Exploration, Inc.: Littleton, CO, USA, 2006; 625p. [Google Scholar]
- van Aswegen, P.C.; van Niekerk, J.; Olivier, W. The BIOX process for the treatment of refractory gold concentrate. In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 1–35. [Google Scholar] [CrossRef]
- Gericke, M.; Neale, J.W.; van Staden, P.J. A Mintek perspective of the past 25 years in minerals bioleaching. J. S. Afr. Inst. Min. Metall 2009, 109, 567–585. [Google Scholar]
- Belyi, A.V.; Chernov, D.V.; Solopova, N.V. Development of BIONORD® technology on Olimpiada deposit refractory arsenic–gold ores treatment in conditions of Extreme North. Hydrometallurgy 2018, 179, 188–191. [Google Scholar] [CrossRef]
- Karavaiko, G.I.; Dubinina, G.A.; Kondrat’eva, T.F. Lithotrophic microorganisms of the oxidative cycles of sulfur and iron. Microbiology 2006, 75, 512–545. [Google Scholar] [CrossRef]
- Rawlings, D.E.; Coram, N.J.; Gardner, M.N.; Deane, S.M. Thiobacillus caldus and Leptospirillum ferrooxidans are widely distributed in continuous flow biooxidation tanks used to treat a variety of metal containing ores and concentrates. In Biohydrometallurgy and the Environment: Toward the Mining of the 21st Century. Part A.; Amils, R., Ballester, A., Eds.; Elsevier: Amsterdam, The Netherlands, 1999; pp. 777–786. [Google Scholar] [CrossRef]
- Coram, N.J.; Rawlings, D.E. Molecular relationship between two groups of the genus Leptospirillum and the finding that Leptospirillum ferriphilum sp. nov. dominates South African commercial biooxidation tanks that operate at 40 °C. Appl. Environ. Microbiol. 2002, 68, 838–845. [Google Scholar] [CrossRef]
- Okibe, N.; Gericke, M.; Hallberg, K.B.; Johnson, D.B. Enumeration and characterization of acidophilic microorganisms isolated from a pilot plant stirred-tank bioleaching operation. Appl. Environ. Microbiol. 2003, 69, 1936–1943. [Google Scholar] [CrossRef]
- Dopson, M.; Lindstrom, E.B. Analysis of community composition during moderately thermophilic bioleaching of pyrite, arsenical pyrite, and chalcopyrite. Micr. Ecol. 2004, 48, 19–28. [Google Scholar] [CrossRef]
- Morin, D.H.R.; d’Hugues, P. Bioleaching of a Cobalt-Containing Pyrite in Stirred Reactors: A Case Study from Laboratory Scale to Industrial Application In Biomining; Rawlings, D.E., Johnson, D.B., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 35–55. [Google Scholar] [CrossRef]
- Spolaore, P.; Joulian, C.; Gouin, J.; Ibáñez, A.; Auge, T.; Morin, D.; d’Hugues, P. Bioleaching of an organic–rich polymetallic concentrate using stirred–tank technology. Hydrometallurgy 2009, 99, 137–143. [Google Scholar] [CrossRef]
- Spolaore, P.; Joulian, C.; Gouin, J.; Morin, D.; d’Hugues, P. Relationship between bioleaching performance, bacterial community structure and mineralogy in the bioleaching of a copper concentrate in stirred–tank reactors. Appl. Microbiol. Biotechnol. 2011, 89, 441–448. [Google Scholar] [CrossRef]
- Zeng, W.; Qiu, G.; Zhou, H.; Peng, J.; Chen, M.; Tan, S.N.; Chao, W.; Liu, X.; Zhang, Y. Community structure and dynamics of the free and attached microorganisms during moderately thermophilic bioleaching of chalcopyrite concentrate. Bioresour. Technol. 2010, 101, 7068–7075. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Su, L.; Zhang, L.; Zeng, W.; Wu, J.; Wan, L.; Qiu, G.; Chen, X.; Zhou, H. Bioleaching of chalcopyrite by defined mixed moderately thermophilic consortium including a marine acidophilic halotolerant bacterium. Bioresour. Technol. 2012, 121, 348–354. [Google Scholar] [CrossRef] [PubMed]
- van Hille, R.P.; van Wyk, N.; Froneman, T.; Harrison, S.T.L. Dynamic evolution of the microbial community in BIOX leaching tanks. Adv. Mater. Res. 2013, 825, 331–334. [Google Scholar] [CrossRef]
- Kondrat’eva, T.F.; Pivovarova, T.A.; Bulaev, A.G.; Moshchanetskii, P.V.; Tsaplina, I.A.; Grigor’eva, N.V.; Zhuravleva, A.E.; Melamud, V.S.; Belyi, A.V. Selection of a community of acidochemolithotrophic microorganisms with a high oxidation rate of pyrrhotite–containing sulphide ore flotation concentrate. Appl. Biochem. Microbiol. 2013, 49, 495–501. [Google Scholar] [CrossRef]
- Muravyov, M.I.; Bulaev, A.G. Two–step oxidation of a refractory gold–bearing sulfidic concentrate and the effect of organic nutrients on its biooxidation. Miner. Eng. 2013, 45, 108–114. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, W.; Qiu, G.; Chen, X.; Zhou, H. A moderately thermophilic mixed microbial culture for bioleaching of chalcopyrite concentrate at high pulp density. Appl. Environ. Microbiol. 2014, 80, 741–750. [Google Scholar] [CrossRef]
- Hedrich, S.; Guézennec, A.-G.; Charron, M.; Schippers, A.; Joulian, C. Quantitative monitoring of microbial species during bioleaching of a copper concentrate. Front. Microbiol. 2016, 7, 20441. [Google Scholar] [CrossRef] [PubMed]
- Smart, M.; Huddy, R.J.; Edward, C.J.; Fourie, C.; Shumba, T.; Iron, J.; Harrison, S.T.L. Linking microbial community dynamics in biox® leaching tanks to process conditions: Integrating lab and commercial experience. Solid State Phenom. 2017, 262, 38–42. [Google Scholar] [CrossRef]
- Bulaev, A.; Belyi, A.; Panyushkina, A.; Solopova, N.; Pivovarova, T. Microbial population of industrial biooxidation reactors. Solid State Phenom. 2017, 262, 48–52. [Google Scholar] [CrossRef]
- Bulaev, A.; Melamud, V.; Boduen, A. Bioleaching of non-ferrous metals from arsenic-bearing sulfide concentrate. Solid State Phenom. 2020, 299, 1064–1068. [Google Scholar] [CrossRef]
- Edward, C.J.; Kotsiopoulos, A.; Harrison, S.T.L. Ferrous iron oxidation kinetics of Acidiplasma cupricumulans, a key archaeon in the mineral biooxidation consortium: Impact of nutrient availability, ferric iron and thiocyanate. Hydrometallurgy 2022, 211, 105890. [Google Scholar] [CrossRef]
- Gonzalez-Tori, E.; Llobet-Brossa, E.; Casamayor, E.O.; Amann, R.; Amils, R. Microbial ecology of an extreme acidic environment, the Tinto River. Appl. Environ. Microbiol. 2003, 69, 4853–4865. [Google Scholar] [CrossRef] [PubMed]
- Druschel, G.K.; Baker, B.J.; Gihring, T.H.; Banfield, J.F. Acid mine drainage biogeochemistry at Iron Mountain, California. Geochem. Trans. 2004, 5, 13–32. [Google Scholar] [CrossRef]
- Schnaitman, C.; Lundgren, D.G. Organic compounds in the spent medium of Ferrobacillus ferrooxidans. Can. J. Microbiol. 1965, 11, 23–27. [Google Scholar] [CrossRef]
- Borichewski, R.M. Keto acids as growth-limiting factors in autotrophic growth of Thiobacillus thiooxidans. J. Bacteriol. 1967, 93, 597–599. [Google Scholar] [CrossRef]
- Okibe, N.; Johnson, D.B. Biooxidation of pyrite by defined mixed cultures of moderately thermophilic acidophiles in pH-controlled bioreactors: Significance of microbial interactions. Biotechnol. Bioeng. 2004, 87, 574–583. [Google Scholar] [CrossRef] [PubMed]
- Nancucheo, I.; Johnson, D.B. Production of glycolic acid by chemolithotrophic iron- and sulfur-oxidizing bacteria and its role in delineating and sustaining acidophilic sulfide mineral-oxidizing consortia. Appl. Environ. Microbiol. 2010, 76, 461–467. [Google Scholar] [CrossRef]
- Bulaev, A.; Nechaeva, A.; Elkina, Y.; Melamud, V. Effect of Carbon Sources on Pyrite-Arsenopyrite Concentrate Bio-Oxidation and Growth of Microbial Population in Stirred Tank Reactors. Microorganisms 2021, 9, 2350. [Google Scholar] [CrossRef]
- Bulaev, A.; Boduen, A. Carbon Sources as a Factor Determining the Activity of Microbial Oxidation of Sulfide Concentrate at Elevated Temperature. Minerals 2022, 12, 110. [Google Scholar] [CrossRef]
- Elkina, Y.; Nechaeva, A.; Artykova, A.; Kolosoff, A.; Bugubaeva, A.; Melamud, V.; Mardanov, A.; Bulaev, A. Continuous Bioleaching of Arsenic-Containing Copper-Zinc Concentrate and Shift of Microbial Population under Various Conditions. Minerals 2022, 12, 592. [Google Scholar] [CrossRef]
- Bulaev, A.G. Biooxidation of refractory pyrite-arsenopyrite gold bearing sulfide concentrate. Int. Multidiscip. Sci. GeoConference SGEM 2019, 19, 67–74. [Google Scholar] [CrossRef]
- Reznikov, A.A.; Mulikovskaya, E.P.; Sokolov, I.Y. Metody Analiza Prirodnykh vod (Methods for Analysis of Natural Waters); Nedra: Moscow, Russia, 1970; 140p. (In Russian) [Google Scholar]
- Surovskaya, I.A.; Titov, V.I.; Brodskaya, V.M.; Vasil’ev, P.I.; Lipshits, B.M.; Elentukh, B.M. Tekhnicheskii Analiz Tsvetnoi Metallurgii (Technical Analysis in Nonferrous Metallurgy); Metallurgizdat: Moscow, Russia, 1957; p. 567. (In Russian) [Google Scholar]
- Filippova, N.A. Fazovyi Analiz Rud i Produktov ikh Pererabotki (Phase Analysis of Ores and Products of Their Processing); Khimiya: Moscow, Russia, 1975; 280p. (In Russian) [Google Scholar]
- Zelenov, V.I. Metodika Issledovania of Zoloto- and Serebrosodegaschich Rud (Methods for the Analysis of Gold- and Silver-Bearing Ores); Nedra: Moscow, Russia, 1989; 302p. (In Russian) [Google Scholar]
- Frey, B.; Rime, T.; Phillips, M.; Stierli, B.; Hajdas, I.; Widmer, F.; Hartmann, M. Microbial diversity in European alpine permafrost and active layers. FEMS Microbiol. Ecol. 2016, 92, fiw018. [Google Scholar] [CrossRef] [PubMed]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ. 2016, 4, e2584. [Google Scholar] [CrossRef]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10–12. [Google Scholar] [CrossRef]
- Nurk, S.; Meleshko, D.; Korobeynikov, A.; Pevzner, P.A. MetaSPAdes: A new versatile metagenomic assembler. Genome Res. 2017, 27, 824–834. [Google Scholar] [CrossRef]
- Kang, D.D.; Froula, J.; Egan, R.; Wang, Z. MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ. 2015, 3, e1165. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- Cao, M.D.; Nguyen, S.H.; Ganesamoorthy, D.; Elliott, A.G.; Cooper, M.A.; Coin, L.J. Scaffolding and completing genome assemblies in real-time with nanopore sequencing. Nat. Commun. 2017, 8, 14515. [Google Scholar] [CrossRef]
- Kolmogorov, M.; Yuan, J.; Lin, Y.; Pevzner, P.A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 2019, 37, 540–546. [Google Scholar] [CrossRef] [PubMed]
- Walker, B.J.; Abeel, T.; Shea, T.; Priest, M.; Abouelliel, A.; Sakthikumar, S.; Cuomo, C.A.; Zeng, Q.; Wortman, J.; Young, S.K.; et al. Pilon: An integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 2014, 9, e112963. [Google Scholar] [CrossRef] [PubMed]
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Imelfort, M.; Skennerton, C.T.; Hugenholtz, P.; Tyson, G.W. CheckM: Assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015, 25, 1043–1055. [Google Scholar] [CrossRef]
- Chaumeil, P.A.; Mussig, A.J.; Hugenholtz, P.; Parks, D.H. GTDB-Tk: A toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 2020, 36, 1925–1927. [Google Scholar] [CrossRef] [PubMed]
- Tatusova, T.; DiCuccio, M.; Badretdin, A.; Chetvernin, V.; Nawrocki, E.P.; Zaslavsky, L.; Lomsadze, A.; Pruitt, K.D.; Borodovsky, M.; Ostell, J. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016, 44, 6614–6624. [Google Scholar] [CrossRef]
- Aziz, R.K.; Bartels, D.; Best, A.A.; DeJongh, M.; Disz, T.; Edwards, R.A.; Formsma, K.; Gerdes, S.; Glass, E.M.; Kubal, M.; et al. The RAST Server: Rapid annotations using subsystems technology. BMC Genom. 2008, 9, 1–75. [Google Scholar] [CrossRef]
- Zhou, Z.; Tran, P.Q.; Breister, A.M.; Liu, Y.; Kieft, K.; Cowley, E.S.; Karaoz, U.; Anantharaman, K. METABOLIC: High-throughput profiling of microbial genomes for functional traits, metabolism, biogeochemistry, and community-scale functional networks. Microbiome 2022, 10, 33. [Google Scholar] [CrossRef]
- Rodriguez, R.L.M.; Konstantinidis, K.T. The enveomics collection: A toolbox for specialized analyses of microbial genomes and metagenomes. PeerJ Prepr. 2016, 4, e1900v1. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef]
- Deng, S.; Gu, G.; He, G.; Li, L. Catalytic effect of pyrite on the leaching of arsenopyrite in sulfuric acid and acid culture medium. Electrochim. Acta 2018, 263, 8–16. [Google Scholar] [CrossRef]
- Xu, J.-N.; Shi, W.-G.; Ma, P.-C.; Lu, L.-S.; Chen, G.-M.; Yang, H.-Y. Corrosion behavior of a pyrite and arsenopyrite galvanic pair in the presence of sulfuric acid, ferric ions and HQ0211 bacterial strain. Minerals 2019, 9, 169. [Google Scholar] [CrossRef]
- Hallberg, K.B.; Lindström, E.B. Characterization of Thiobacillus caldus sp. nov., a moderately thermophilic acidophile. Microbiology 1994, 140, 3451–3456. [Google Scholar] [CrossRef] [PubMed]
- Valdes, J.; Quatrini, R.; Hallberg, K.; Dopson, M.; Valenzuela, P.D.; Holmes, D.S. Draft genome sequence of the extremely acidophilic bacterium Acidithiobacillus caldus ATCC 51756 reveals metabolic versatility in the genus Acidithiobacillus. J. Bacteriol. 2009, 191, 5877–5878. [Google Scholar] [CrossRef] [PubMed]
- Castelle, C.; Guiral, M.; Malarte, G.; Ledgham, F.; Leroy, G.; Brugna, M.; Giudici-Orticoni, M.-T. A new iron-oxidizing/O-2-reducing supercomplex spanning both inner and outer membranes, isolated from the extreme acidophile Acidithiobacillus ferrooxidans. J. Biol. Chem. 2008, 283, 25803–25811. [Google Scholar] [CrossRef]
- Ferguson, S.J.; Ingledew, W.J. Energetic problems faced by microorganisms growing or surviving on parsimonious energy sources and at acidic pH: I. Acidithiobacillus ferrooxidans as a paradigm. Biochim. Biophys. Acta. 2008, 1777, 1471–1479. [Google Scholar]
- Bulaev, A.G.; Kanygina, A.V.; Manolov, A.I. Genome analysis of Acidiplasma sp. MBA-1, a polyextremophilic archaeon predominant in the microbial community of a bioleaching reactor. Microbiology 2017, 86, 89–95. [Google Scholar] [CrossRef]
- Parks, D.H.; Rinke, C.; Chuvochina, M.; Chaumeil, P.A.; Woodcroft, B.J.; Evans, P.N.; Hugenholtz, P.; Tyson, G.W. Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life. Nat. Microbiol. 2017, 2, 1533–1542. [Google Scholar] [CrossRef]
- Yelton, A.P.; Comolli, L.R.; Justice, N.B.; Castelle, C.; Denef, V.J.; Thomas, B.C.; Banfield, J.F. Comparative genomics in acid mine drainage biofilm communities reveals metabolic and structural differentiation of co-occurring archaea. BMC Genom. 2013, 14, 485. [Google Scholar] [CrossRef]
- Dick, G.J.; Andersson, A.F.; Baker, B.J.; Simmons, S.L.; Thomas, B.C.; Yelton, A.P.; Banfield, J.F. Community-wide analysis of microbial genome sequence signatures. Genome Biol. 2009, 10, R85. [Google Scholar] [CrossRef]
- Gao, S.; Paez-Espino, D.; Li, J.; Ai, H.; Liang, J.; Luo, Z.; Zheng, J.; Chen, H.; Shu, W.; Huang, L. Patterns and ecological drivers of viral communities in acid mine drainage sediments across Southern China. Nat. Commun. 2022, 13, 2389. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.H.; Li, Q.; Chen, N.; Tang, L.Y.; Liao, B.; Yang, T.T.; Huang, L.N. Genome-resolved metagenomics reveals depth-related patterns of microbial community structure and functions in a highly stratified, AMD overlaying mine tailings. J. Hazard Mater. 2023, 447, 130774. [Google Scholar] [CrossRef] [PubMed]
- Johnson, D.B.; Hallberg, K.B. The microbiology of acidic mine waters. Res. Microbiol. 2003, 154, 466–473. [Google Scholar] [CrossRef]
- Fowler, T.A.; Holmes, P.R.; Crundwell, F.K. Mechanism of pyrite dissolution in the presence of Thiobacillus ferrooxidans. Appl. Environ. Microbiol. 1999, 65, 2987–2993. [Google Scholar] [CrossRef]
- Cobley, J.G.; Haddock, B.A. Respiratory chain of Thiobacillus ferrooxidans: Reduction of cytochromes by Fe2+ and preliminary characterization of rusticyanin, a novel blue copper protein. FEBS Lett. 1975, 60, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Giudici-Orticoni, M.T.; Guerlesquin, F.; Bruschi, M.; Nitschke, W. Interaction-induced redox switch in the electron transfer complex rusticyanin-cytochrome c(4). J. Biol. Chem. 1999, 274, 30365–30369. [Google Scholar] [CrossRef] [PubMed]
- Carlos, C.; Reis, F.C.; Vicentini, R.; Madureira, D.J.; Ottoboni, L.M.M. The rus operon genes are differentially regulated when Acidithiobacillus ferrooxidans LR is kept in contact with metal sulfides. Curr. Microbiol. 2008, 57, 375–380. [Google Scholar] [CrossRef]
- Giudici-Orticoni, M.T.; Leroy, G.; Nitschke, W.; Bruschi, M. Characterization of a new dihemic c(4)-type cytochrome isolated from Thiobacillus ferrooxidans. Biochemistry 2000, 39, 7205–7211. [Google Scholar] [CrossRef]
- Yarzabal, A.; Appia-Ayme, C.; Ratouchniak, J.; Bonnefoy, V. Regulation of the expression of the Acidithiobacillus ferrooxidans rus operon encoding two cytochromes c, a cytochrome oxidase and rusticyanin. Microbiology 2004, 150, 2113–2123. [Google Scholar] [CrossRef]
- Quatrini, R.; Appia-Ayme, C.; Denis, Y.; Ratouchniak, J.; Veloso, F.; Valdes, J.; Lefimil, C.; Silver, S.; Roberto, F.; Orellana, O. Insights into the iron and sulfur energetic metabolism of Acidithiobacillus ferrooxidans by microarray transcriptome profiling. Hydrometallurgy 2006, 83, 263–272. [Google Scholar] [CrossRef]
- Holmes, D.S.; Bonnefoy, V. Genetic and bioinformatic insights into iron and sulfur oxidation mechanisms of bioleaching organisms. In Biomining; Rawlings, D.E., Johnson, B.D., Eds.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 281–307. [Google Scholar]
- Allen, E.E.; Tyson, G.W.; Whitaker, R.J.; Detter, J.C.; Richardson, P.M.; Banfield, J.F. Genome dynamics in a natural archaeal population. Proc. Natl. Acad Sci. USA 2007, 104, 1883–1888. [Google Scholar] [CrossRef] [PubMed]
- Dopson, M.; Baker-Austin, C.; Bond, P.L. Analysis of differential protein expression during growth states of Ferroplasma strains and insights into electron transport for iron oxidation. Microbiology 2005, 151, 4127–4137. [Google Scholar] [CrossRef] [PubMed]
- Nichols, D.G.; Ferguson, S.J. Bioenergetics, 4th ed.; Academic Press: London, UK, 2013; 288p. [Google Scholar]
- Borisov, V.B.; Gennis, R.B.; Hemp, J.; Verkhovsky, M.I. The cytochrome bd respiratory oxygen reductases. Biochim. Biophys. Acta 2011, 1807, 1398–1413. [Google Scholar] [CrossRef]
- Kozubal, M.A.; Dlakić, M.; Macur, R.E.; Inskeep, W.P. Terminal Oxidase Diversity and Function in “Metallosphaera yellowstonensis”: Gene Expression and Protein Modeling Suggest Mechanisms of Fe (II) Oxidation in the Sulfolobales. Appl. Environ. Microbiol. 2011, 77, 1844–1853. [Google Scholar] [CrossRef]
- Baker, B.J.; Banfield, J.F. Microbial communities in acid mine drainage. FEMS Microbiol. Ecol. 2003, 44, 139–152. [Google Scholar] [CrossRef] [PubMed]
- Baker, B.J.; Tyson, G.W.; Webb, R.I.; Flanagan, J.; Hugenholtz, P.; Allen, E.E.; Banfield, J.F. Lineages of acidophilic archaea revealed by community genomic analysis. Science 2006, 314, 1933–1935. [Google Scholar] [CrossRef] [PubMed]
- Golyshina, O.V.; Lünsdorf, H.; Kublanov, I.V.; Goldenstein, N.I.; Hinrichs, K.U.; Golyshin, P.N. The novel, extremely acidophilic, cell wall-deficient archaeon Cuniculiplasma divulgatum gen. nov., sp. nov. represents a new family of Cuniculiplasmataceae fam. nov., order Thermoplasmatales. Int. J. Syst. Evol. Microbiol. 2016, 66, 332–340. [Google Scholar] [CrossRef]
- Golyshina, O.V.; Bargiela, R.; Golyshin, P.N. Cuniculiplasmataceae, their ecogenomic and metabolic patterns, and interactions with ‘ARMAN’. Extremophiles 2019, 23, 1–7. [Google Scholar] [CrossRef]
- Golyshina, O.V.; Bargiela, R.; Toshchakov, S.V.; Chernyh, N.A.; Ramayah, S.; Korzhenkov, A.A.; Kublanov, I.V.; Golyshin, P.N. Diversity of “Ca. Micrarchaeota” in two distinct types of acidic environments and their associations with Thermoplasmatales. Genes 2019, 10, 461. [Google Scholar] [CrossRef]
- Korzhenkov, A.A.; Toshchakov, S.V.; Bargiela, R.; Gibbard, H.; Ferrer, M.; Teplyuk, A.V.; Jones, D.L.; Kublanov, I.V.; Golyshin, P.N.; Golyshina, O.V. Archaea dominate the microbial community in an ecosystem with low-to-moderate temperature and extreme acidity. Microbiome 2019, 7, 11. [Google Scholar] [CrossRef]
- Bargiela, R.; Korzhenkov, A.A.; McIntosh, O.A.; Toshchakov, S.V.; Yakimov, M.M.; Golyshin, P.N.; Golyshina, O.V. Evolutionary patterns of archaea predominant in acidic environment. Environ. Microbiome 2023, 18, 61. [Google Scholar] [CrossRef] [PubMed]
- Artykova, A.; Elkina, Y.; Nechaeva, A.; Melamud, V.; Boduen, A.; Bulaev, A. Options for Increasing the Rate of Bioleaching of Arsenic Containing Copper Concentrate. Microbiol. Res. 2022, 13, 466–479. [Google Scholar] [CrossRef]
- Konstantinidis, K.T.; Rosselló-Móra, R. Classifying the uncultivated microbial majority: A place for metagenomic data in the Candidatus proposal. Syst. Appl. Microbiol. 2015, 38, 223–230. [Google Scholar] [CrossRef] [PubMed]
- Konstantinidis, K.T.; Rosselló-Móra, R.; Amann, R. Uncultivated microbes in need of their own taxonomy. ISME J. 2017, 11, 2399–2406. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.B.; Sutcliffe, I.C.; Rossello-Mora, R. Proposal for changes in the International Code of Nomenclature of Prokaryotes: Granting priority to Candidatus names. ISME J. 2019, 69, 2174–2175. [Google Scholar] [CrossRef]
- Oren, A. Nomenclature of prokaryotic ‘Candidatus’ taxa: Establishing order in the current chaos. New Microbes New Infect. 2021, 13, 100932. [Google Scholar] [CrossRef]
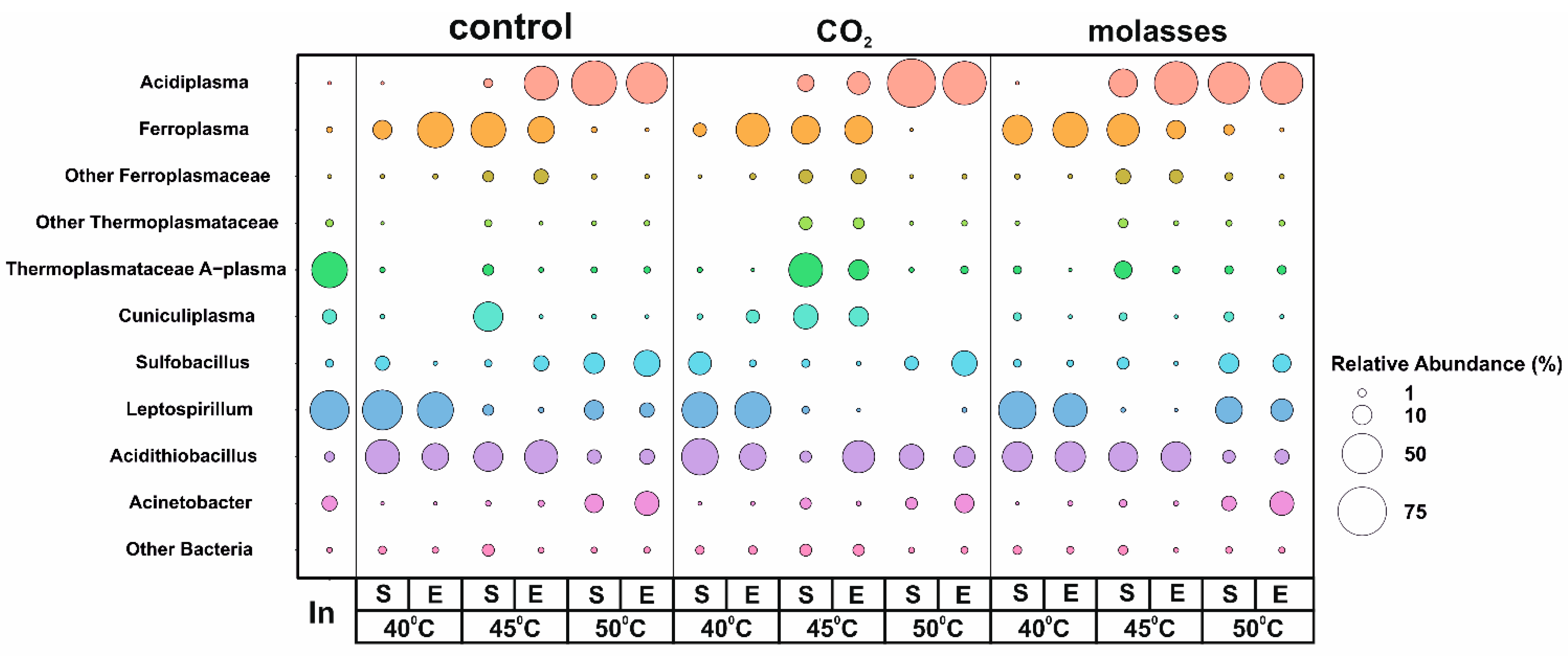
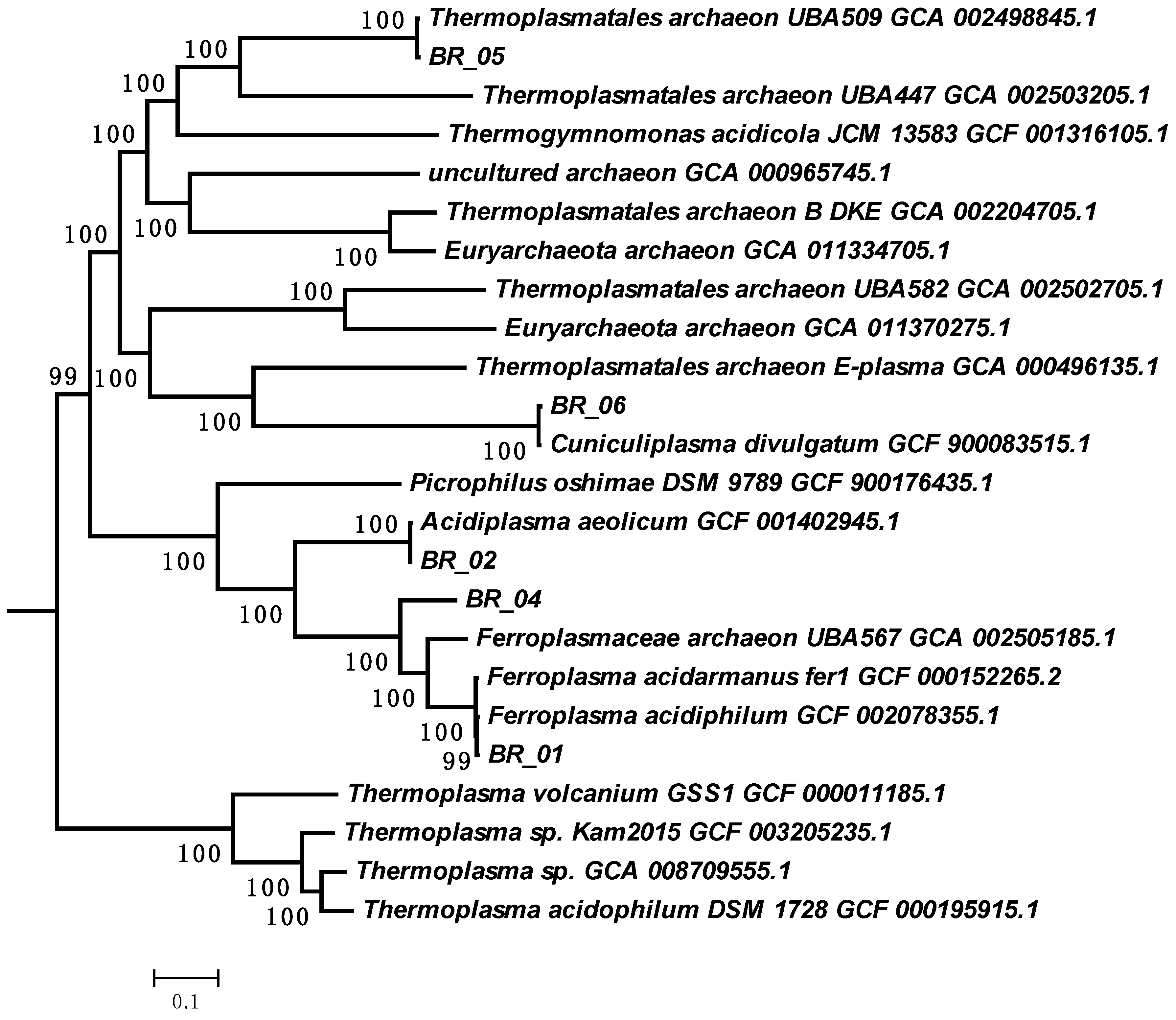
| T, °C | Carbon Source | Stage of the Experiment | pH | Eh, mV | Concentration, g/L | Cell Number, Cell/mL ×107 | |
|---|---|---|---|---|---|---|---|
| Fe3+ | Fe2+ | ||||||
| 40 | control | start | 1.21 | 725 | 1.05 | 0.77 | 5 |
| end | 0.59 | 866 | 23.87 | 0.35 | 225 | ||
| CO2 | start | 1.22 | 724 | 1.26 | 0.63 | 5 | |
| end | 0.61 | 862 | 26.95 | 0.49 | 260 | ||
| molasses | start | 1.23 | 731 | 0.98 | 0.7 | 5 | |
| end | 0.74 | 878 | 21.56 | 0 | 203 | ||
| 45 | control | start | 1.28 | 723 | 4.76 | 1.26 | 59 |
| end | 0.68 | 826 | 24.36 | 0 | 163 | ||
| CO2 | start | 1.33 | 713 | 4.06 | 1.05 | 58 | |
| end | 0.78 | 905 | 31.92 | 0 | 122 | ||
| molasses | start | 1.27 | 725 | 4.48 | 1.12 | 66 | |
| end | 0.63 | 825 | 26.46 | 0 | 112 | ||
| 50 | control | start | 1.24 | 719 | 1.89 | 0.56 | 10 |
| end | 0.76 | 748 | 16.24 | 1.68 | 15 | ||
| CO2 | start | 1.26 | 741 | 4.2 | 0.84 | 19 | |
| end | 0.76 | 776 | 22.96 | 0.56 | 30 | ||
| molasses | start | 1.26 | 727 | 1.75 | 0.77 | 14 | |
| end | 0.82 | 735 | 9.94 | 2.38 | 10 | ||
| T °C | Carbon Source | pH | Eh, mV | Concentration, g/L | Cell Number, Cell/mL ×107 | CaCO3 Consumption, kg/t | ||
|---|---|---|---|---|---|---|---|---|
| Fe3+ | Fe2+ | As | ||||||
| 40 | control | 0.75 ± 0.15 | 877 ± 44 | 28.3 ± 0.4 | ND * | 7.1 ± 0.1 | 359 ± 22 | 133 |
| CO2 | 0.76 ± 0.14 | 831 ± 21 | 24.7 ± 0.5 | ND | 6.6 ± 0.2 | 370 ± 25 | 133 | |
| molasses | 0.78 ± 0.13 | 845 ± 41 | 26.9 ± 1.5 | 0.01 ± 0.01 | 6.9 ± 0.3 | 399 ± 17 | 143 | |
| 45 | control | 1.07 ± 0.06 | 785 ± 18 | 14.1 ± 1.5 | 0.5 ± 0.1 | 4.6 ± 0.1 | 118 ± 42 | 17 |
| CO2 | 0.92 ± 0.02 | 855 ± 36 | 27.7 ± 0.7 | ND | 4.7 ± 0.2 | 245 ± 29 | 114 | |
| molasses | 1.12 ± 0.07 | 777 ± 16 | 10.4 ± 1.0 | 0.6 ± 0.2 | 4.3 ± 0.1 | 117 ± 23 | 0 | |
| 50 | control | 1.11 ± 0.03 | 621 ± 6 | 7.7 ± 0.4 | 1.2 ± 0.2 | 4.2 ± 0.4 | 29 ± 9 | 0 |
| CO2 | 0.92 ± 0.12 | 670 ± 8 | 16.7 ± 2.2 | 0.23 ± 0.18 | 4.7 ± 0.1 | 71 ± 11 | 80 | |
| molasses | 1.05 ± 0.05 | 620 ± 7 | 6.0 ± 0.3 | 0.87 ± 0.06 | 3.1 ± 0.2 | 24 ± 4 | 0 | |
| T, °C | Carbon Source | Mass Yield, % | Oxidation, % | Au Extraction Rate, % | |
|---|---|---|---|---|---|
| Pyrite | Arsenopyrite | ||||
| Concentrate | - | - | - | - | 31 |
| 40 | control | 42.6 | 89 | 99 | 80 |
| CO2 | 41.4 | 90 | 99 | 72 | |
| molasses | 46.0 | 91 | 99 | 58 | |
| 45 | control | 54.5 | 61 | 93 | 92 |
| CO2 | 40.7 | 89 | 98 | 91 | |
| molasses | 52.5 | 62 | 93 | 92 | |
| 50 | control | 73.5 | 35 | 89 | 89 |
| CO2 | 61.6 | 53 | 95 | 92 | |
| molasses | 80.0 | 36 | 82 | 87 | |
| Bin Id | Genome Size (b.p.) | Share in the Metagenome % | Taxonomy According to GTDB |
|---|---|---|---|
| BR_02 | 1,718,531 | 18.7 | Archaea; Thermoplasmatota; Thermoplasmata; Thermoplasmatales; Thermoplasmataceae; Acidiplasma; Acidiplasma sp. |
| BR_06 | 1,931,334 | 13.6 | Archaea; Thermoplasmatota; Thermoplasmata; Thermoplasmatales; Thermoplasmataceae; Cuniculiplasma; Cuniculiplasma divulgatum |
| BR_04 | 1,859,113 | 3.7 | Archaea; Thermoplasmatota; Thermoplasmata; Thermoplasmatales; Thermoplasmataceae; Ferroplasma; |
| BR_01 | 1,844,541 | 4.1 | Archaea; Thermoplasmatota; Thermoplasmata; Thermoplasmatales; Thermoplasmataceae; Ferroplasma; Ferroplasma acidiphilum |
| BR_05 | 2,006,340 | 24.4 | Archaea; Thermoplasmatota; Thermoplasmata; Thermoplasmatales; Thermoplasmataceae; UBA509; UBA509 sp002498845 |
| BR_03 | 2,900,829 | 23.7 | Bacteria; Proteobacteria; Gammaproteobacteria; Acidithiobacillales; Acidithiobacillaceae; Acidithiobacillus_A; Acidithiobacillus; A caldus |
| Organism/Genome | GenBank | Total Length Scaffolds, bp | Genome Completeness, % | Number Scaffolds | Median Scaffold Length (N50), bp | Protein-Coding Genes | AAI | ANI | Reference |
|---|---|---|---|---|---|---|---|---|---|
| Thermoplasmatales archaeon UBA509 * | GCA_002498845.1 | 1,811,892 | 98.79 | 65 | 40,222 | 1919 | 97.01 | 98.37 | [67] |
| Thermoplsmatales archaeon UBA574 | GCA_002497065 | 1,711,194 | 98.79 | 86 | 27,819 | 1797 | 97.79 | 98.58 | [67] |
| Thermoplsmatales archaeon UBA263 | GCA_002496665 | 1,729,665 | 96.1 | 90 | 27,683 | 1849 | 97.86 | 98.48 | [67] |
| Thermoplsmatales archaeon UBA517 | GCA_002499245 | 1,863,948 | 96.06 | 97 | 25,054 | 1966 | 97.22 | 98.36 | [67] |
| Thermoplsmatales archaeon UBA612 | GCA_002505105 | 1,602,081 | 95.3 | 99 | 21,298 | 1703 | 98.01 | 98.58 | [67] |
| Thermoplsmatales archaeon UBA617 | GCA_002507365 | 1,808,499 | 98.79 | 111 | 24,240 | 1903 | 97.51 | 98.61 | [67] |
| Thermoplsmatales archaeon UBA565 | GCA_002506245 | 1,687,753 | 94.35 | 119 | 23,108 | 1977 | 97.55 | 98.27 | [67] |
| Thermoplsmatales archaeon UBA565 | GCA_002507555 | 1,600,814 | 94.35 | 106 | 20,202 | 1693 | 97.93 | 98.6 | [67] |
| Thermoplsmatales archaeon UBA512 | GCA_002502885 | 1,424,627 | 91.94 | 86 | 20,724 | 1514 | 98.55 | 98.66 | [67] |
| Thermoplasmatales archaeon A-plasma | GCA_000447225 | 1,989,604 | 97.18 | 118 | 46,831 | 2277 | 97.62 | 98.65 | [68] |
| Thermoplsmatales archaeon UBA580 | GCA_002497605 | 1,632,468 | 91.53 | 84 | 28,963 | 1765 | 97.41 | 98.66 | [67] |
| Thermoplsmatales archaeon UBA568 | GCA_002498865 | 1,524,549 | 85.49 | 147 | 14,312 | 1984 | 97.63 | 98.38 | [67] |
| Thermoplsmatales archaeon UBA571 | GCA_002506945 | 1,736,188 | 89.83 | 157 | 15,245 | 2174 | 97.16 | 98.4 | [67] |
| Thermoplasmata archaeon | GCA_021797595 | 1,282,202 | 84.62 | 106 | 15,487 | 1388 | 97.16 | 96.15 | Unpublished |
| Thermoplsmatales archaeon UBA504 | GCA_002499625 | 1,441,013 | 83.03 | 51 | 39,963 | 1499 | 97.81 | 98.67 | [67] |
| Thermoplasmatales archaeon UBA578 | GCA_002506955 | 1,430,970 | 86.38 | 169 | 10,758 | 1848 | 97.41 | 98.36 | [67] |
| Thermoplasmatales archaeon UBA521 | GCA_002501735 | 1,736,577 | 94.35 | 157 | 27,197 | 1830 | 97.94 | 98.56 | [67] |
| Thermoplasmatales archaeon UBA269 | GCA_002505615 | 1,323,754 | 79.44 | 65 | 28,427 | 1402 | 98.55 | 98.8 | [67] |
| Candidatus Thermoplasmatota archaeon | GCA_023381335 | 1,233,498 | 70.7 | 127 | 12,404 | 1332 | 96.9 | 97.06 | Unpublished |
| Candidatus Thermoplasmatota archaeon | GCA_023484265 | 1,207,223 | 68.25 | 137 | 11,603 | 1347 | 96.72 | 97.28 | Unpublished |
| Thermoplasmatales archaeon A-plasma | GCA_009387835 | 1,957,458 | 92.45 | 103 | 46,831 | 2263 | 97.35 | 98.68 | [69] |
| Thermoplasmata archaeon | GCA_021797355 | 1,736,203 | 99.02 | 15 | 205,936 | 1837 | 71.26 | 84.07 | [70] |
| Thermoplasmata archaeon | GCA_021802255 | 897,991 | 55.44 | 49 | 35,795 | 966 | 74.72 | 87.06 | [70] |
| Thermoplasmata archaeon | GCA_021799185 | 1,681,583 | 90.55 | 163 | 14,860 | 1792 | 85.64 | 80.81 | [70] |
| Thermoplasmata archaeon | GCA_021819605 | 1,414,939 | 91.67 | 146 | 12,230 | 1537 | 94.05 | 90.2 | [71] |
| Thermoplasmata archaeon | GCA_021787155 | 2,053,782 | 82.6 | 205 | 13,296 | 2164 | 93.11 | 89.78 | [70] |
| Candidatus Thermoplasmatota archaeon | GCA_023379345 | 1,063,747 | 77.22 | 115 | 11,683 | 1137 | 91.2 | 88.21 | [71] |
| Thermoplasmatales archaeon BR_05 | CP133596 | 2,006,340 | 99.6 | 1 | 2,006,340 | 1993 | This work |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bulaev, A.; Kadnikov, V.; Elkina, Y.; Beletsky, A.; Melamud, V.; Ravin, N.; Mardanov, A. Shifts in the Microbial Populations of Bioleach Reactors Are Determined by Carbon Sources and Temperature. Biology 2023, 12, 1411. https://doi.org/10.3390/biology12111411
Bulaev A, Kadnikov V, Elkina Y, Beletsky A, Melamud V, Ravin N, Mardanov A. Shifts in the Microbial Populations of Bioleach Reactors Are Determined by Carbon Sources and Temperature. Biology. 2023; 12(11):1411. https://doi.org/10.3390/biology12111411
Chicago/Turabian StyleBulaev, Aleksandr, Vitaliy Kadnikov, Yulia Elkina, Aleksey Beletsky, Vitaliy Melamud, Nikolai Ravin, and Andrey Mardanov. 2023. "Shifts in the Microbial Populations of Bioleach Reactors Are Determined by Carbon Sources and Temperature" Biology 12, no. 11: 1411. https://doi.org/10.3390/biology12111411
APA StyleBulaev, A., Kadnikov, V., Elkina, Y., Beletsky, A., Melamud, V., Ravin, N., & Mardanov, A. (2023). Shifts in the Microbial Populations of Bioleach Reactors Are Determined by Carbon Sources and Temperature. Biology, 12(11), 1411. https://doi.org/10.3390/biology12111411







