Medulloblastoma: From TP53 Mutations to Molecular Classification and Liquid Biopsy
Abstract
:Simple Summary
Abstract
1. Introduction
| Year | Author | Probe | Method | Tumor | Milestone |
|---|---|---|---|---|---|
| 1910 | Wright [8] | Tumor (Autopsy and Biopsy/ Operation) | Histology | Neurocytoma or neuroblastoma (before the creation of the term for medulloblastoma) | A pathologist described CNS tumors differing from most others, later named medulloblastoma. Described as (pseudo-) rosettes, until today referred to as “Homer-Wright” rosettes. |
| 1925 | Cushing and Bailey [5] | Neurosurgically removed posterior fossa tumors | Histology | Medulloblastoma | Introduced the term medulloblastoma |
| 1953 | Paterson and Farr [9] | Clinical study | Irradiation: 5000 cGy posterior fossa 3500 cGy neuraxis | Reached 65% 3-year survival of medulloblastoma | Irradiation treatment of the whole CNS |
| 1969 | Chang et al. [10] | Clinical study | Staging | Medulloblastoma | Staging system |
| 1973 | Hart and Earle [11] | Classification | Histology | PNET | Introduced term PNET, regardless of location within CNS |
| (1950-)1980s | Various authors [12] | Experimental and clinical studies | Development of different chemotherapies and combinations thereof | Brain tumors, incl. medulloblastoma | Introduction of antineoplastic agents for different types of cell cycle, incl. alkylating agents |
| 1991 | Eibl and Wiestler [13,14] | Experimentally induced tumors and derived cell lines | Retrovirus-mediated gene transfer of SV40 LT into neural transplants | PNET (indistinguishable from medulloblastoma morphology) | Rat tumor model, histologically identical to human medulloblastoma (neuroblastic rosettes, bipotential differentiation), triggered medulloblastoma research in Bonn and Heidelberg, Germany |
| 1991 | Ohgaki, Eibl et al. [1] | Primary tumor tissue | SSCP-PCR, direct sequencing | Medulloblastoma | First detection of p53 mutations in primary medulloblastoma tissue by Eibl, supporting Eibl’s earlier tumor model of the inactivation of p53, also triggered medulloblastoma research, incl. molecular profiling leading to current WHO classification |
| 2001 | Reya et al. [15] | Cancer stem cell (CSC) | Compared self-renewal of hematopoetic stem cells with heterogeneity of cancer cells | Migratory cancer stem cells | Established CSC theory (Weissman/Clarke) |
| 2014 | Bettegowda et al. [16] | ctDNA | Digital PCR, sequencing | 14 tumor types, incl. medulloblastoma | ctDNA detectable for most tumors outside the brain |
| 2016 | Louis et al. [2] | Tissue biopsy | Molecular genetics | Medulloblastoma | WHO classification introduced four medulloblastoma groups based on molecular profile (transcriptome) |
| 2018 | Garzia et al. [17] | CTC | Parabiotic xenograft model | Medulloblastoma | Hematogenous spread of metastasis to leptomeninges by chemokine-chemokine receptor |
| 2021 | Louis et al. [3] | Tissue biopsy | Molecular profile, incl. methylation profile | Medulloblastoma | WHO introduced methylome to further classify medulloblastoma groups |
| 2022 | Smith et al. [18] | Normal and tumor tissue | Multi-omics, molecular signatures, expression profiles | Medulloblastoma, groups 3 and 4 | Identification of “Cell of origin” in groups 3 and 4 derived from rhombic lip nodulus in developing cerebellum |
| 2022 | Hendrikse et al. [19] | Transcriptomics, mutations upstream of CBFA: CBFA2T2, CBFA2T3, PRDM6, UTX, OTX2 | Medulloblastoma group 4 | Identification of medulloblastoma group 4 progenitor cells in rhombic lip |
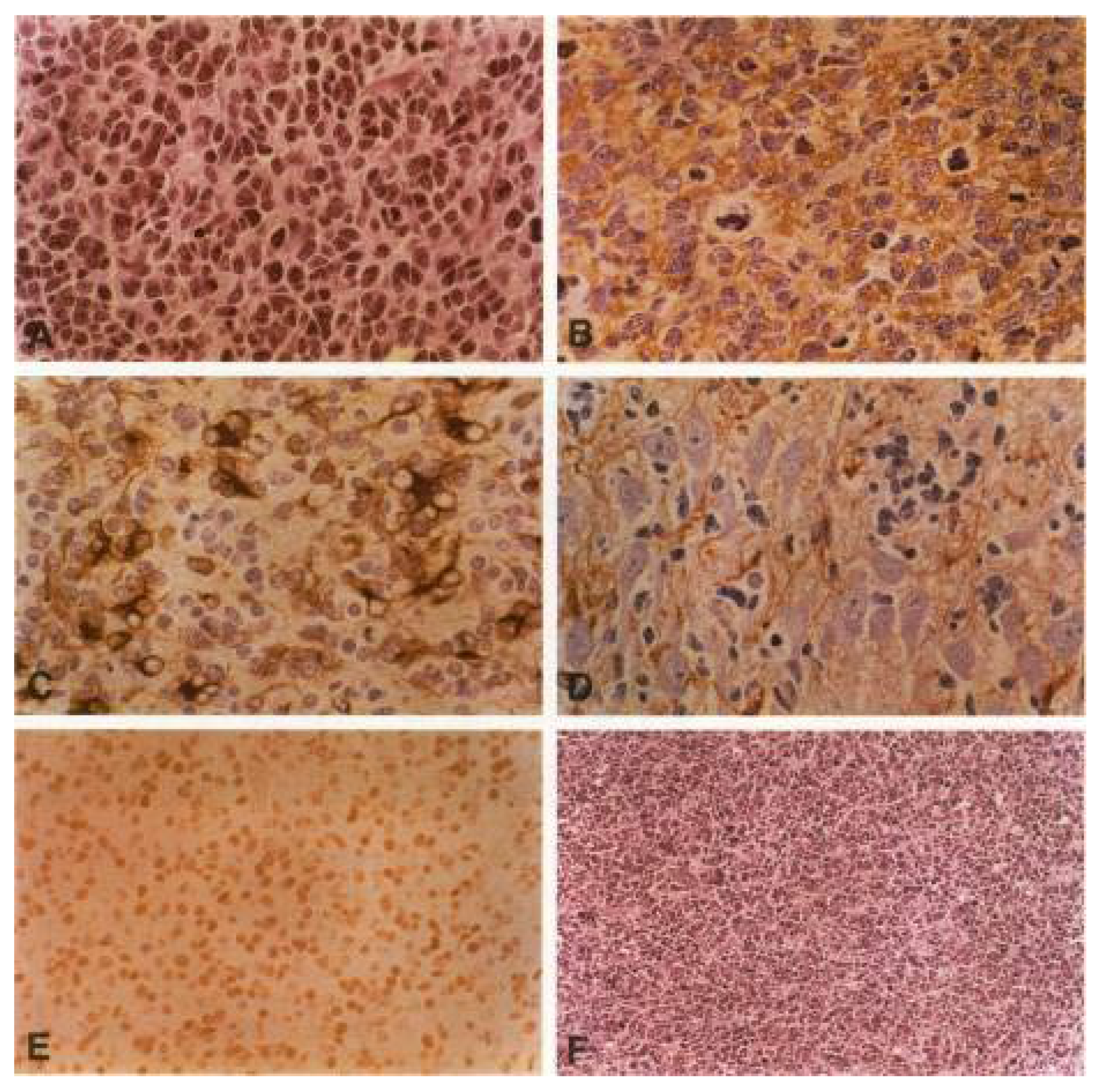
2. Diagnosis—A Century of Debates: Does Medulloblastoma Per Se Exist?
2.1. The New WHO Diagnostic Classification: Activated Oncogenic Signaling Pathways
2.2. Cell of Origin
2.3. Animal Models
2.4. First TP53 Mutations
3. Liquid Biopsy


| Year | Author | Tumor | Method | Findings |
|---|---|---|---|---|
| 2020 | Escudero et al. [65] | MB | WES, CNVs | ctDNA from CSF sufficient for diagnosis of MB-subgroups, risk stratification, and monitoring (proof of concept study) |
| 2020 | Li et al. [66] | Pediatric MB | Whole genome methylation sequencing | High specificity and sensitivity to monitor treatment response of epigenetic signatures in ctDNA from CSF, potential diagnostic, and prognostic value |
| 2021 | Liu et al. [67] | MB | WGS | ctDNA from serial CSF samples as prospective marker for MRD in half of patients before radiographic progression |
| 2021 | Sun et al. [68] | Pediatric MB | Deep sequencing/NGS, ctDNA in CSF | More alterations detectable in ctDNA from CSF than from primary tumor: a superior monitoring technique when ctDNA is detected from CSF |
| 2022 | Lee et al. [69] | MB | RT-PCR sequencing | Circular RNA circ_463 as candidate biomarker |
| 2022 | Pagès et al. [70] | Pediatric CNS tumors, incl. MB | ULP-WGS, deep sequencing of specific mutations and fusions | ctDNA is detectable better in CSF than in blood and not in urine. Molecular profiling feasible for small subset of high-grade tumors (incl. MB). Liquid biopsy remains a major challenge for such tumors with low clonal aberrations |
| 2019–2024 | NCT03936465 [71] ongoing Phase I study, 66 patients | Pediatric cancer, incl. brain tumors | ctDNA | Clinical toxicity study: ctDNA markers in blood and CSF monitoring treatment response |
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ohgaki, H.; Eibl, R.H.; Wiestler, O.D.; Yasargil, M.G.; Newcomb, E.W.; Kleihues, P. P53 Mutations in Nonastrocytic Human Brain Tumors. Cancer Res. 1991, 51, 6202–6205. [Google Scholar] [PubMed]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: A Summary. Acta Neuropathol. 2016, 131, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Louis, D.N.; Perry, A.; Wesseling, P.; Brat, D.J.; Cree, I.A.; Figarella-Branger, D.; Hawkins, C.; Ng, H.K.; Pfister, S.M.; Reifenberger, G.; et al. The 2021 WHO Classification of Tumors of the Central Nervous System: A Summary. Neuro-Oncology 2021, 23, 1231–1251. [Google Scholar] [CrossRef]
- Cotter, J.A.; Hawkins, C. Medulloblastoma: WHO 2021 and Beyond. Pediatr. Dev. Pathol. 2022, 25, 23–33. [Google Scholar] [CrossRef] [PubMed]
- Bailey, P.; Cushing, H. Medulloblastoma Cerebelli: A Common Type of Midcerebellar Glioma of the Childhood. Arch. Neurol. Psychiatry 1925, 14, 192–224. [Google Scholar] [CrossRef]
- Bailey, P.; Cushing, H. A Classification of the Tumours of the Glioma Group on a Histogenetic Basis, with a Correlated Study of Prognosis; J. B. Lippincott Company: Philadelphia, PA, USA; London, UK; Montreal, QC, Canada, 1926. [Google Scholar]
- Martin, A.M.; Raabe, E.; Eberhart, C.; Cohen, K.J. Management of Pediatric and Adult Patients with Medulloblastoma. Curr. Treat. Options Oncol. 2014, 15, 581–594. [Google Scholar] [CrossRef]
- Wright, J.H. Neurocytoma or neuroblastoma, a kind of tumor not generally recognized. J. Exp. Med. 1910, 12, 556–561. [Google Scholar] [CrossRef]
- Paterson, E.; Farr, R.F. Cerebellar Medulloblastoma: Treatment by Irradiation of the Whole Central Nervous System. Acta Radiol. 1953, 39, 323–336. [Google Scholar] [CrossRef]
- Chang, C.H.; Housepian, E.M.; Herbert, C. An Operative Staging System and a Megavoltage Radiotherapeutic Technic for Cerebellar Medulloblastomas. Radiology 1969, 93, 1351–1359. [Google Scholar] [CrossRef]
- Hart, M.N.; Earle, K.M. Primitive Neuroectodermal Tumors of the Brain in Children. Cancer 1973, 32, 890–897. [Google Scholar] [CrossRef]
- Broder, L.E.; Rall, D.P. Chemotherapy of Brain Tumors. Prog. Exp. Tumor Res. 1972, 17, 373–399. [Google Scholar] [CrossRef] [PubMed]
- Eibl, R.H.; Wiestler, O.D. Induction of Primitive Neuroectodermal Tumors Following Retrovirus-Mediated Transfer of SV40 Large T Antigen into Neural Transplants. In Proceedings of the Clinical Neuropathology; Dustri: Rockledge, FL, USA, 1991; p. 245. [Google Scholar]
- Eibl, R.H.; Kleihues, P.; Jat, P.S.; Wiestler, O.D. A Model for Primitive Neuroectodermal Tumors in Transgenic Neural Transplants Harboring the SV40 Large T Antigen. Am. J. Pathol. 1994, 144, 556–564. [Google Scholar]
- Reya, T.; Morrison, S.J.; Clarke, M.F.; Weissman, I.L. Stem Cells, Cancer, and Cancer Stem Cells. Nature 2001, 414, 105–111. [Google Scholar] [CrossRef] [PubMed]
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of Circulating Tumor DNA in Early- and Late-Stage Human Malignancies. Sci. Transl. Med. 2014, 6, 224ra24. [Google Scholar] [CrossRef]
- Garzia, L.; Kijima, N.; Morrissy, A.S.; Antonellis, P.D.; Guerreiro-Stucklin, A.; Holgado, B.L.; Wu, X.; Wang, X.; Parsons, M.; Zayne, K.; et al. A Hematogenous Route for Medulloblastoma Leptomeningeal Metastases. Cell 2018, 172, 1050–1062.e14. [Google Scholar] [CrossRef]
- Smith, K.S.; Bihannic, L.; Gudenas, B.L.; Haldipur, P.; Tao, R.; Gao, Q.; Li, Y.; Aldinger, K.A.; Iskusnykh, I.Y.; Chizhikov, V.V.; et al. Unified Rhombic Lip Origins of Group 3 and Group 4 Medulloblastoma. Nature 2022, 609, 1012–1020. [Google Scholar] [CrossRef]
- Hendrikse, L.D.; Haldipur, P.; Saulnier, O.; Millman, J.; Sjoboen, A.H.; Erickson, A.W.; Ong, W.; Gordon, V.; Coudière-Morrison, L.; Mercier, A.L.; et al. Failure of Human Rhombic Lip Differentiation Underlies Medulloblastoma Formation. Nature 2022, 609, 1021–1028. [Google Scholar] [CrossRef]
- Ostrom, Q.T.; Cioffi, G.; Waite, K.; Kruchko, C.; Barnholtz-Sloan, J.S. CBTRUS Statistical Report: Primary Brain and Other Central Nervous System Tumors Diagnosed in the United States in 2014–2018. Neuro-Oncol. 2021, 23, iii1–iii105. [Google Scholar] [CrossRef]
- Siegel, R.L.; Miller, K.D.; Wagle, N.S.; Jemal, A. Cancer Statistics, 2023. CA. Cancer J. Clin. 2023, 73, 17–48. [Google Scholar] [CrossRef] [PubMed]
- Eibl, R.H. From Leukocyte Trafficking to Tumor Cell Metastasis—A New Approach for the Development of Antimetastatic Drugs. Biochem. Soc. Trans. 2000, 28, A350. [Google Scholar] [CrossRef]
- Eibl, R.H.; Höfler, H. The Multistep Homing Model of Tumor Metastasis: From Cell Adhesion Molecules to Chemokine Receptors. Scand. J. Immunol. 2001, 54, 50. [Google Scholar] [CrossRef]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy for Monitoring Medulloblastoma. Extracell. Vesicles Circ. Nucleic Acids 2022, 3, 263–274. [Google Scholar] [CrossRef]
- Gibson, P.; Tong, Y.; Robinson, G.; Thompson, M.C.; Currle, D.S.; Eden, C.; Kranenburg, T.A.; Hogg, T.; Poppleton, H.; Martin, J.; et al. Subtypes of Medulloblastoma Have Distinct Developmental Origins. Nature 2010, 468, 1095–1099. [Google Scholar] [CrossRef]
- Zhukova, N.; Ramaswamy, V.; Remke, M.; Pfaff, E.; Shih, D.J.H.; Martin, D.C.; Castelo-Branco, P.; Baskin, B.; Ray, P.N.; Bouffet, E.; et al. Subgroup-Specific Prognostic Implications of TP53 Mutation in Medulloblastoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 2927–2935. [Google Scholar] [CrossRef]
- Biegel, J.A.; Fogelgren, B.; Zhou, J.Y.; James, C.D.; Janss, A.J.; Allen, J.C.; Zagzag, D.; Raffel, C.; Rorke, L.B. Mutations of the INI1 Rhabdoid Tumor Suppressor Gene in Medulloblastomas and Primitive Neuroectodermal Tumors of the Central Nervous System. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2000, 6, 2759–2763. [Google Scholar]
- Rorke, L.B. Experimental Production of Primitive Neuroectodermal Tumors and Its Relevance to Human Neuro-Oncology. Am. J. Pathol. 1994, 144, 444–448. [Google Scholar]
- Brinster, R.L.; Chen, H.Y.; Messing, A.; van Dyke, T.; Levine, A.J.; Palmiter, R.D. Transgenic Mice Harboring SV40 T-Antigen Genes Develop Characteristic Brain Tumors. Cell 1984, 37, 367–379. [Google Scholar] [CrossRef]
- Fung, K.M.; Chikaraishi, D.M.; Suri, C.; Theuring, F.; Messing, A.; Albert, D.M.; Lee, V.M.; Trojanowski, J.Q. Molecular Phenotype of Simian Virus 40 Large T Antigen-Induced Primitive Neuroectodermal Tumors in Four Different Lines of Transgenic Mice. Lab. Investig. J. Tech. Methods Pathol. 1994, 70, 114–124. [Google Scholar]
- Fung, K.M.; Trojanowski, J.Q. Animal Models of Medulloblastomas and Related Primitive Neuroectodermal Tumors. A Review. J. Neuropathol. Exp. Neurol. 1995, 54, 285–296. [Google Scholar] [CrossRef]
- Marcus, D.M.; Carpenter, J.L.; O’Brien, J.M.; Kivela, T.; Brauner, E.; Tarkkanen, A.; Virtanen, I.; Albert, D.M. Primitive Neuroectodermal Tumor of the Midbrain in a Murine Model of Retinoblastoma. Invest. Ophthalmol. Vis. Sci. 1991, 32, 293–301. [Google Scholar]
- Kivelä, T.; Virtanen, I.; Marcus, D.M.; O’Brien, J.M.; Carpenter, J.L.; Brauner, E.; Tarkkanen, A.; Albert, D.M. Neuronal and Glial Properties of a Murine Transgenic Retinoblastoma Model. Am. J. Pathol. 1991, 138, 1135–1148. [Google Scholar] [PubMed]
- Korf, H.-W.; Götz, W.; Herken, R.; Theuring, F.; Gruss, P.; Schachenmayr, W. S-Antigen and Rod-Opsin Immunoreactions in Midline Brain Neoplasms of Transgenic Mice: Similarities to Pineal Cell Tumors and Certain Medulloblastomas in Man. J. Neuropathol. Exp. Neurol. 1990, 49, 424–437. [Google Scholar] [CrossRef] [PubMed]
- Brüstle, O.; Jones, K.N.; Learish, R.D.; Karram, K.; Choudhary, K.; Wiestler, O.D.; Duncan, I.D.; McKay, R.D. Embryonic Stem Cell-Derived Glial Precursors: A Source of Myelinating Transplants. Science 1999, 285, 754–756. [Google Scholar] [CrossRef] [PubMed]
- Carroll, R.B.; Gurney, E.G. Time-Dependent Maturation of the Simian Virus 40 Large T Antigen-P53 Complex Studied by Using Monoclonal Antibodies. J. Virol. 1982, 44, 565–573. [Google Scholar] [CrossRef]
- Wiestler, O.D.; Aguzzi, A.; Schneemann, M.; Eibl, R.; von Deimling, A.; Kleihues, P. Oncogene Complementation in Fetal Brain Transplants. Cancer Res. 1992, 52, 3760–3767. [Google Scholar]
- Wiestler, O.D.; Brüstle, O.; Eibl, R.H.; Radner, H.; Aguzzi, A.; Kleihues, P. Retrovirus-Mediated Oncogene Transfer into Neural Transplants. Brain Pathol. Zurich Switz. 1992, 2, 47–59. [Google Scholar]
- Wiestler, O.D.; Aguzzi, A.; Brüstle, O.; Eibl, R.; Radner, H.; Kleihues, P. Oncogene Complementation in Transgenic Neural Transplants. Neuropathology 1991, 4, 304–309. [Google Scholar]
- Radner, H.; el-Shabrawi, Y.; Eibl, R.H.; Brüstle, O.; Kenner, L.; Kleihues, P.; Wiestler, O.D. Tumor Induction by Ras and Myc Oncogenes in Fetal and Neonatal Brain: Modulating Effects of Developmental Stage and Retroviral Dose. Acta Neuropathol. 1993, 86, 456–465. [Google Scholar] [CrossRef]
- Wiestler, O.D.; Brüstle, O.; Eibl, R.H.; Radner, H.; Aguzzi, A.; Kleihues, P. Oncogene Transfer into the Brain. Recent Results Cancer Res. Fortschr. Krebsforsch. Prog. Dans Rech. Sur Cancer 1994, 135, 55–66. [Google Scholar] [CrossRef]
- Saylors, R.L.; Sidransky, D.; Friedman, H.S.; Bigner, S.H.; Bigner, D.D.; Vogelstein, B.; Brodeur, G.M. Infrequent P53 Gene Mutations in Medulloblastomas. Cancer Res. 1991, 51, 4721–4723. [Google Scholar]
- Ohgaki, H.; Eibl, R.H.; Schwab, M.; Reichel, M.B.; Mariani, L.; Gehring, M.; Petersen, I.; Höll, T.; Wiestler, O.D.; Kleihues, P. Mutations of the P53 Tumor Suppressor Gene in Neoplasms of the Human Nervous System. Mol. Carcinog. 1993, 8, 74–80. [Google Scholar] [CrossRef] [PubMed]
- von Deimling, A.; Eibl, R.H.; Ohgaki, H.; Louis, D.N.; von Ammon, K.; Petersen, I.; Kleihues, P.; Chung, R.Y.; Wiestler, O.D.; Seizinger, B.R. P53 Mutations Are Associated with 17p Allelic Loss in Grade II and Grade III Astrocytoma. Cancer Res. 1992, 52, 2987–2990. [Google Scholar] [PubMed]
- Romero, D. Tracking Cancer in Liquid Biopsies. Available online: https://www.nature.com/articles/d42859-020-00070-z (accessed on 3 March 2022).
- Eibl, R.H.; Schneemann, M. Cell-Free DNA as a Biomarker in Cancer. Extracell. Vesicles Circ. Nucleic Acids 2022, 3, 178–198. [Google Scholar] [CrossRef]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy and Primary Brain Tumors. Cancers 2021, 13, 5429. [Google Scholar] [CrossRef] [PubMed]
- Eibl, R.H.; Schneemann, M. Liquid Biopsy and Glioblastoma. Explor. Target. Anti-Tumor Ther. 2023, 4. [Google Scholar]
- Eibl, R.H.; Pietsch, T.; Moll, J.; Skroch-Angel, P.; Heider, K.H.; von Ammon, K.; Wiestler, O.D.; Ponta, H.; Kleihues, P.; Herrlich, P. Expression of Variant CD44 Epitopes in Human Astrocytic Brain Tumors. J. Neurooncol. 1995, 26, 165–170. [Google Scholar] [CrossRef]
- Eibl, R.H.; Benoit, M. Molecular Resolution of Cell Adhesion Forces. IEE Proc. Nanobiotechnol. 2004, 151, 128–132. [Google Scholar] [CrossRef]
- Eibl, R.H.; Moy, V.T. AFM-Based Adhesion Measurements of Single Receptor-Ligand Bonds on Living Cells. In Recent Research Developments in Biophysics; Pandalai, S.G., Ed.; Transworld Research Network: Trivandrum, India, 2004; pp. 235–246. ISBN 81-7895-130-4. [Google Scholar]
- Eibl, R.H.; Moy, V.T. Atomic Force Microscopy Measurements of Protein-Ligand Interactions on Living Cells. Methods Mol. Biol. Clifton NJ 2005, 305, 439–450. [Google Scholar] [CrossRef]
- Eibl, R.H. Direct Force Measurements of Receptor–Ligand Interactions on Living Cells. In Applied Scanning Probe Methods XII: Characterization; Bhushan, B., Fuchs, H., Eds.; NanoScience and Technology; Springer: Berlin/Heidelberg, Germany, 2009; pp. 1–31. ISBN 978-3-540-85039-7. [Google Scholar]
- Eibl, R.H. Cell Adhesion Receptors Studied by AFM-Based Single-Molecule Force Spectroscopy. In Scanning Probe Microscopy in Nanoscience and Nanotechnology 2; Bhushan, B., Ed.; NanoScience and Technology; Springer: Berlin/Heidelberg, Germany, 2011; pp. 197–215. ISBN 978-3-642-10497-8. [Google Scholar]
- Eibl, R.H. Single-Molecule Studies of Integrins by AFM-Based Force Spectroscopy on Living Cells. In Scanning Probe Microscopy in Nanoscience and Nanotechnology 3; Bhushan, B., Ed.; NanoScience and Technology; Springer: Berlin/Heidelberg, Germany, 2013; pp. 137–169. ISBN 978-3-642-25414-7. [Google Scholar]
- Eibl, R.H. Comment on “A Method to Measure Cellular Adhesion Utilizing a Polymer Micro-Cantilever” [Appl. Phys. Lett. 103, 123702 (2013)]. Appl. Phys. Lett. 2014, 104, 236103. [Google Scholar] [CrossRef]
- Di Santo, R.; Romanò, S.; Mazzini, A.; Jovanović, S.; Nocca, G.; Campi, G.; Papi, M.; De Spirito, M.; Di Giacinto, F.; Ciasca, G. Recent Advances in the Label-Free Characterization of Exosomes for Cancer Liquid Biopsy: From Scattering and Spectroscopy to Nanoindentation and Nanodevices. Nanomaterials 2021, 11, 1476. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, T.; Huang, J.; Dong, H.; Xie, J.; Jia, L. Biostable Double-Strand Circular Aptamers Conjugated Onto Dendrimers for Specific Capture and Inhibition of Circulating Leukemia Cells. OncoTargets Ther. 2021, Volume 13, 13465–13477. [Google Scholar] [CrossRef]
- Deliorman, M.; Glia, A.; Qasaimeh, M.A. Characterizing Circulating Tumor Cells Using Affinity-Based Microfluidic Capture and AFM-Based Biomechanics. STAR Protoc. 2022, 3, 101433. [Google Scholar] [CrossRef]
- Chowdhury, T.; Cressiot, B.; Parisi, C.; Smolyakov, G.; Thiébot, B.; Trichet, L.; Fernandes, F.M.; Pelta, J.; Manivet, P. Circulating Tumor Cells in Cancer Diagnostics and Prognostics by Single-Molecule and Single-Cell Characterization. ACS Sens. 2023. [Google Scholar] [CrossRef]
- Pax, M.; Rieger, J.; Eibl, R.H.; Thielemann, C.; Johannsmann, D. Measurements of Fast Fluctuations of Viscoelastic Properties with the Quartz Crystal Microbalance. The Analyst 2005, 130, 1474–1477. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Beltran, A.N.; Masuzzo, P.; Ampe, C.; Bakker, G.-J.; Besson, S.; Eibl, R.H.; Friedl, P.; Gunzer, M.; Kittisopikul, M.; Dévédec, S.E.L.; et al. Community Standards for Open Cell Migration Data. GigaScience 2020, 9. [Google Scholar] [CrossRef] [PubMed]
- SMART. Available online: https://smart.servier.com/ (accessed on 31 October 2022).
- Creative Commons — Attribution 3.0 Unported — CC BY 3.0. Available online: https://creativecommons.org/licenses/by/3.0/ (accessed on 31 October 2022).
- Escudero, L.; Llort, A.; Arias, A.; Diaz-Navarro, A.; Martínez-Ricarte, F.; Rubio-Perez, C.; Mayor, R.; Caratù, G.; Martínez-Sáez, E.; Vázquez-Méndez, É.; et al. Circulating Tumour DNA from the Cerebrospinal Fluid Allows the Characterisation and Monitoring of Medulloblastoma. Nat. Commun. 2020, 11, 5376. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhao, S.; Lee, M.; Yin, Y.; Li, J.; Zhou, Y.; Ballester, L.Y.; Esquenazi, Y.; Dashwood, R.H.; Davies, P.J.A.; et al. Reliable Tumor Detection by Whole-Genome Methylation Sequencing of Cell-Free DNA in Cerebrospinal Fluid of Pediatric Medulloblastoma. Sci. Adv. 2020, 6, eabb5427. [Google Scholar] [CrossRef]
- Liu, A.P.Y.; Smith, K.S.; Kumar, R.; Paul, L.; Bihannic, L.; Lin, T.; Maass, K.K.; Pajtler, K.W.; Chintagumpala, M.; Su, J.M.; et al. Serial Assessment of Measurable Residual Disease in Medulloblastoma Liquid Biopsies. Cancer Cell 2021, 39, 1519–1530.e4. [Google Scholar] [CrossRef]
- Sun, Y.; Li, M.; Ren, S.; Liu, Y.; Zhang, J.; Li, S.; Gao, W.; Gong, X.; Liu, J.; Wang, Y.; et al. Exploring Genetic Alterations in Circulating Tumor DNA from Cerebrospinal Fluid of Pediatric Medulloblastoma. Sci. Rep. 2021, 11, 5638. [Google Scholar] [CrossRef]
- Lee, B.; Mahmud, I.; Pokhrel, R.; Murad, R.; Yuan, M.; Stapleton, S.; Bettegowda, C.; Jallo, G.; Eberhart, C.G.; Garrett, T.; et al. Medulloblastoma Cerebrospinal Fluid Reveals Metabolites and Lipids Indicative of Hypoxia and Cancer-Specific RNAs. Acta Neuropathol. Commun. 2022, 10, 25. [Google Scholar] [CrossRef]
- Pagès, M.; Rotem, D.; Gydush, G.; Reed, S.; Rhoades, J.; Ha, G.; Lo, C.; Fleharty, M.; Duran, M.; Jones, R.; et al. Liquid Biopsy Detection of Genomic Alterations in Pediatric Brain Tumors from Cell-Free DNA in Peripheral Blood, CSF, and Urine. Neuro-Oncology 2022, 24, noab299. [Google Scholar] [CrossRef] [PubMed]
- DuBois, S. Phase 1 Study of the Bromodomain (BRD) and Extra-Terminal Domain (BET) Inhibitors BMS-986158 and BMS-986378 (CC-90010) in Pediatric Cancer. Available online: https://clinicaltrials.gov/ct2/show/NCT03936465 (accessed on 14 November 2022).


| Medulloblastoma, Molecularly Defined | Pathway |
|---|---|
| Group 1 | WNT-activated |
| Group 2 | SHH-activated and TP53-wildtype |
| SHH-activated and TP53-mutant | |
| Group 3 | (non-WNT/non-SHH) |
| Group 4 | (non-WNT/non-SHH) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Eibl, R.H.; Schneemann, M. Medulloblastoma: From TP53 Mutations to Molecular Classification and Liquid Biopsy. Biology 2023, 12, 267. https://doi.org/10.3390/biology12020267
Eibl RH, Schneemann M. Medulloblastoma: From TP53 Mutations to Molecular Classification and Liquid Biopsy. Biology. 2023; 12(2):267. https://doi.org/10.3390/biology12020267
Chicago/Turabian StyleEibl, Robert H., and Markus Schneemann. 2023. "Medulloblastoma: From TP53 Mutations to Molecular Classification and Liquid Biopsy" Biology 12, no. 2: 267. https://doi.org/10.3390/biology12020267
APA StyleEibl, R. H., & Schneemann, M. (2023). Medulloblastoma: From TP53 Mutations to Molecular Classification and Liquid Biopsy. Biology, 12(2), 267. https://doi.org/10.3390/biology12020267





