The Function and Therapeutic Potential of lncRNAs in Cardiac Fibrosis
Abstract
:Simple Summary
Abstract
1. Introduction
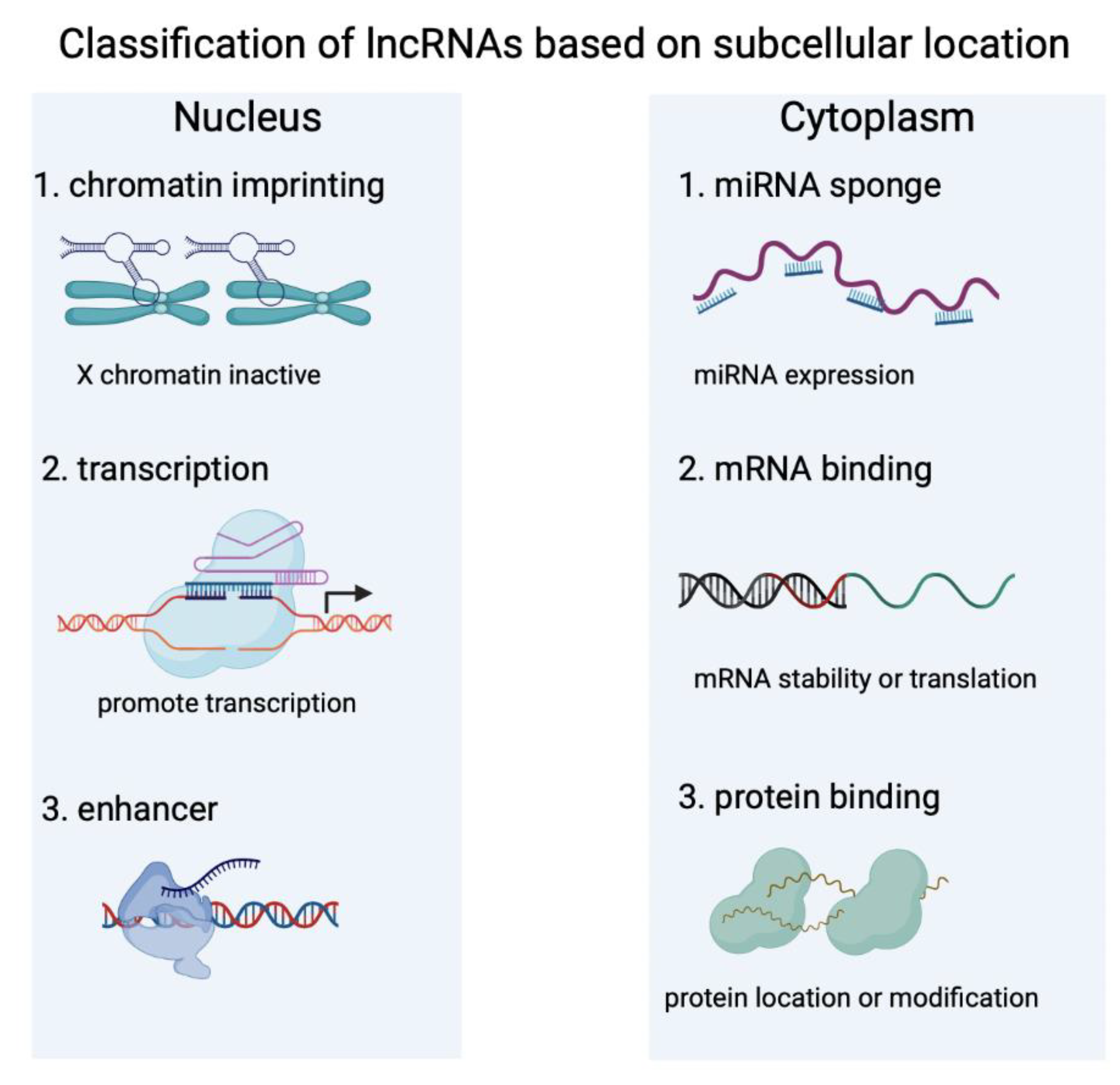
2. Characteristics of lncRNAs
2.1. LncRNAs Serve as ceRNAs in Controlling Cardiac Fibrosis
2.2. LncRNAs Regulate Cardiac Fibrosis through TGF-β Signaling Pathways
2.3. LncRNAs Control Cardiac Fibrosis by Regulating ECM Gene Expression
2.4. Exosome-Derived LncRNAs Regulate Cardiac Fibrosis
2.5. The Way of LncRNAs Secreted into Exosomes
2.6. LncRNAs Serve as Potential Therapeutic Targets
3. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López, B.; Ravassa, S.; Moreno, M.U.; José, G.S.; Beaumont, J.; González, A.; Díez, J. Diffuse myocardial fibrosis: Mechanisms, diagnosis and therapeutic approaches. Nat. Rev. Cardiol. 2021, 18, 479–498. [Google Scholar] [CrossRef]
- Tallquist, M.D. Cardiac Fibroblast Diversity. Annu. Rev. Physiol. 2020, 82, 63–78. [Google Scholar] [CrossRef] [Green Version]
- Weber, K.T.; Pick, R.; Jalil, J.E.; Janicki, J.S.; Carroll, E.P. Patterns of myocardial fibrosis. J. Mol. Cell. Cardiol. 1989, 21 (Suppl. 5), 121–131. [Google Scholar] [CrossRef] [PubMed]
- Kong, P.; Christia, P.; Frangogiannis, N. The pathogenesis of cardiac fibrosis. Cell. Mol. Life Sci. 2014, 71, 549–574. [Google Scholar] [CrossRef] [Green Version]
- Horckmans, M.; Bianchini, M.; Santovito, D.; Megens, R.T.A.; Springael, J.Y.; Negri, I.; Vacca, M.; Di Eusanio, M.; Moschetta, A.; Weber, C.; et al. Pericardial Adipose Tissue Regulates Granulopoiesis, Fibrosis, and Cardiac Function after Myocardial Infarction. Circulation 2018, 137, 948–960. [Google Scholar] [CrossRef]
- Prabhu, S.D.; Frangogiannis, N.G. The Biological Basis for Cardiac Repair after Myocardial Infarction: From Inflammation to Fibrosis. Circ. Res. 2016, 119, 91–112. [Google Scholar] [CrossRef]
- Epelman, S.; Liu, P.; Mann, D. Role of innate and adaptive immune mechanisms in cardiac injury and repair. Nat. Rev. Immunol. 2015, 15, 117–129. [Google Scholar] [CrossRef]
- Plehn, J.F.; Biederman, R. Diffuse Myocardial Fibrosis in Dilated Cardiomyopathy: Risk Marker, Risk Factor, or Does it Matter? JACC Cardiovasc. Imaging 2022, 15, 591–593. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Sui, W.; Xing, Y.; Cheng, J.; Cheng, C.; Xue, F.; Zhang, J.; Wang, X.; Zhang, C.; Hao, P.; et al. Angiotensin IV attenuates diabetic cardiomyopathy via suppressing FoxO1-induced excessive autophagy, apoptosis and fibrosis. Theranostics 2021, 11, 8624–8639. [Google Scholar] [CrossRef] [PubMed]
- Marques, F.Z.; Nelson, E.; Chu, P.Y.; Horlock, D.; Fiedler, A.; Ziemann, M.; Tan, J.K.; Kuruppu, S.; Rajapakse, N.W.; El-Osta, A.; et al. High-Fiber Diet and Acetate Supplementation Change the Gut Microbiota and Prevent the Development of Hypertension and Heart Failure in Hypertensive Mice. Circulation 2017, 135, 964–977. [Google Scholar] [CrossRef]
- Maisch, B. Extracellular matrix and cardiac interstitium: Restriction is not a restricted phenomenon. Herz 1995, 20, 75–80. [Google Scholar]
- Tallquist, M.D.; Molkentin, J. Redefining the identity of cardiac fibroblasts. Nat. Rev. Cardiol. 2017, 14, 484–491. [Google Scholar] [CrossRef]
- Spinale, F.G. Matrix metalloproteinases: Regulation and dysregulation in the failing heart. Circ. Res. 2002, 90, 520–530. [Google Scholar] [CrossRef] [Green Version]
- Powell, D.W.; Mifflin, R.C.; Valentich, J.D.; Crowe, S.E.; Saada, J.I.; West, A.B. Myofibroblasts. I. Paracrine cells important in health and disease. Am. J. Physiol. Cell Physiol. 1999, 277, C1–C19. [Google Scholar] [CrossRef] [PubMed]
- Tomasek, J.J.; Gabbiani, G.; Hinz, B.; Chaponnier, C.; Brown, R.A. Myofibroblasts and mechano: Regulation of connective tissue remodelling. Nat. Rev. Mol. Cell Biol. 2002, 3, 349–363. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Nkennor, B.; Mastikhina, O.; Soon, K.; Nunes, S.S. Endothelium-mediated contributions to fibrosis. Semin. Cell Dev. Biol. 2020, 101, 78–86. [Google Scholar] [CrossRef]
- Howard, C.M.; Baudino, T. Dynamic cell-cell and cell-ECM interactions in the heart. J. Mol. Cell. Cardiol. 2014, 70, 19–26. [Google Scholar] [CrossRef]
- Deb, A. Cell-cell interaction in the heart via Wnt/β-catenin pathway after cardiac injury. Cardiovasc. Res. 2014, 102, 214–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tan, Y.T.; Lin, J.F.; Li, T.; Li, J.J.; Xu, R.H.; Ju, H.Q. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 2021, 41, 109–120. [Google Scholar] [CrossRef]
- Nojima, T.; Proudfoot, N. Mechanisms of lncRNA biogenesis as revealed by nascent transcriptomics. Nat. Rev. Mol. Cell Biol. 2022, 23, 389–406. [Google Scholar] [CrossRef]
- Ilieva, M.; Uchida, S. Long Non-Coding RNAs in Cardiac and Pulmonary Fibroblasts and Fibrosis. Noncoding RNA 2022, 8, 53. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Jiang, S.; Shang, J.; Jiang, Y.; Dai, Y.; Xu, B.; Yu, Y.; Liang, Z.; Yang, Y. LncRNA: Shedding light on mechanisms and opportunities in fibrosis and aging. Ageing Res. Rev. 2019, 52, 17–31. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Zhang, L.; Song, J.; Wang, Z.; Huang, X.; Guo, Z.; Chen, F.; Zhao, X. Long noncoding RNA MALAT1 mediates cardiac fibrosis in experimental postinfarct myocardium mice model. J. Cell Physiol. 2019, 234, 2997–3006. [Google Scholar] [CrossRef] [PubMed]
- ENCODE Project Consortium. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elkon, R.; Agami, R. Characterization of noncoding regulatory DNA in the human genome. Nat. Biotechnol. 2017, 35, 732–746. [Google Scholar] [CrossRef] [PubMed]
- Mattick, J.S. Non-coding RNAs: The architects of eukaryotic complexity. EMBO Rep. 2001, 2, 986–991. [Google Scholar] [CrossRef] [Green Version]
- Yip, C.W.; Sivaraman, D.M.; Prabhu, A.V.; Shin, J.W. Functional annotation of lncRNA in high-throughput screening. Essays Biochem. 2021, 65, 761–773. [Google Scholar]
- Maeda, N.; Kasukawa, T.; Oyama, R.; Gough, J.; Frith, M.; Engström, P.G.; Lenhard, B.; Aturaliya, R.N.; Batalov, S.; Beisel, K.W.; et al. Transcript annotation in FANTOM3: Mouse gene catalog based on physical cDNAs. PLoS Genet. 2006, 2, e62. [Google Scholar] [CrossRef] [Green Version]
- Derrien, T.; Johnson, R.; Bussotti, G.; Tanzer, A.; Djebali, S.; Tilgner, H.; Guernec, G.; Martin, D.; Merkel, A.; Knowles, D.G.; et al. The GENCODE v7 catalog of human long noncoding RNAs: Analysis of their gene structure, evolution, and expression. Genome Res. 2012, 22, 1775–1789. [Google Scholar] [CrossRef] [Green Version]
- Fang, S.; Zhang, L.; Guo, J.; Niu, Y.; Wu, Y.; Li, H.; Zhao, L.; Li, X.; Teng, X.; Sun, X.; et al. NONCODEV5: A comprehensive annotation database for long non-coding RNAs. Nucleic Acids Res. 2018, 46, D308–D314. [Google Scholar] [CrossRef]
- Kopp, F.; Mendell, J. Functional Classification and Experimental Dissection of Long Noncoding RNAs. Cell 2018, 172, 393–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousefi, H.; Maheronnaghsh, M.; Molaei, F.; Mashouri, L.; Reza Aref, A.; Momeny, M.; Alahari, S.K. Long noncoding RNAs and exosomal lncRNAs: Classification, and mechanisms in breast cancer metastasis and drug resistance. Oncogene 2020, 39, 953–974. [Google Scholar] [CrossRef] [PubMed]
- Mondal, T.; Rasmussen, M.; Pandey, G.K.; Isaksson, A.; Kanduri, C. Characterization of the RNA content of chromatin. Genome Res. 2010, 20, 899–907. [Google Scholar] [CrossRef] [Green Version]
- Mao, Y.S.; Zhang, B.; Spector, D. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Sun, X. The functions of LncRNA in the heart. Diabetes Res. Clin. Pract. 2020, 168, 108249. [Google Scholar] [CrossRef]
- Bhan, A.; Soleimani, M.; Mandal, S. Long Noncoding RNA and Cancer: A New Paradigm. Cancer Res. 2017, 77, 3965–3981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, Y.; Rong, X.; Luo, Y.; Li, P.; Han, Q.; Wei, L.; Wang, E. A novel long non-coding RNA LINC00355 promotes proliferation of lung adenocarcinoma cells by down-regulating miR-195 and up-regulating the expression of CCNE1. Cell Signal. 2020, 66, 109462. [Google Scholar] [CrossRef]
- Qian, X.; Zhao, J.; Yeung, P.Y.; Zhang, Q.C.; Kwok, C.K. Revealing lncRNA Structures and Interactions by Sequencing-Based Approaches. Trends Biochem. Sci. 2019, 44, 33–52. [Google Scholar] [CrossRef]
- Schmitz, S.U.; Grote, P.; Herrmann, B. Mechanisms of long noncoding RNA function in development and disease. Cell. Mol. Life Sci. 2016, 73, 2491–2509. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.L. Linking Long Noncoding RNA Localization and Function. Trends Biochem. Sci. 2016, 41, 761–772. [Google Scholar] [CrossRef]
- Johnson, S.J.; Cooper, T. Overlapping mechanisms of lncRNA and expanded microsatellite RNA. Wiley Interdiscip. Rev. RNA 2021, 12, e1634. [Google Scholar] [CrossRef] [PubMed]
- Brockdorff, N.; Bowness, J.; Wei, G. Progress toward understanding chromosome silencing by Xist RNA. Genes Dev. 2020, 34, 733–744. [Google Scholar] [CrossRef]
- Yu, B.; Qi, Y.; Li, R.; Shi, Q.; Satpathy, A.T.; Chang, H.Y. B cell-specific XIST complex enforces X-inactivation and restrains atypical B cells. Cell 2021, 184, 1790–1803. [Google Scholar] [CrossRef] [PubMed]
- Engreitz, J.M.; Pandya-Jones, A.; McDonel, P.; Shishkin, A.; Sirokman, K.; Surka, C.; Kadri, S.; Xing, J.; Goren, A.; Lander, E.S.; et al. The Xist lncRNA exploits three-dimensional genome architecture to spread across the X chromosome. Science 2013, 341, 1237973. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.; Kopp, F.; Chang, T.C.; Sataluri, A.; Chen, B.; Sivakumar, S.; Yu, H.; Xie, Y. Noncoding RNA NORAD Regulates Genomic Stability by Sequestering PUMILIO Proteins. Cell 2016, 164, 69–80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elguindy, M.M.; Mendell, J. NORAD-induced Pumilio phase separation is required for genome stability. Nature 2021, 595, 303–308. [Google Scholar] [CrossRef]
- Wu, H.; Qin, W.; Lu, S.; Wang, X.; Zhang, J.; Sun, T.; Hu, X.; Li, Y.; Chen, Q.; Wang, Y.; et al. Long noncoding RNA ZFAS1 promoting small nucleolar RNA-mediated 2’-O-methylation via NOP58 recruitment in colorectal cancer. Mol. Cancer 2020, 19, 95. [Google Scholar] [CrossRef]
- Wang, Y.; Lu, J.H.; Wu, Q.N.; Jin, Y.; Wang, D.S.; Chen, Y.X.; Liu, J.; Luo, X.J.; Meng, Q.; Pu, H.Y.; et al. LncRNA LINRIS stabilizes IGF2BP2 and promotes the aerobic glycolysis in colorectal cancer. Mol. Cancer 2019, 18, 174. [Google Scholar] [CrossRef] [Green Version]
- Thomson, D.W.; Dinger, M. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 2016, 17, 272–283. [Google Scholar] [CrossRef]
- Zhang, Z.K.; Li, J.; Guan, D.; Liang, C.; Zhuo, Z.; Liu, J.; Lu, A.; Zhang, G.; Zhang, B.T. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J. Cachexia Sarcopenia Muscle 2018, 9, 613–626. [Google Scholar] [CrossRef] [Green Version]
- Liang, H.; Su, X.; Wu, Q.; Shan, H.; Lv, L.; Yu, T.; Zhao, X.; Sun, J.; Yang, R.; Zhang, L.; et al. LncRNA 2810403D21Rik/Mirf promotes ischemic myocardial injury by regulating autophagy through targeting Mir26a. Autophagy 2020, 16, 1077–1091. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.J.; Chang, H. Unique features of long non-coding RNA biogenesis and function. Nat. Rev. Genet. 2016, 17, 47–62. [Google Scholar] [CrossRef] [PubMed]
- Nelson, B.R.; Makarewich, C.A.; Anderson, D.M.; Winders, B.R.; Troupes, C.D.; Wu, F.; Reese, A.L.; McAnally, J.R.; Chen, X.; Kavalali, E.T.; et al. A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 2016, 351, 271–275. [Google Scholar] [CrossRef] [Green Version]
- Lai, Y.; He, S.; Ma, L.; Lin, H.; Ren, B.; Ma, J.; Zhu, X.; Zhuang, S. HOTAIR functions as a competing endogenous RNA to regulate PTEN expression by inhibiting miR-19 in cardiac hypertrophy. Mol. Cell. Biochem. 2017, 432, 179–187. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.H.; Yuan, Y.X.; Rao, S.L.; Wang, P. LncRNA MIAT enhances cardiac hypertrophy partly through sponging miR-150. Eur. Rev. Med. Pharmacol. Sci. 2016, 20, 3653–3660. [Google Scholar] [PubMed]
- Liu, L.; An, X.; Li, Z.; Song, Y.; Li, L.; Zuo, S.; Liu, N.; Yang, G.; Wang, H.; Cheng, X.; et al. The H19 long noncoding RNA is a novel negative regulator of cardiomyocyte hypertrophy. Cardiovasc. Res. 2016, 111, 56–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greco, S.; Salgado Somoza, A.; Devaux, Y.; Martelli, F. Long Noncoding RNAs and Cardiac Disease. Antioxid. Redox Signal. 2018, 29, 880–901. [Google Scholar] [CrossRef]
- Lozano-Vidal, N.; Bink, D.; Boon, R. Long noncoding RNA in cardiac aging and disease. J. Mol. Cell Biol. 2019, 11, 860–867. [Google Scholar] [CrossRef]
- Scheuermann, J.C.; Boyer, L. Getting to the heart of the matter: Long non-coding RNAs in cardiac development and disease. EMBO J. 2013, 32, 1805–1816. [Google Scholar] [CrossRef]
- Li, H.; Trager, L.E.; Liu, X.; Hastings, M.H.; Xiao, C.; Guerra, J.; To, S.; Li, G.; Yeri, A.; Rodosthenous, R.; et al. lncExACT1 and DCHS2 Regulate Physiological and Pathological Cardiac Growth. Circulation 2022, 145, 1218–1233. [Google Scholar] [CrossRef]
- Viereck, J.; Bührke, A.; Foinquinos, A.; Chatterjee, S.; Kleeberger, J.A.; Xiao, K.; Janssen-Peters, H.; Batkai, S.; Ramanujam, D.; Kraft, T.; et al. Targeting muscle-enriched long non-coding RNA H19 reverses pathological cardiac hypertrophy. Eur. Heart J. 2020, 41, 3462–3474. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Huo, X.; Yang, X.R.; He, J.; Cheng, L.; Wang, N.; Deng, X.; Jin, H.; Wang, N.; Wang, C.; et al. STAT3-mediated upregulation of lncRNA HOXD-AS1 as a ceRNA facilitates liver cancer metastasis by regulating SOX4. Mol. Cancer 2017, 16, 136. [Google Scholar] [CrossRef] [PubMed]
- Ghafouri-Fard, S.; Abak, A.; Talebi, S.F.; Shoorei, H.; Branicki, W.; Taheri, M.; Akbari Dilmaghani, N. Role of miRNA and lncRNAs in organ fibrosis and aging. Biomed. Pharmacother. 2021, 143, 112132. [Google Scholar] [CrossRef]
- Tanwar, V.S.; Reddy, M.A.; Natarajan, R. Emerging Role of Long Non-Coding RNAs in Diabetic Vascular Complications. Front. Endocrinol. 2021, 12, 665811. [Google Scholar] [CrossRef] [PubMed]
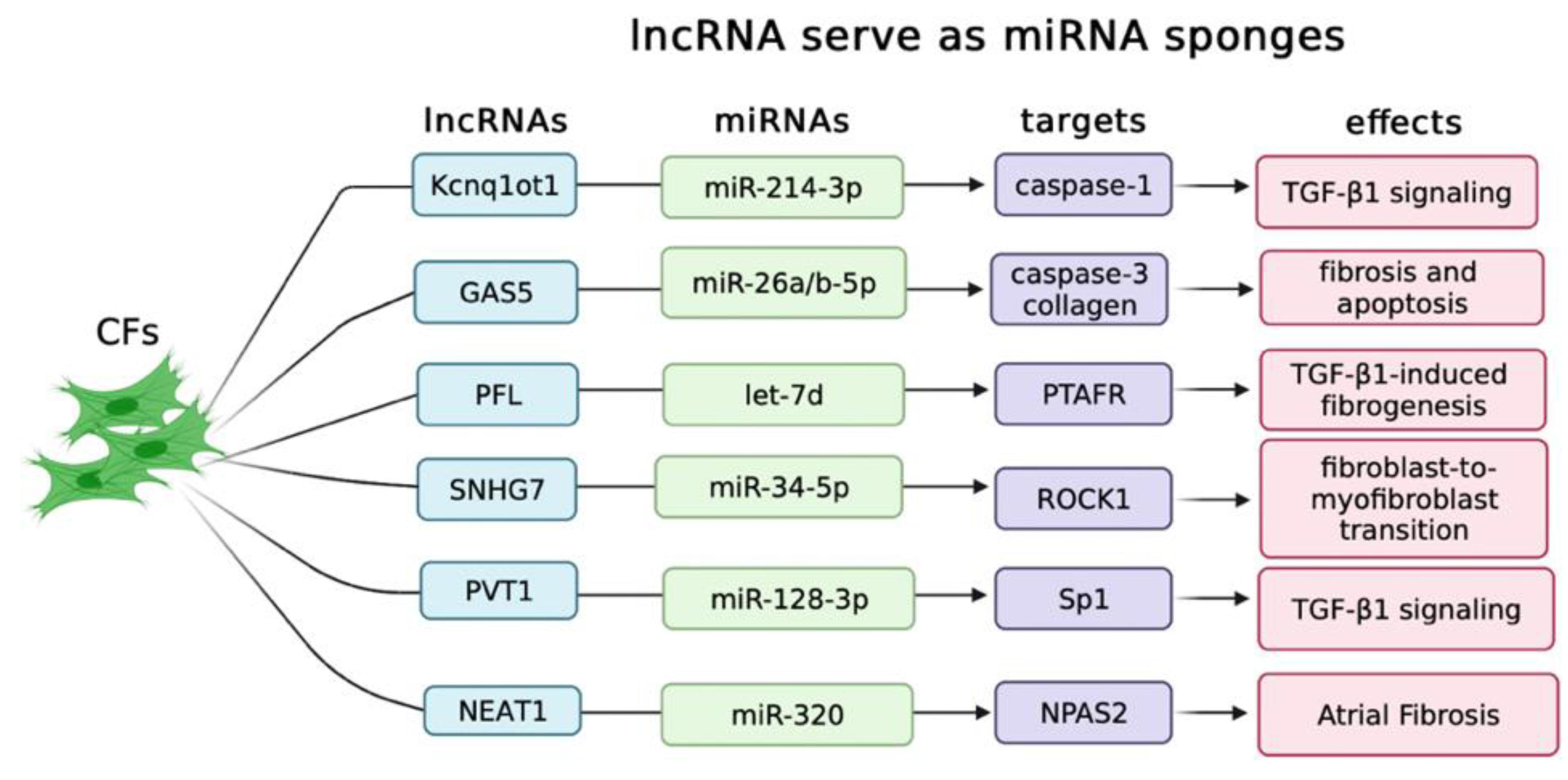
- Yang, F.; Qin, Y.; Lv, J.; Wang, Y.; Che, H.; Chen, X.; Jiang, Y.; Li, A.; Sun, X.; Yue, E.; et al. Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis. 2018, 9, 1000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, C.; Zhang, H.; Wei, D.; Sun, Z. Silencing lncRNA GAS5 alleviates apoptosis and fibrosis in diabetic cardiomyopathy by targeting miR-26a/b-5p. Acta Diabetol. 2021, 58, 1491–1501. [Google Scholar] [CrossRef]
- Varzideh, F.; Kansakar, U.; Donkor, K.; Wilson, S.; Jankauskas, S.S.; Mone, P.; Wang, X.; Lombardi, A.; Santulli, G. Cardiac Remodeling After Myocardial Infarction: Functional Contribution of microRNAs to Inflammation and Fibrosis. Front. Cardiovasc. Med. 2022, 9, 863238. [Google Scholar] [CrossRef]
- Liang, H.; Pan, Z.; Zhao, X.; Liu, L.; Sun, J.; Su, X.; Xu, C.; Zhou, Y.; Zhao, D.; Xu, B.; et al. LncRNA PFL contributes to cardiac fibrosis by acting as a competing endogenous RNA of let-7d. Theranostics 2018, 8, 1180–1194. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Zhang, S.; Li, X.; Gong, M. LncRNA SNHG7 promotes cardiac remodeling by upregulating ROCK1 via sponging miR-34-5p. Aging 2020, 12, 10441–10456. [Google Scholar] [CrossRef]
- Micheletti, R.; Plaisance, I.; Abraham, B.J.; Sarre, A.; Ting, C.C.; Alexanian, M.; Maric, D.; Maison, D.; Nemir, M.; Young, R.A.; et al. The long noncoding RNA Wisper controls cardiac fibrosis and remodeling. Sci. Transl. Med. 2017, 9, eaai9118. [Google Scholar] [CrossRef] [Green Version]
- Cao, F.; Li, Z.; Ding, W.M.; Yan, L.; Zhao, Q.Y. LncRNA PVT1 regulates atrial fibrosis via miR-128-3p-SP1-TGF-β1-Smad axis in atrial fibrillation. Mol Med. 2019, 25, 7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, H.; Zhao, N.; Liu, H.; Zheng, Y.; Zhao, L. LncRNA Nuclear-Enriched Abundant Transcript 1 Regulates Atrial Fibrosis via the miR-320/NPAS2 Axis in Atrial Fibrillation. Front. Pharmacol. 2021, 12, 647124. [Google Scholar] [CrossRef] [PubMed]
- Qi, X.; Zhang, D.H.; Wu, N.; Xiao, J.H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Xu, J.; Liu, X.; Jiang, J. The role of lncRNA-mediated ceRNA regulatory networks in pancreatic cancer. Cell Death Discov. 2022, 8, 287. [Google Scholar] [CrossRef]
- Chiu, H.S.; Martínez, M.R.; Komissarova, E.V.; Llobet-Navas, D.; Bansal, M.; Paull, E.O.; Silva, J.; Yang, X.; Sumazin, P.; Califano, A. The number of titrated microRNA species dictates ceRNA regulation. Nucleic Acids Res. 2018, 46, 4354–4369. [Google Scholar] [CrossRef] [Green Version]
- Saadat, S.; Noureddini, M.; Mahjoubin-Tehran, M.; Nazemi, S.; Shojaie, L.; Aschner, M.; Maleki, B.; Abbasi-Kolli, M.; Rajabi Moghadam, H.; Alani, B.; et al. Pivotal Role of TGF-β/Smad Signaling in Cardiac Fibrosis: Non-coding RNAs as Effectual Players. Front. Cardiovasc. Med. 2020, 7, 588347. [Google Scholar] [CrossRef]
- Hu, H.H.; Chen, D.Q.; Wang, Y.N.; Feng, Y.L.; Cao, G.; Vaziri, N.D.; Zhao, Y.Y. New insights into TGF-β/Smad signaling in tissue fibrosis. Chem. Biol. Interact. 2018, 292, 76–83. [Google Scholar] [CrossRef] [Green Version]
- Khalil, H.; Kanisicak, O.; Prasad, V.; Correll, R.N.; Fu, X.; Schips, T.; Vagnozzi, R.J.; Liu, R.; Huynh, T.; Lee, S.J.; et al. Fibroblast-specific TGF-β-Smad2/3 signaling underlies cardiac fibrosis. J. Clin. Investig. 2017, 127, 3770–3783. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wang, H.L.; Liu, T.T.; Lan, H.Y. TGF-Beta as a Master Regulator of Diabetic Nephropathy. Int. J. Mol. Sci. 2021, 22, 7881. [Google Scholar] [CrossRef]
- Yoshida, K.; Matsuzaki, K.; Murata, M.; Yamaguchi, T.; Suwa, K.; Okazaki, K. Clinico-Pathological Importance of TGF-β/Phospho-Smad Signaling during Human Hepatic Fibrocarcinogenesis. Cancers 2018, 10, 183. [Google Scholar] [CrossRef] [Green Version]
- Györfi, A.H.; Matei, A.; Distler, J. Targeting TGF-β signaling for the treatment of fibrosis. Matrix Biol. 2018, 68, 8–27. [Google Scholar] [CrossRef] [PubMed]
- Wei, Y.; Kim, T.J.; Peng, D.H.; Duan, D.; Gibbons, D.L.; Yamauchi, M.; Jackson, J.R.; Le Saux, C.J.; Calhoun, C.; Peters, J.; et al. Fibroblast-specific inhibition of TGF-β1 signaling attenuates lung and tumor fibrosis. J. Clin Investig. 2017, 127, 3675–3688. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Yu, Y.; Zhang, P.; Chen, Y.; Li, C.; Chen, J.; Wang, Y.; Li, Y. The crucial role of activin A/ALK4 pathway in the pathogenesis of Ang-II-induced atrial fibrosis and vulnerability to atrial fibrillation. Basic Res. Cardiol. 2017, 112, 47. [Google Scholar] [CrossRef]
- Chen, X.; Long, L.; Cheng, Y.; Chu, J.; Shen, Z.; Liu, L.; Li, J.; Xie, Q.; Liu, H.; Wu, M.; et al. Qingda granule attenuates cardiac fibrosis via suppression of the TGF-β1/Smad2/3 signaling pathway in vitro and in vivo. Biomed. Pharmacother. 2021, 137, 111318. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Shen, A.; Chen, X.; Wu, X.; Chu, J.; Cheng, Y.; Peng, M.; Chen, Y.; Weygant, N.; Wu, M.; et al. Huoxin pill attenuates myocardial infarction-induced apoptosis and fibrosis via suppression of p53 and TGF-β1/Smad2/3 pathways. Biomed. Pharmacother. 2020, 130, 110618. [Google Scholar] [CrossRef]
- Yousefi, F.; Shabaninejad, Z.; Vakili, S.; Derakhshan, M.; Movahedpour, A.; Dabiri, H.; Ghasemi, Y.; Mahjoubin-Tehran, M.; Nikoozadeh, A.; Savardashtaki, A.; et al. TGF-β and WNT signaling pathways in cardiac fibrosis: Non-coding RNAs come into focus. Cell Commun Signal. 2020, 18, 87. [Google Scholar] [CrossRef]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-β promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef] [Green Version]
- Pachera, E.; Assassi, S.; Salazar, G.A.; Stellato, M.; Renoux, F.; Wunderlin, A.; Blyszczuk, P.; Lafyatis, R.; Kurreeman, F.; de Vries-Bouwstra, J.; et al. Long noncoding RNA H19X is a key mediator of TGF-β-driven fibrosis. J. Clin. Investig. 2020, 130, 4888–4905. [Google Scholar] [CrossRef]
- Janakiraman, H.; House, R.P.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H.; Palanisamy, V. The Long (lncRNA) and Short (miRNA) of It: TGFβ-Mediated Control of RNA-Binding Proteins and Noncoding RNAs. Mol. Cancer Res. 2018, 16, 567–579. [Google Scholar] [CrossRef] [Green Version]
- Meng, X.M.; Nikolic-Paterson, D.; Lan, H. TGF-β: The master regulator of fibrosis. Nat. Rev. Nephrol. 2016, 12, 325–338. [Google Scholar] [CrossRef]
- Ge, Z.; Yin, C.; Li, Y.; Tian, D.; Xiang, Y.; Li, Q.; Tang, Y.; Zhang, Y. Long noncoding RNA NEAT1 promotes cardiac fibrosis in heart failure through increased recruitment of EZH2 to the Smad7 promoter region. J. Transl. Med. 2022, 20, 7. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Lei, W.; Wu, H.; Wu, J.; Yang, Z.; Yan, S.; Lu, X.A.; Li, J.; Xia, X.; Han, X.; et al. LncRNA-Safe contributes to cardiac fibrosis through Safe-Sfrp2-HuR complex in mouse myocardial infarction. Theranostics 2019, 9, 7282–7297. [Google Scholar] [CrossRef]
- Derynck, R.; Zhang, Y. Smad-dependent and Smad-independent pathways in TGF-beta family signalling. Nature 2003, 425, 577–584. [Google Scholar] [CrossRef] [PubMed]
- Weng, J.; Tu, M.; Wang, P.; Zhou, X.; Wang, C.; Wan, X.; Zhou, Z.; Wang, L.; Zheng, X.; Li, J.; et al. Amiodarone induces cell proliferation and myofibroblast differentiation via ERK1/2 and p38 MAPK signaling in fibroblasts. Biomed. Pharmacother. 2019, 115, 108889. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, Y.Y.; Li, T.T.; Wang, J.; Jiang, Y.; Zhao, Y.; Jin, X.X.; Xue, G.L.; Yang, Y.; Zhang, X.F.; et al. Ablation of interleukin-17 alleviated cardiac interstitial fibrosis and improved cardiac function via inhibiting long non-coding RNA-AK081284 in diabetic mice. J. Mol. Cell. Cardiol. 2018, 115, 64–72. [Google Scholar] [CrossRef]
- Zhang, F.; Fu, X.; Kataoka, M.; Liu, N.; Wang, Y.; Gao, F.; Liang, T.; Dong, X.; Pei, J.; Hu, X.; et al. Long noncoding RNA Cfast regulates cardiac fibrosis. Mol. Ther. Nucleic Acids 2021, 23, 377–392. [Google Scholar] [CrossRef]
- Tang, R.; Wang, Y.C.; Mei, X.; Shi, N.; Sun, C.; Ran, R.; Zhang, G.; Li, W.; Staveley-O’Carroll, K.F.; Li, G.; et al. LncRNA GAS5 attenuates fibroblast activation through inhibiting Smad3 signaling. Am. J. Physiol. Cell Physiol. 2020, 319, C105–C115. [Google Scholar] [CrossRef]
- Li, L.; Zhao, Q.; Kong, W. Extracellular matrix remodeling and cardiac fibrosis. Matrix Biol. 2018, 68, 490–506. [Google Scholar] [CrossRef] [PubMed]
- Chaudhuri, O.; Cooper-White, J.; Janmey, P.A.; Mooney, D.J.; Shenoy, V.B. Effects of extracellular matrix viscoelasticity on cellular behaviour. Nature 2020, 584, 535–546. [Google Scholar] [CrossRef]
- Huang, Z.P.; Ding, Y.; Chen, J.; Wu, G.; Kataoka, M.; Hu, Y.; Yang, J.H.; Liu, J.; Drakos, S.G.; Selzman, C.H.; et al. Long non-coding RNAs link extracellular matrix gene expression to ischemic cardiomyopathy. Cardiovasc. Res. 2016, 112, 543–554. [Google Scholar] [CrossRef] [Green Version]
- Choong, O.K.; Chen, C.Y.; Zhang, J.; Lin, J.H.; Lin, P.J.; Ruan, S.C.; Kamp, T.J.; Hsieh, P.C.H. Hypoxia-induced H19/YB-1 cascade modulates cardiac remodeling after infarction. Theranostics 2019, 9, 6550–6567. [Google Scholar] [CrossRef] [PubMed]
- Qu, X.; Du, Y.; Shu, Y.; Gao, M.; Sun, F.; Luo, S.; Yang, T.; Zhan, L.; Yuan, Y.; Chu, W.; et al. MIAT Is a Pro-fibrotic Long Non-coding RNA Governing Cardiac Fibrosis in Post-Infarct Myocardium. Sci. Rep. 2017, 7, 42657. [Google Scholar] [CrossRef]
- Piccoli, M.T.; Gupta, S.K.; Viereck, J.; Foinquinos, A.; Samolovac, S.; Kramer, F.L.; Garg, A.; Remke, J.; Zimmer, K.; Batkai, S.; et al. Inhibition of the Cardiac Fibroblast-Enriched lncRNA Meg3 Prevents Cardiac Fibrosis and Diastolic Dysfunction. Circ. Res. 2017, 121, 575–583. [Google Scholar] [CrossRef] [PubMed]
- Barile, L.; Moccetti, T.; Marban, E.; Vassalli, G. Roles of exosomes in cardioprotection. Eur. Heart J. 2017, 38, 1372–1379. [Google Scholar] [CrossRef] [Green Version]
- Mathiyalagan, P.; Sahoo, S. Exosomes-Based Gene Therapy for MicroRNA Delivery. Methods Mol. Biol. 2017, 1521, 139–152. [Google Scholar]
- Ruan, Y.; Lin, N.; Ma, Q.; Chen, R.; Zhang, Z.; Wen, W.; Chen, H.; Sun, J. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function. Cell. Physiol. Biochem. 2018, 46, 335–350. [Google Scholar] [CrossRef]
- Tao, S.C.; Rui, B.Y.; Wang, Q.Y.; Zhou, D.; Zhang, Y.; Guo, S.C. Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018, 25, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Thery, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef] [Green Version]
- Chaturvedi, P.; Kalani, A.; Medina, I.; Familtseva, A.; Tyagi, S.C. Cardiosome mediated regulation of MMP9 in diabetic heart: Role of mir29b and mir455 in exercise. J. Cell. Mol. Med. 2015, 19, 2153–2161. [Google Scholar] [CrossRef] [Green Version]
- Kenneweg, F.; Bang, C.; Xiao, K.; Boulanger, C.M.; Loyer, X.; Mazlan, S.; Schroen, B.; Hermans-Beijnsberger, S.; Foinquinos, A.; Hirt, M.N.; et al. Long Noncoding RNA-Enriched Vesicles Secreted by Hypoxic Cardiomyocytes Drive Cardiac Fibrosis. Mol. Ther. Nucleic Acids 2019, 18, 363–374. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cao, X.; Yan, L.; Zheng, Y.; Yu, J.; Sun, F.; Lian, Z.; Sun, L. Exosome-derived long non-coding RNA ZFAS1 controls cardiac fibrosis in chronic kidney disease. Aging 2021, 13. [Google Scholar] [CrossRef]
- Chen, Y.; Chen, X.; Li, H.; Li, Y.; Cheng, D.; Tang, Y.; Sang, H. Serum extracellular vesicles containing MIAT induces atrial fibrosis, inflammation and oxidative stress to promote atrial remodeling and atrial fibrillation via blockade of miR-485-5p-mediated CXCL10 inhibition. Clin. Transl. Med. 2021, 11, e482. [Google Scholar] [CrossRef] [PubMed]
- Raposo, G.; Stahl, P. Extracellular vesicles: A new communication paradigm? Nat. Rev. Mol. Cell Biol. 2019, 20, 509–510. [Google Scholar] [CrossRef]
- Villarroya-Beltri, C.; Gutierrez-Vazquez, C.; Sanchez-Cabo, F.; Perez-Hernandez, D.; Vazquez, J.; Martin-Cofreces, N.; Martinez-Herrera, D.J.; Pascual-Montano, A.; Mittelbrunn, M.; Sanchez-Madrid, F. Sumoylated hnRNPA2B1 controls the sorting of miRNAs into exosomes through binding to specific motifs. Nat. Commun. 2013, 4, 2980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, S.; Li, Y.; Chen, B.; Zhao, J.; Yu, S.; Tang, Y.; Zheng, Q.; Li, Y.; Wang, P.; He, X.; et al. exoRBase: A database of circRNA, lncRNA and mRNA in human blood exosomes. Nucleic Acids Res. 2018, 46, D106–D112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs modulate microRNA sorting to exosomes and transfer to acceptor cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppers-Lalic, D.; Hackenberg, M.; Bijnsdorp, I.V.; van Eijndhoven, M.A.J.; Sadek, P.; Sie, D.; Zini, N.; Middeldorp, J.M.; Ylstra, B.; de Menezes, R.X.; et al. Nontemplated nucleotide additions distinguish the small RNA composition in cells from exosomes. Cell Rep. 2014, 8, 1649–1658. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Hanson, P. Membrane budding and scission by the ESCRT machinery: It’s all in the neck. Nat. Rev. Mol. Cell Biol. 2010, 11, 556–566. [Google Scholar] [CrossRef]
- Puri, N.; Roche, P. Ternary SNARE complexes are enriched in lipid rafts during mast cell exocytosis. Traffic 2006, 7, 1482–1494. [Google Scholar] [CrossRef]
- Yang, L.; Peng, X.; Li, Y.; Zhang, X.; Ma, Y.; Wu, C.; Fan, Q.; Wei, S.; Li, H.; Liu, J. Long non-coding RNA HOTAIR promotes exosome secretion by regulating RAB35 and SNAP23 in hepatocellular carcinoma. Mol. Cancer 2019, 18, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooke, S.T.; Witztum, J.L.; Bennett, C.F.; Baker, B.F. RNA-Targeted Therapeutics. Cell Metab. 2018, 27, 714–739. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Crooke, S.T. Molecular Mechanisms of Antisense Oligonucleotides. Nucleic Acid Ther. 2017, 27, 70–77. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.H.; Han, Z.; Jeon, H.Y.; Kach, J.; Jing, E.; Weyn-Vanhentenryck, S.; Downs, M.; Corrionero, A.; Oh, R.; Scharner, J.; et al. Antisense oligonucleotide modulation of non-productive alternative splicing upregulates gene expression. Nat. Commun. 2020, 11, 3501. [Google Scholar] [CrossRef]
- Modarresi, F.; Faghihi, M.A.; Lopez-Toledano, M.A.; Fatemi, R.P.; Magistri, M.; Brothers, S.P.; van der Brug, M.P.; Wahlestedt, C. Inhibition of natural antisense transcripts in vivo results in gene-specific transcriptional upregulation. Nat. Biotechnol. 2012, 30, 453–459. [Google Scholar] [CrossRef] [Green Version]
- Lima, J.F.; Cerqueira, L.; Figueiredo, C.; Oliveira, C.; Azevedo, N.F. Anti-miRNA oligonucleotides: A comprehensive guide for design. RNA Biol. 2018, 15, 338–352. [Google Scholar] [CrossRef] [Green Version]
- Kluiver, J.; Slezak-Prochazka, I.; Smigielska-Czepiel, K.; Halsema, N.; Kroesen, B.J.; van den Berg, A. Generation of miRNA sponge constructs. Methods 2012, 58, 113–117. [Google Scholar] [CrossRef]
- Wronska, A. The Role of microRNA in the Development, Diagnosis, and Treatment of Cardiovascular Disease: Recent Developments. J. Pharmacol. Exp. Ther. 2023, 384, 123–132. [Google Scholar] [CrossRef]
- Das, S.; Kohr, M.; Dunkerly-Eyring, B.; Lee, D.I.; Bedja, D.; Kent, O.A.; Leung, A.K.; Henao-Mejia, J.; Flavell, R.A.; Steenbergen, C. Divergent Effects of miR-181 Family Members on Myocardial Function Through Protective Cytosolic and Detrimental Mitochondrial microRNA Targets. J. Am. Heart Assoc. 2017, 6, e004694. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Gregorevic, P.; Ritchie, R.H.; McMullen, J.R. Generation of MicroRNA-34 Sponges and Tough Decoys for the Heart: Developments and Challenges. Front. Pharmacol. 2018, 9, 1090. [Google Scholar] [CrossRef] [Green Version]
- Esau, C.; Davis, S.; Murray, S.F.; Yu, X.X.; Pandey, S.K.; Pear, M.; Watts, L.; Booten, S.L.; Graham, M.; McKay, R.; et al. miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab. 2006, 3, 87–98. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockey, D.C.; Bell, P.; Hill, J. Fibrosis—A common pathway to organ injury and failure. N. Engl. J. Med. 2015, 372, 1138–1149. [Google Scholar] [CrossRef] [PubMed]
- Zhang, K.; Han, X.; Zhang, Z.; Zheng, L.; Hu, Z.; Yao, Q.; Cui, H.; Shu, G.; Si, M.; Li, C.; et al. The liver-enriched lnc-LFAR1 promotes liver fibrosis by activating TGFβ and Notch pathways. Nat. Commun. 2017, 8, 144. [Google Scholar] [CrossRef]
- Bang, C.; Antoniades, C.; Antonopoulos, A.S.; Eriksson, U.; Franssen, C.; Hamdani, N.; Lehmann, L.; Moessinger, C.; Mongillo, M.; Muhl, L.; et al. Intercellular communication lessons in heart failure. Eur. J. Heart Fail. 2015, 17, 1091–1103. [Google Scholar] [CrossRef] [PubMed]
- Zanotti, S.; Gibertini, S.; Blasevich, F.; Bragato, C.; Ruggieri, A.; Saredi, S.; Fabbri, M.; Bernasconi, P.; Maggi, L.; Mantegazza, R.; et al. Exosomes and exosomal miRNAs from muscle-derived fibroblasts promote skeletal muscle fibrosis. Matrix Biol. 2018, 74, 77–100. [Google Scholar] [CrossRef]

| LncRNA Name | Expression | Experimental Model | Targeted Genes | Effects | Exosomes |
|---|---|---|---|---|---|
| Kcnq1ot1 | Up-regulated | DCM | miR-214-3/Caspase1 | TGF-β1 signaling | No |
| GAS5 | Up-regulated | DCM | miR-26a/b-5p/ Caspase3 | Fibrosis and apoptosis | No |
| PFL | Up-regulated | MI | Let-7d/PTAFR | TGF-β1-induced fibrogenesis | No |
| SNHG7 | Up-regulated | MI | miR-34-5p/ROCK1 | Fibroblast-to-myofibroblast transition | No |
| PVT1 | Up-regulated | AF | miR-128-3p/SP1 | TGF-β1 signaling | No |
| NEAT1 | Up-regulated | AF | miR-320/NPAS2 | Atrial fibrosis | No |
| AK081284 | Up-regulated | DCM | TGF-β1 | Collagen I and III production | No |
| CFAST | Up-regulated | MI | COTL1 | Enhances TGF-β signaling | No |
| SAFE | Up-regulated | MI | SFRP2 | Fibroblast to myofibroblast transition | No |
| H19 | Up-regulated | MI | YB-1 | Reduction of collagen 1A expression | No |
| Wisper | Up-regulated | MI | LH2 | CF behavior and survival | No |
| Meg3 | Down-regulated | MMP-2 | Diastolic performance | No | |
| ZFAS1 | Up-regulated | DCM | miR-4711-5p | Wnt4/β-catenin signal pathway | Yes |
| MIAT | Up-regulated | AF | miR-485-5p | Atrial remodeling | Yes |
| Neat1 | Up-regulated | MI | CDK1 | Fibroblast and cardiomyocyte survival | Yes |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nie, X.; Fan, J.; Wang, D.W. The Function and Therapeutic Potential of lncRNAs in Cardiac Fibrosis. Biology 2023, 12, 154. https://doi.org/10.3390/biology12020154
Nie X, Fan J, Wang DW. The Function and Therapeutic Potential of lncRNAs in Cardiac Fibrosis. Biology. 2023; 12(2):154. https://doi.org/10.3390/biology12020154
Chicago/Turabian StyleNie, Xiang, Jiahui Fan, and Dao Wen Wang. 2023. "The Function and Therapeutic Potential of lncRNAs in Cardiac Fibrosis" Biology 12, no. 2: 154. https://doi.org/10.3390/biology12020154
APA StyleNie, X., Fan, J., & Wang, D. W. (2023). The Function and Therapeutic Potential of lncRNAs in Cardiac Fibrosis. Biology, 12(2), 154. https://doi.org/10.3390/biology12020154





