Single-Cell Labeling Strategies to Dissect Neuronal Structures and Local Functions
Abstract
:Simple Summary
Abstract
1. Introduction
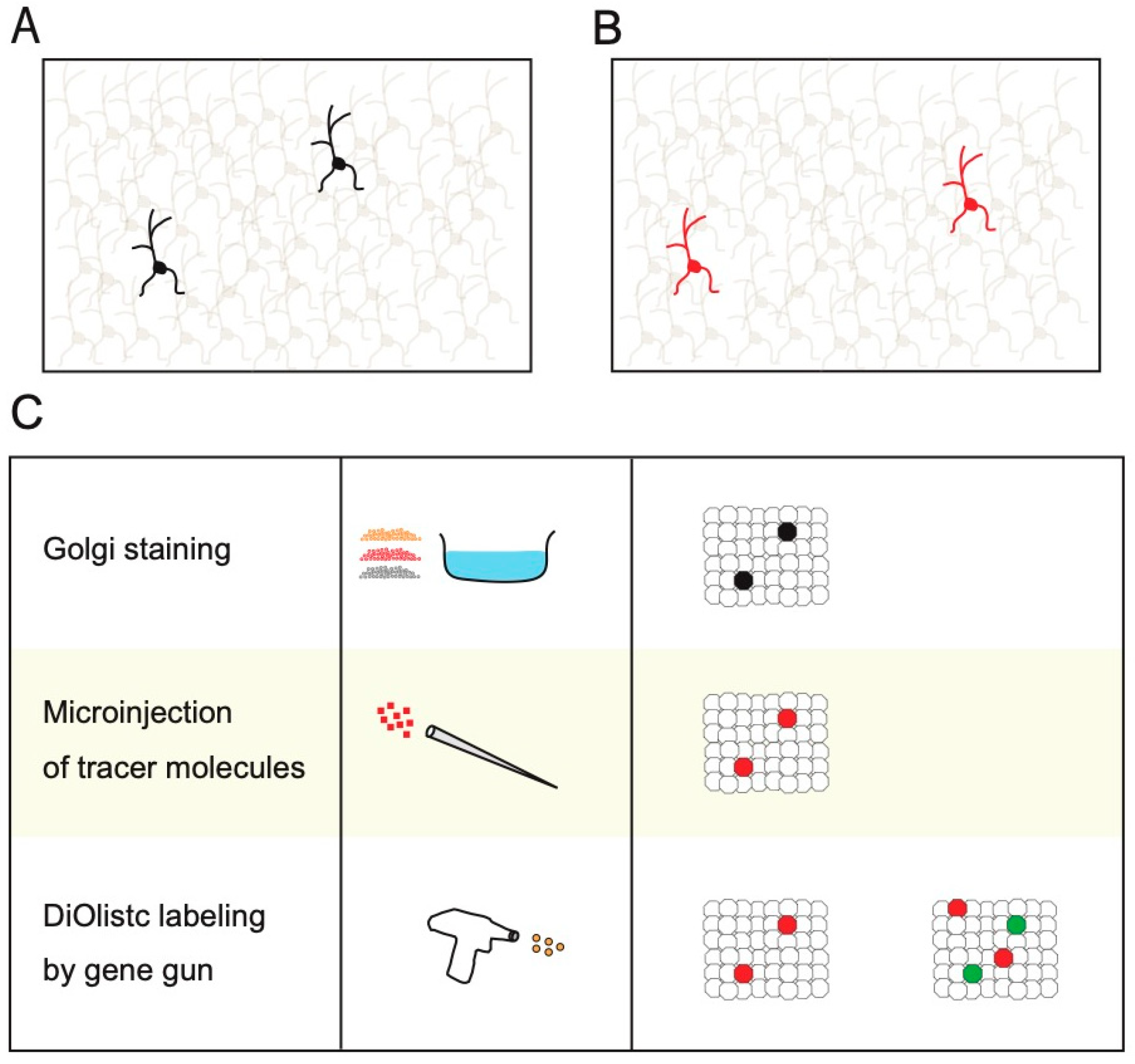
2. Non-Transgenic Methods for Sparse Single-Cell Labeling
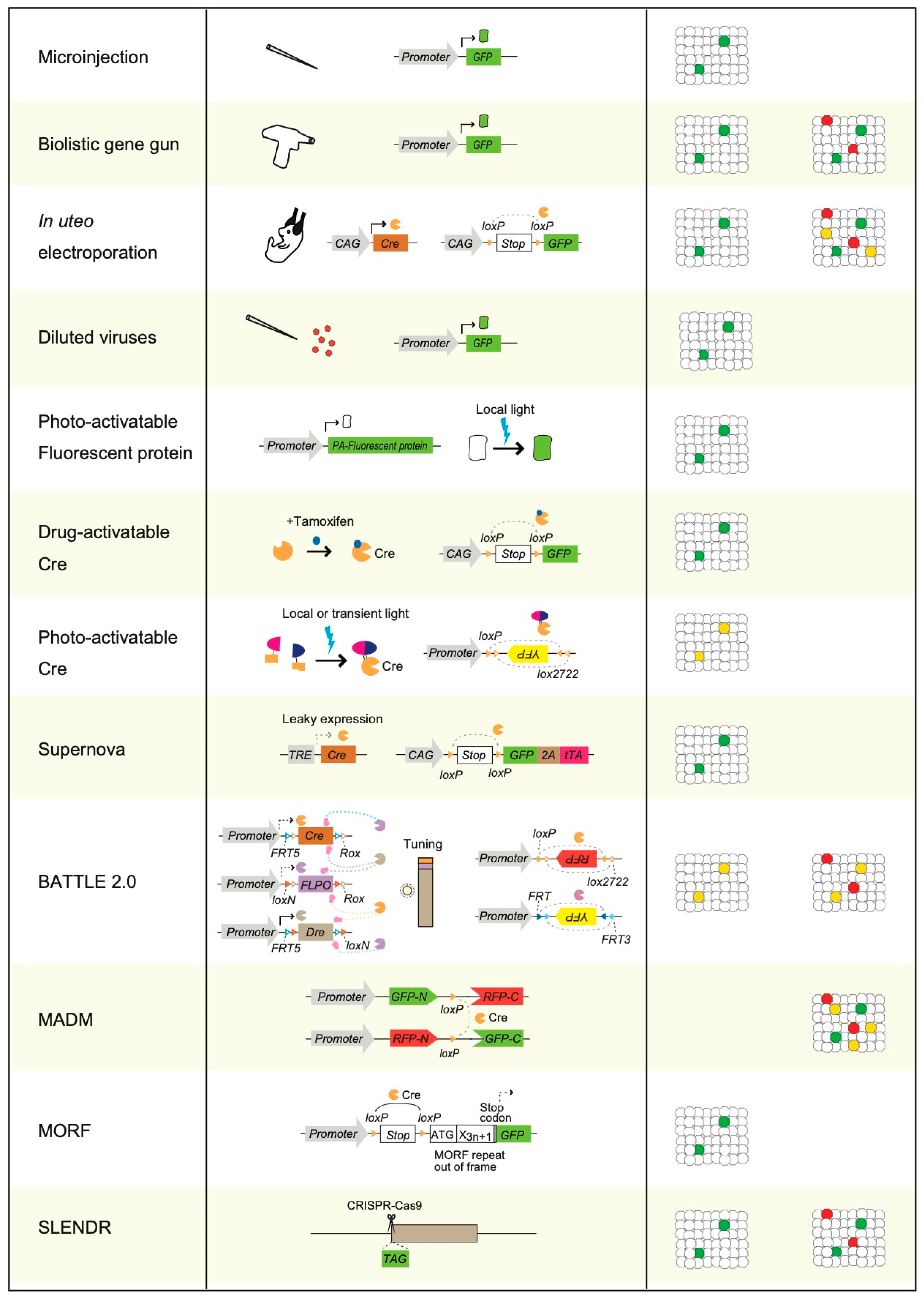
3. Physical Transgenic Methods for Sparse Single-Cell Labeling
4. Sparse Single-Cell Labeling Using Electroporation Methods and by Injection of Diluted Viruses
5. Photoactivatable Fluorescent Proteins Mediated Sparse Single-Cell Labeling
6. Photoactivatable and Drug-Activatable Cre Recombinase Mediated Sparse Single-Cell Labeling
7. A Leaky Expression-Dependent Sparse Single-Cell Labeling Method: Supernova
8. A multicolor Sparse Single-Cell Labeling Method: BATTLE 2.0
9. MADM, MORF, and SPARC
10. SLENDR
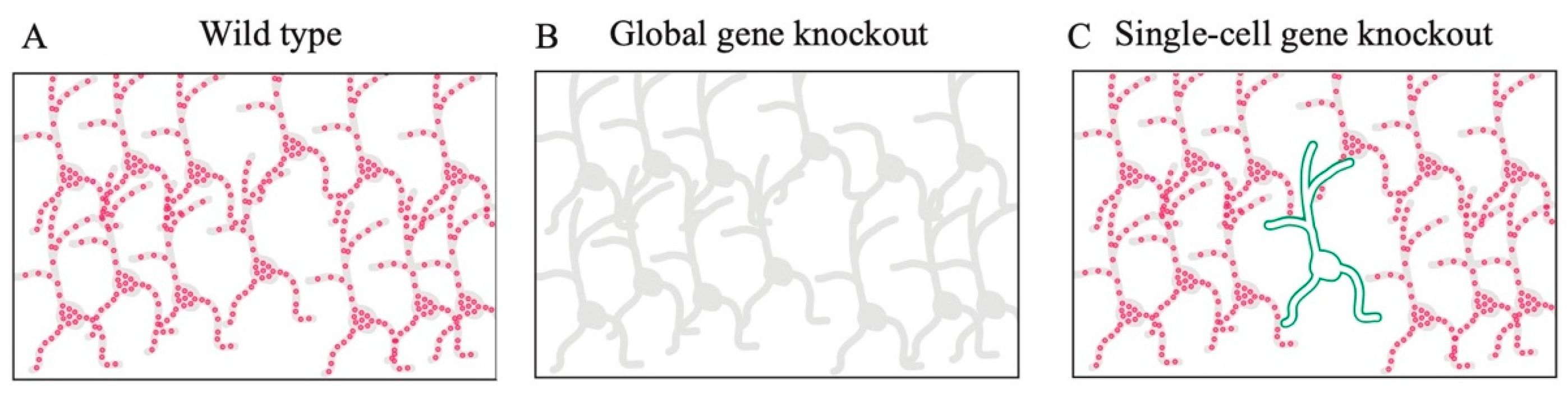
11. Functional Analysis Using Single-Cell Gene Knockout
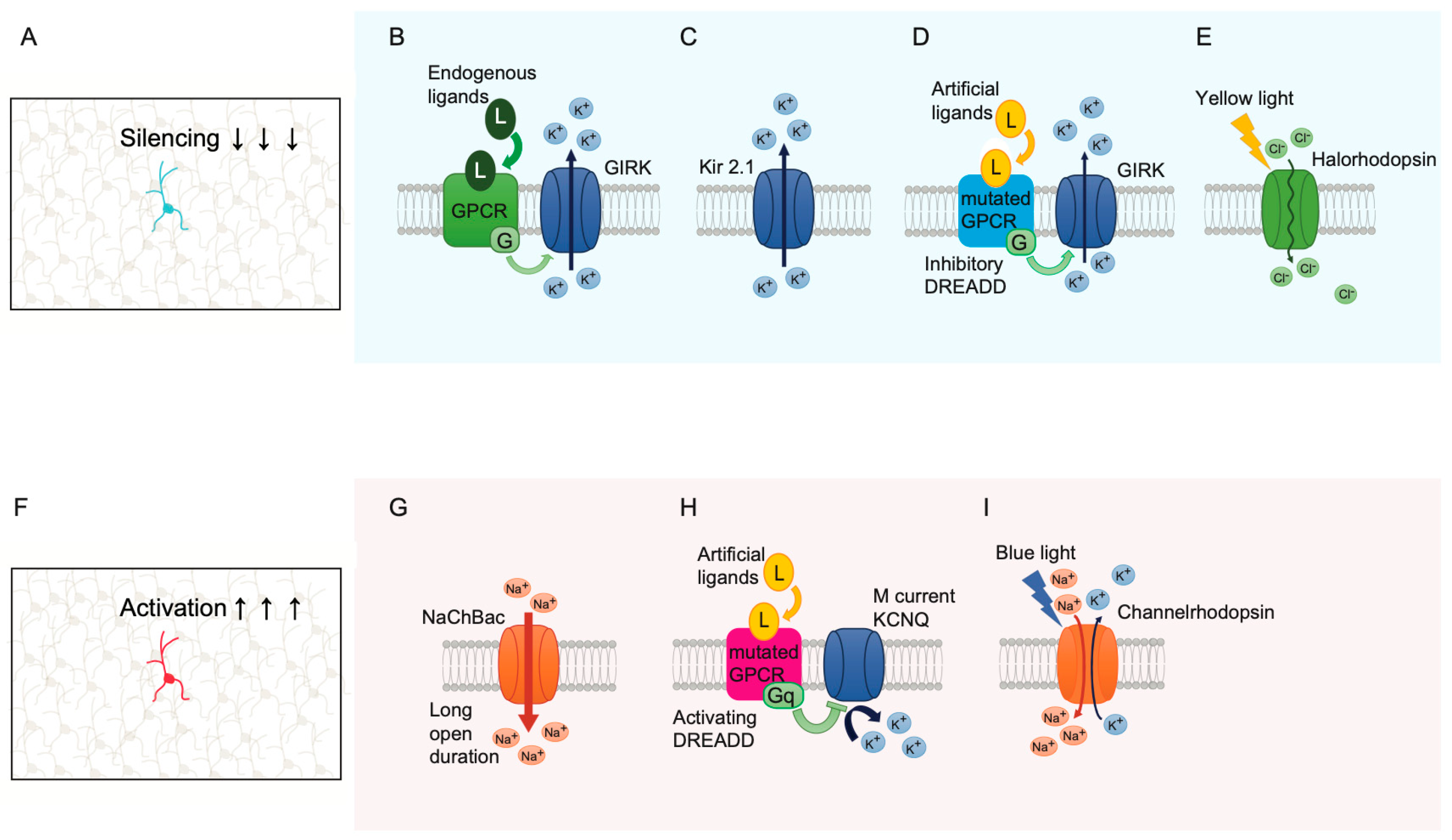
12. Single-Cell Silencing and Activation
13. Discussion
14. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bentivoglio, M.; Cotrufo, T.; Ferrari, S.; Tesoriero, C.; Mariotto, S.; Bertini, G.; Berzero, A.; Mazzarello, P. The Original Histological Slides of Camillo Golgi and His Discoveries on Neuronal Structure. Front. Neuroanat. 2019, 13, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Glickstein, M. Golgi and Cajal: The Neuron Doctrine and the 100th Anniversary of the 1906 Nobel Prize. Curr. Biol. 2006, 16, R147–R151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zaqout, S.; Kaindl, A.M. Golgi-Cox Staining Step by Step. Front. Neuroanat. 2016, 10, 38. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morange, M. History of Molecular Biology; Harvard University Press: Cambridge, MA, USA, 2000. [Google Scholar]
- Jaenisch, R.; Mintz, B. Simian Virus 40 DNA Sequences in DNA of Healthy Adult Mice Derived from Preimplantation Blastocysts Injected with Viral DNA. Proc. Natl. Acad. Sci. USA 1974, 71, 1250–1254. [Google Scholar] [CrossRef] [Green Version]
- Capecchi, M.R. High efficiency transformation by direct microinjection of DNA into cultured mammalian cells. Cell 1980, 22, 479–488. [Google Scholar] [CrossRef]
- Shimomura, O. Structure of the chromophore of Aequorea green fluorescent protein. FEBS Lett. 1979, 104, 220–222. [Google Scholar] [CrossRef] [Green Version]
- Tsien, R.Y. The green fluorescent protein. Annu. Rev. Biochem. 1998, 67, 509–544. [Google Scholar] [CrossRef]
- Alberts, B. Molecular Biology of the Cell; W.W. Norton and Company: New York, NY, USA, 2022. [Google Scholar]
- Weissman, T.A.; Pan, Y.A. Brainbow: New Resources and Emerging Biological Applications for Multicolor Genetic Labeling and Analysis. Genetics 2015, 199, 293–306. [Google Scholar] [CrossRef] [Green Version]
- Tashiro, A.; Zhao, C.; Gage, F.H. Retrovirus-mediated single-cell gene knockout technique in adult newborn neurons in vivo. Nat. Protoc. 2006, 1, 3049–3055. [Google Scholar] [CrossRef]
- Kohara, K.; Yasuda, H.; Huang, Y.; Adachi, N.; Sohya, K.; Tsumoto, T. A Local Reduction in Cortical GABAergic Synapses after a Loss of Endogenous Brain-Derived Neurotrophic Factor, as Revealed by Single-Cell Gene Knock-Out Method. J. Neurosci. 2007, 27, 7234–7244. [Google Scholar] [CrossRef] [Green Version]
- Martini, F.J.; Guillamón-Vivancos, T.; Moreno-Juan, V.; Valdeolmillos, M.; López-Bendito, G. Spontaneous Activity in Developing Thalamic and Cortical Sensory Networks. Neuron 2021, 109, 2519–2534. [Google Scholar] [CrossRef]
- Tamamaki, N.; Abe, K.; Nojyo, Y. Three-dimensional analysis of the whole axonal arbors originating from single CA2 pyramidal neurons in the rat hippocampus with the aid of a computer graphic technique. Brain Res. 1988, 452, 255–272. [Google Scholar] [CrossRef]
- Horikawa, K.; Armstrong, W.E. A Versatile Means of Intracellular Labeling: Injection of Biocytin and Its Detection with Avidin Conjugates. J. Neurosci. Methods 1988, 25, 1–11. [Google Scholar] [CrossRef]
- Rockland, K.S. Visual Cortical Organization at the Single Axon Level: A Beginning. Neurosci. Res. 2002, 42, 155–166. [Google Scholar] [CrossRef]
- Kohara, K.; Kitamura, A.; Adachi, N.; Nishida, M.; Itami, C.; Nakamura, S.; Tsumoto, T. Inhibitory but Not Excitatory Cortical Neurons Require Presynaptic Brain-Derived Neurotrophic Factor for Dendritic Development, as Revealed by Chimera Cell Culture. J. Neurosci. 2003, 23, 6123–6131. [Google Scholar] [CrossRef] [Green Version]
- Hanani, M. Lucifer Yellow—An Angel Rather than the Devil. J. Cell. Mol. Med. 2011, 16, 22–31. [Google Scholar] [CrossRef] [Green Version]
- Kitai, S.T.; Kocsis, J.D.; Preston, R.J.; Sugimori, M. Monosynaptic Inputs to Caudate Neurons Identified by Intracellular Injection of Horseradish Peroxidase. Brain Res. 1976, 109, 601–606. [Google Scholar] [CrossRef]
- Cullheim, S.; Kellerth, J.-O. Combined Light and Electron Microscopic Tracing of Neurons, Including Axons and Synaptic Terminals, after Intracellular Injection of Horseradish Peroxidase. Neurosci. Lett. 1976, 2, 307–313. [Google Scholar] [CrossRef]
- Snow, P.J.; Rose, P.K.; Brown, A.G. Tracing Axons and Axon Collaterals of Spinal Neurons Using Intracellular Injection of Horseradish Peroxidase. Science 1976, 191, 312–313. [Google Scholar] [CrossRef] [Green Version]
- Klausberger, T.; Magill, P.J.; Márton, L.F.; Roberts, J.D.; Cobden, P.M.; Buzsáki, G.; Somogyi, P. Brain-State- and Cell-Type-Specific Firing of Hippocampal Interneurons in Vivo. Nature 2003, 421, 844–848. [Google Scholar] [CrossRef]
- Klausberger, T.; Márton, L.F.; Baude, A.; Roberts, J.D.; Magill, P.J.; Somogyi, P. Spike Timing of Dendrite-Targeting Bistratified Cells during Hippocampal Network Oscillations In Vivo. Nat. Neurosci. 2003, 7, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Klausberger, T. Complementary Roles of Cholecystokinin- and Parvalbumin-Expressing GABAergic Neurons in Hippocampal Network Oscillations. J. Neurosci. 2005, 25, 9782–9793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Somogyi, P.; Klausberger, T. Defined Types of Cortical Interneurone Structure Space and Spike Timing in the Hippocampus. J. Physiol. 2004, 562, 9–26. [Google Scholar] [CrossRef] [PubMed]
- Pinault, D. The Juxtacellular Recording-Labeling Technique. Electrophysiol. Rec. Tech. 2010, 54, 41–75. [Google Scholar]
- Gan, W.-B.; Grutzendler, J.; Wong, W.T.; Wong, R.O.L.; Lichtman, J.W. Multicolor “Diolistic” Labeling of the Nervous System Using Lipophilic Dye Combinations. Neuron 2000, 27, 219–225. [Google Scholar] [CrossRef] [Green Version]
- McAllister, A.K. Biolistic Transfection of Neurons. Sci. STKE 2000, 2000, pl1. [Google Scholar] [CrossRef]
- Kohara, K.; Kitamura, A.; Morishima, M.; Tsumoto, T. Activity-Dependent Transfer of Brain-Derived Neurotrophic Factor to Postsynaptic Neurons. Science 2001, 291, 2419–2423. [Google Scholar] [CrossRef] [Green Version]
- Adachi, N.; Kohara, K.; Tsumoto, T. Difference in Trafficking of Brain-Derived Neurotrophic Factor between Axons and Dendrites of Cortical Neurons, Revealed by Live-Cell Imaging. BMC Neurosci. 2005, 6, 42. [Google Scholar] [CrossRef] [Green Version]
- Lappe-Siefke, C.; Maas, C.; Kneussel, M. Microinjection into Cultured Hippocampal Neurons: A Straightforward Approach for Controlled Cellular Delivery of Nucleic Acids, Peptides and Antibodies. J. Neurosci. Methods 2008, 175, 88–95. [Google Scholar] [CrossRef]
- Tabata, H.; Nakajima, K. Efficient in Utero Gene Transfer System to the Developing Mouse Brain Using Electroporation: Visualization of Neuronal Migration in the Developing Cortex. Neuroscience 2001, 103, 865–872. [Google Scholar] [CrossRef]
- Fukuchi-Shimogori, T.; Grove, E.A. Neocortex Patterning by the Secreted Signaling Molecule FGF8. Science 2001, 294, 1071–1074. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, T.; Cepko, C.L. Controlled Expression of Transgenes Introduced by in Vivo Electroporation. Proc. Natl. Acad. Sci. USA 2007, 104, 1027–1032. [Google Scholar] [CrossRef] [Green Version]
- Luo, W.; Mizuno, H.; Iwata, R.; Nakazawa, S.; Yasuda, K.; Itohara, S.; Iwasato, T. Supernova: A Versatile Vector System for Single-Cell Labeling and Gene Function Studies in vivo. Sci. Rep. 2016, 6, 35747. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, H.; Hirano, T.; Tagawa, Y. Evidence for Activity-Dependent Cortical Wiring: Formation of Interhemispheric Connections in Neonatal Mouse Visual Cortex Requires Projection Neuron Activity. J. Neurosci. 2007, 27, 6760–6770. [Google Scholar] [CrossRef]
- Takeo, Y.H.; Kakegawa, W.; Miura, E.; Yuzaki, M. ROR Regulates Multiple Aspects of Dendrite Development in Cerebellar Purkinje Cells in Vivo. J. Neurosci. 2015, 35, 12518–12534. [Google Scholar] [CrossRef] [Green Version]
- Shinmyo, Y.; Kawasaki, H. CRISPR/Cas9-Mediated Gene Knockout in the Mouse Brain Using in Utero Electroporation. Curr. Protoc. Neurosci. 2017, 79, 3.32.1–3.32.11. [Google Scholar] [CrossRef] [Green Version]
- Ako, R.; Wakimoto, M.; Ebisu, H.; Tanno, K.; Hira, R.; Kasai, H.; Matsuzaki, M.; Kawasaki, H. Simultaneous Visualization of Multiple Neuronal Properties with Single-Cell Resolution in the Living Rodent Brain. Mol. Cell. Neurosci. 2011, 48, 246–257. [Google Scholar] [CrossRef]
- Dhande, O.S.; Hua, E.W.; Guh, E.; Yeh, J.; Bhatt, S.; Zhang, Y.; Ruthazer, E.S.; Feller, M.B.; Crair, M.C. Development of Single Retinofugal Axon Arbors in Normal and β2 Knock-out Mice. J. Neurosci. 2011, 31, 3384–3399. [Google Scholar] [CrossRef] [Green Version]
- Haas, K.; Sin, W.-C.; Javaherian, A.; Li, Z.; Cline, H.T. Single-Cell Electroporationfor Gene Transfer in Vivo. Neuron 2001, 29, 583–591. [Google Scholar] [CrossRef] [Green Version]
- Oyama, K.; Ohara, S.; Sato, S.; Karube, F.; Fujiyama, F.; Isomura, Y.; Mushiake, H.; Iijima, T.; Tsutsui, K.-I. Long-Lasting Single-Neuron Labeling by in Vivo Electroporation without Microscopic Guidance. J. Neurosci. Methods 2013, 218, 139–147. [Google Scholar] [CrossRef]
- Schohl, A.; Chorghay, Z.; Ruthazer, E.S. A Simple and Efficient Method for Visualizing Individual Cells in Vivo by Cre-Mediated Single-Cell Labeling by Electroporation (CREMSCLE). Front. Neural Circuits 2020, 14, 47. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, S.; Sugi, J.; Iijima, T.; Hou, X. Single-Cell Visualization Deep in Brain Structures by Gene Transfer. Front. Neural Circuits 2020, 14, 586043. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, Y.; Namikawa, T.; Yamauchi, K.; Kawaguchi, Y. Cortical Divergent Projections in Mice Originate from Two Sequentially Generated, Distinct Populations of Excitatory Cortical Neurons with Different Initial Axonal Outgrowth Characteristics. Cereb. Cortex 2015, 26, 2257–2270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuta, T.; Tomioka, R.; Taki, K.; Nakamura, K.; Tamamaki, N.; Kaneko, T. In Vivo Transduction of Central Neurons Using Recombinant Sindbis Virus. J. Histochem. Cytochem. 2001, 49, 1497–1507. [Google Scholar] [CrossRef]
- Watakabe, A.; Takaji, M.; Kato, S.; Kobayashi, K.; Mizukami, H.; Ozawa, K.; Ohsawa, S.; Matsui, R.; Watanabe, D.; Yamamori, T. Simultaneous Visualization of Extrinsic and Intrinsic Axon Collaterals in Golgi-like Detail for Mouse Corticothalamic and Corticocortical Cells: A Double Viral Infection Method. Front. Neural Circuits 2014, 8, 110. [Google Scholar] [CrossRef]
- Economo, M.N.; Clack, N.G.; Lavis, L.D.; Gerfen, C.R.; Svoboda, K.; Myers, E.W.; Chandrashekar, J. A Platform for Brain-Wide Imaging and Reconstruction of Individual Neurons. eLife 2016, 5, e10566. [Google Scholar] [CrossRef]
- Kuramoto, E. Method for Labeling and Reconstruction of Single Neurons Using Sindbis Virus Vectors. J. Chem. Neuroanat. 2019, 100, 101648. [Google Scholar] [CrossRef]
- Jia, F.; Zhu, X.; Lv, P.; Hu, L.; Liu, Q.; Jin, S.; Xu, F. Rapid and Sparse Labeling of Neurons Based on the Mutant Virus-like Particle of Semliki Forest Virus. Neurosci. Bull. 2019, 35, 378–388. [Google Scholar] [CrossRef]
- Ohno, S.; Kuramoto, E.; Furuta, T.; Hioki, H.; Tanaka, Y.R.; Fujiyama, F.; Sonomura, T.; Uemura, M.; Sugiyama, K.; Kaneko, T. A Morphological Analysis of Thalamocortical Axon Fibers of Rat Posterior Thalamic Nuclei: A Single Neuron Tracing Study with Viral Vectors. Cereb. Cortex 2011, 22, 2840–2857. [Google Scholar] [CrossRef] [Green Version]
- Lukyanov, K.A.; Chudakov, D.M.; Lukyanov, S.; Verkhusha, V.V. Photoactivatable Fluorescent Proteins. Nat. Rev. Mol. Cell Biol. 2005, 6, 885–890. [Google Scholar] [CrossRef]
- Patterson, G.H.; Lippincott-Schwartz, J. A Photoactivatable GFP for Selective Photolabeling of Proteins and Cells. Science 2002, 297, 1873–1877. [Google Scholar] [CrossRef]
- Ando, R.; Hama, H.; Yamamoto-Hino, M.; Mizuno, H.; Miyawaki, A. An Optical Marker Based on the UV-Induced Green-to-Red Photoconversion of a Fluorescent Protein. Proc. Natl. Acad. Sci. USA 2002, 99, 12651–12656. [Google Scholar] [CrossRef] [Green Version]
- McKinney, S.A.; Murphy, C.S.; Hazelwood, K.L.; Davidson, M.W.; Looger, L.L. A Bright and Photostable Photoconvertible Fluorescent Protein. Nat. Methods 2009, 6, 131–133. [Google Scholar] [CrossRef] [Green Version]
- Nowotschin, S.; Hadjantonakis, A.-K. Use of KikGR a photoconvertible green-to-red fluorescent protein for cell labeling and lineage analysis in ES cells and mouse embryos. BMC Dev. Biol. 2009, 9, 49. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, E.A.; Campbell, R.E.; Lin, J.Y.; Lin, M.Z.; Miyawaki, A.; Palmer, A.E.; Shu, X.; Zhang, J.; Tsien, R.Y. The Growing and Glowing Toolbox of Fluorescent and Photoactive Proteins. Trends Biochem. Sci. 2017, 42, 111–129. [Google Scholar] [CrossRef] [Green Version]
- Tomura, M.; Yoshida, N.; Tanaka, J.; Karasawa, S.; Miwa, Y.; Miyawaki, A.; Kanagawa, O. Monitoring Cellular Movement in Vivo with Photoconvertible Fluorescence Protein “Kaede” Transgenic Mice. Proc. Natl. Acad. Sci. USA 2008, 105, 10871–10876. [Google Scholar] [CrossRef] [Green Version]
- Schindler, S.E.; McCall, J.G.; Yan, P.; Hyrc, K.L.; Li, M.; Tucker, C.L.; Lee, J.-M.; Bruchas, M.R.; Diamond, M.I. Photo-activatable Cre recombinase regulates gene expression in vivo. Sci. Rep. 2015, 5, 13627. [Google Scholar] [CrossRef] [Green Version]
- Kawano, F.; Okazaki, R.; Yazawa, M.; Sato, M. A photoactivatable Cre–loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 2016, 12, 1059–1064. [Google Scholar] [CrossRef]
- Taslimi, A.; Zoltowski, B.; Miranda, J.G.; Pathak, G.P.; Hughes, R.M.; Tucker, C.L. Optimized second-generation CRY2–CIB dimerizers and photoactivatable Cre recombinase. Nat. Chem. Biol. 2016, 12, 425–430. [Google Scholar] [CrossRef] [Green Version]
- Morikawa, K.; Furuhashi, K.; de Sena-Tomas, C.; Garcia-Garcia, A.L.; Bekdash, R.; Klein, A.D.; Gallerani, N.; Yamamoto, H.E.; Park, S.-H.E.; Collins, G.S.; et al. Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat. Commun. 2020, 11, 2141. [Google Scholar] [CrossRef]
- Yao, S.; Yuan, P.; Ouellette, B.; Zhou, T.; Mortrud, M.; Balaram, P.; Chatterjee, S.; Wang, Y.; Daigle, T.L.; Tasic, B.; et al. RecV recombinase system for in vivo targeted optogenomic modifications of single cells or cell populations. Nat. Methods 2020, 17, 422–429. [Google Scholar] [CrossRef] [PubMed]
- Yoshimi, K.; Yamauchi, Y.; Tanaka, T.; Shimada, T.; Sato, M.; Mashimo, T. Photoactivatable Cre knock-in mice for spatiotemporal control of genetic engineering in vivo. Lab. Investig. 2021, 101, 125–135. [Google Scholar] [CrossRef] [PubMed]
- Duplus-Bottin, H.; Spichty, M.; Triqueneaux, G.; Place, C.; Mangeot, P.E.; Ohlmann, T.; Vittoz, F.; Yvert, G. A single-chain and fast-responding light-inducible Cre recombinase as a novel optogenetic switch. eLife 2021, 10, e61268. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wu, Y.; Qiu, Y.; Li, X.; Guan, Y.; Cao, X.; Liu, M.; Zhang, D.; Huang, S.; Lin, L.; et al. Stable Transgenic Mouse Strain with Enhanced Photoactivatable Cre Recombinase for Spatiotemporal Genome Manipulation. Adv. Sci. 2022, 9, 2201352. [Google Scholar] [CrossRef]
- Young, P.; Qiu, L.; Wang, D.; Zhao, S.; Gross, J.; Feng, G. Single-neuron labeling with inducible Cre-mediated knockout in transgenic mice. Nat. Neurosci. 2008, 11, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Badea, T.C.; Hua, Z.L.; Smallwood, P.M.; Williams, J.; Rotolo, T.; Ye, X.; Nathans, J. New Mouse Lines for the Analysis of Neuronal Morphology Using Creer(t)/Loxp-Directed Sparse Labeling. PLoS ONE 2009, 4, e7859. [Google Scholar] [CrossRef]
- Sapkota, D.; Dougherty, J.D. An Inducible CRE Mouse Line to Sparsely Target Nervous System Cells, Including Remak Schwann Cells. Neural Dev. 2020, 15, 2. [Google Scholar] [CrossRef] [Green Version]
- Lin, T.Y.; Taniguchi-Sugiura, Y.; Murawala, P.; Hermann, S.; Tanaka, E.M. Inducible and Tissue-Specific Cell Labeling in Cre-Ert2 Transgenic Xenopus Lines. Dev. Growth Differ. 2022, 64, 243–253. [Google Scholar] [CrossRef]
- Sun, X.; Bernstein, M.J.; Meng, M.; Rao, S.; Sørensen, A.T.; Yao, L.; Zhang, X.; Anikeeva, P.O.; Lin, Y. Functionally Distinct Neuronal Ensembles within the Memory Engram. Cell 2020, 181, 410–423.e17. [Google Scholar] [CrossRef]
- Das, A.T.; Tenenbaum, L.; Berkhout, B. Tet-on Systems for Doxycycline-Inducible Gene Expression. Curr. Gene Ther. 2016, 16, 156–167. [Google Scholar] [CrossRef] [Green Version]
- Kallunki, T.; Barisic, M.; Jäättelä, M.; Liu, B. How to Choose the Right Inducible Gene Expression System for Mammalian Studies? Cells 2019, 8, 796. [Google Scholar] [CrossRef] [Green Version]
- Mizuno, H.; Luo, W.; Tarusawa, E.; Saito, Y.M.; Sato, T.; Yoshimura, Y.; Itohara, S.; Iwasato, T. NMDAR-Regulated Dynamics of Layer 4 Neuronal Dendrites during Thalamocortical Reorganization in Neonates. Neuron 2014, 82, 365–379. [Google Scholar] [CrossRef] [Green Version]
- Lin, R.; Wang, R.; Yuan, J.; Feng, Q.; Zhou, Y.; Zeng, S.; Ren, M.; Jiang, S.; Ni, H.; Zhou, C.; et al. Cell-Type-Specific and Projection-Specific Brain-Wide Reconstruction of Single Neurons. Nat. Methods 2018, 15, 1033–1036. [Google Scholar] [CrossRef]
- Kohara, K.; Inoue, A.; Nakano, Y.; Hirai, H.; Kobayashi, T.; Maruyama, M.; Baba, R.; Kawashima, C. BATTLE: Genetically Engineered Strategies for Split-Tunable Allocation of Multiple Transgenes in the Nervous System. iScience 2020, 23, 101248. [Google Scholar] [CrossRef]
- Inoue, A.; Kobayashi, T.; Hirai, H.; Kanaya, N.; Kohara, K. Protocol for BATTLE-1EX: A High-Resolution Imaging Method to Visualize Whole Synaptic Structures and their Components in the Nervous System. STAR Protoc. 2020, 1, 100166. [Google Scholar] [CrossRef]
- Zong, H.; Espinosa, J.S.; Su, H.H.; Muzumdar, M.D.; Luo, L. Mosaic Analysis with Double Markers in Mice. Cell 2005, 121, 479–492. [Google Scholar] [CrossRef] [Green Version]
- Tasic, B.; Miyamichi, K.; Hippenmeyer, S.; Dani, V.S.; Zeng, H.; Joo, W.; Zong, H.; Chen-Tsai, Y.; Luo, L. Extensions of MADM (Mosaic Analysis with Double Markers) in Mice. PLoS ONE 2012, 7, e33332. [Google Scholar] [CrossRef]
- Laukoter, S.; Amberg, N.; Pauler, F.M.; Hippenmeyer, S. Generation and isolation of single cells from mouse brain with mosaic analysis with double markers-induced uniparental chromosome disomy. STAR Protoc. 2020, 1, 100215. [Google Scholar] [CrossRef]
- Contreras, X.; Amberg, N.; Davaatseren, A.; Hansen, A.H.; Sonntag, J.; Andersen, L.; Bernthaler, T.; Streicher, C.; Heger, A.; Johnson, R.L.; et al. A genome-wide library of MADM mice for single-cell genetic mosaic analysis. Cell Rep. 2021, 35, 109274. [Google Scholar] [CrossRef]
- Hippenmeyer, S.; Youn, Y.H.; Moon, H.M.; Miyamichi, K.; Zong, H.; Wynshaw-Boris, A.; Luo, L. Genetic Mosaic Dissection of LIS1 and NDEL1 in Neuronal Migration. Neuron 2010, 68, 695–709. [Google Scholar] [CrossRef] [Green Version]
- Veldman, M.B.; Park, C.S.; Eyermann, C.M.; Zhang, J.Y.; Zuniga-Sanchez, E.; Hirano, A.A.; Daigle, T.L.; Foster, N.N.; Zhu, M.; Langfelder, P.; et al. Brainwide Genetic Sparse Cell Labeling to Illuminate the Morphology of Neurons and Glia with Cre-Dependent MORF Mice. Neuron 2020, 108, 111–127.e6. [Google Scholar] [CrossRef] [PubMed]
- Isaacman-Beck, J.; Paik, K.C.; Wienecke, C.F.; Yang, H.H.; Fisher, Y.E.; Wang, I.E.; Ishida, I.G.; Maimon, G.; Wilson, R.I.; Clandinin, T.R. SPARC Enables Genetic Manipulation of Precise Proportions of Cells. Nat. Neurosci. 2020, 23, 1168–1175. [Google Scholar] [CrossRef] [PubMed]
- Ran, F.A.; Hsu, P.D.; Wright, J.; Agarwala, V.; Scott, D.A.; Zhang, F. Genome Engineering Using the CRISPR-cas9 System. Nat. Protoc. 2013, 8, 2281–2308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doudna, J.A.; Charpentier, E. The New Frontier of Genome Engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef] [PubMed]
- Mikuni, T.; Nishiyama, J.; Sun, Y.; Kamasawa, N.; Yasuda, R. High-Throughput, High-Resolution Mapping of Protein Localization in Mammalian Brain by in Vivo Genome Editing. Cell 2016, 165, 1803–1817. [Google Scholar] [CrossRef] [Green Version]
- Nishiyama, J.; Mikuni, T.; Yasuda, R. Virus-Mediated Genome Editing via Homology-Directed Repair in Mitotic and Postmitotic Cells in Mammalian Brain. Neuron 2017, 96, 755–768. [Google Scholar] [CrossRef] [Green Version]
- Tsien, J.Z.; Huerta, P.T.; Tonegawa, S. The Essential Role of Hippocampal CA1 NMDA Receptor–Dependent Synaptic Plasticity in Spatial Memory. Cell 1996, 87, 1327–1338. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, K.; Quirk, M.C.; Chitwood, R.A.; Watanabe, M.; Yeckel, M.F.; Sun, L.D.; Kato, A.; Carr, C.A.; Johnston, D.; Wilson, M.A.; et al. Requirement for Hippocampal CA3 NMDA Receptors in Associative Memory Recall. Science 2002, 297, 211–218. [Google Scholar] [CrossRef] [Green Version]
- Kohara, K.; Pignatelli, M.; Rivest, A.J.; Jung, H.-Y.; Kitamura, T.; Suh, J.; Frank, D.; Kajikawa, K.; Mise, N.; Obata, Y.; et al. Cell type–specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci. 2014, 17, 269–279. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, T.; Pignatelli, M.; Suh, J.; Kohara, K.; Yoshiki, A.; Abe, K.; Tonegawa, S. Island Cells Control Temporal Association Memory. Science 2014, 343, 896–901. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Bloodgood, B.L.; Hauser, J.L.; Lapan, A.D.; Koon, A.C.; Kim, T.-K.; Hu, L.S.; Malik, A.N.; Greenberg, M.E. Activity-Dependent Regulation of Inhibitory Synapse Development by NPAS4. Nature 2008, 455, 1198–1204. [Google Scholar] [CrossRef] [Green Version]
- Bloodgood, B.L.; Sharma, N.; Browne, H.A.; Trepman, A.Z.; Greenberg, M.E. The Activity-Dependent Transcription Factor NPAS4 Regulates Domain-Specific Inhibition. Nature 2013, 503, 121–125. [Google Scholar] [CrossRef] [Green Version]
- Adesnik, H.; Li, G.; During, M.J.; Pleasure, S.J.; Nicoll, R.A. NMDA Receptors Inhibit Synapse Unsilencing during Brain Development. Proc. Natl. Acad. Sci. USA 2008, 105, 5597–5602. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Shi, Y.; Jackson, A.C.; Bjorgan, K.; During, M.J.; Sprengel, R.; Seeburg, P.H.; Nicoll, R.A. Subunit Composition of Synaptic AMPA Receptors Revealed by a Single-Cell Genetic Approach. Neuron 2009, 62, 254–268. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Isozaki, K.; Roche, K.W.; Nicoll, R.A. Synaptic Targeting of AMPA Receptors Is Regulated by a CaMKII Site in the First Intracellular Loop of GLUA1. Proc. Natl. Acad. Sci. USA 2010, 107, 22266–22271. [Google Scholar] [CrossRef] [Green Version]
- Herring, B.E.; Shi, Y.; Suh, Y.H.; Zheng, C.-Y.; Blankenship, S.M.; Roche, K.W.; Nicoll, R.A. Cornichon Proteins Determine the Subunit Composition of Synaptic AMPA Receptors. Neuron 2013, 77, 1083–1096. [Google Scholar] [CrossRef] [Green Version]
- Granger, A.J.; Shi, Y.; Lu, W.; Cerpas, M.; Nicoll, R.A. LTP Requires a Reserve Pool of Glutamate Receptors Independent of Subunit Type. Nature 2012, 493, 495–500. [Google Scholar] [CrossRef] [Green Version]
- Odajima, J.; Wills, Z.P.; Ndassa, Y.M.; Terunuma, M.; Kretschmannova, K.; Deeb, T.Z.; Geng, Y.; Gawrzak, S.; Quadros, I.M.; Newman, J.; et al. Cyclin E Constrains CDK5 Activity to Regulate Synaptic Plasticity and Memory Formation. Dev. Cell 2011, 21, 655–668. [Google Scholar] [CrossRef] [Green Version]
- Lu, W.; Bushong, E.A.; Shih, T.P.; Ellisman, M.H.; Nicoll, R.A. The Cell-Autonomous Role of Excitatory Synaptic Transmission in the Regulation of Neuronal Structure and Function. Neuron 2013, 78, 433–439. [Google Scholar] [CrossRef] [Green Version]
- Lovero, K.L.; Fukata, Y.; Granger, A.J.; Fukata, M.; Nicoll, R.A. The LGI1–adam22 Protein Complex Directs Synapse Maturation through Regulation of PSD-95 Function. Proc. Natl. Acad. Sci. USA 2015, 112, E4129–E4137. [Google Scholar] [CrossRef] [Green Version]
- Balice-Gordon, R.J.; Lichtman, J.W. Long-Term Synapse Loss Induced by Focal Blockade of POSTSYNAPTLC Receptors. Nature 1994, 372, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Burrone, J.; O’Byrne, M.; Murthy, V.N. Multiple Forms of Synaptic Plasticity Triggered by Selective Suppression of Activity in Individual Neurons. Nature 2002, 420, 414–418. [Google Scholar] [CrossRef] [PubMed]
- De Marco García, N.V.; Karayannis, T.; Fishell, G. Neuronal Activity Is Required for the Development of Specific Cortical Interneuron Subtypes. Nature 2011, 472, 351–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ehrengruber, M.U.; Doupnik, C.A.; Xu, Y.; Garvey, J.; Jasek, M.C.; Lester, H.A.; Davidson, N. Activation of Heteromeric G Protein-Gated Inward Rectifier K + Channels Overexpressed by Adenovirus Gene Transfer Inhibits the Excitability of Hippocampal Neurons. Proc. Natl. Acad. Sci. USA 1997, 94, 7070–7075. [Google Scholar] [CrossRef] [Green Version]
- Okada, M.; Matsuda, H. Chronic Lentiviral Expression of Inwardly Rectifying K+ Channels (Kir2.1) Reduces Neuronal Activity and Downregulates Voltage-Gated Potassium Currents in Hippocampus. Neuroscience 2008, 156, 289–297. [Google Scholar] [CrossRef]
- Nadeau, H.; Mckinney, S.; Anderson, D.J.; Lester, H.A. ROMK1 (Kir1.1) Causes Apoptosis and Chronic Silencing of Hippocampal Neurons. J. Neurophysiol. 2000, 84, 1062–1075. [Google Scholar] [CrossRef]
- Okada, M.; Andharia, N.; Matsuda, H. Increase in the Titer of Lentiviral Vectors Expressing Potassium Channels by Current Blockade during Viral Vector Production. BMC Neurosci. 2015, 16, 30. [Google Scholar] [CrossRef] [Green Version]
- Meng, D.; Li, H.; Deisseroth, K.; Leutgeb, S.; Spitzer, N.C. Neuronal Activity Regulates Neurotransmitter Switching in the Adult Brain Following Light-Induced Stress. Proc. Natl. Acad. Sci. USA 2018, 115, 5064–5071. [Google Scholar] [CrossRef] [Green Version]
- Lin, C.W.; Sim, S.; Ainsworth, A.; Okada, M.; Kelsch, W.; Lois, C. Genetically Increased Cell-Intrinsic Excitability Enhances Neuronal Integration into Adult Brain Circuits. Neuron 2010, 65, 32–39. [Google Scholar] [CrossRef] [Green Version]
- Roth, B.L. DREADDs for Neuroscientists. Neuron 2016, 89, 683–694. [Google Scholar] [CrossRef] [Green Version]
- Fenno, L.; Yizhar, O.; Deisseroth, K. The Development and Application of Optogenetics. Annu. Rev. Neurosci. 2011, 34, 389. [Google Scholar] [CrossRef]
- Mizuno, H.; Ikezoe, K.; Nakazawa, S.; Sato, T.; Kitamura, K.; Iwasato, T. Patchwork-Type Spontaneous Activity in Neonatal Barrel Cortex Layer 4 Transmitted via Thalamocortical Projections. Cell Rep. 2018, 22, 123–135. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, S.; Mizuno, H.; Iwasato, T. Differential Dynamics of Cortical Neuron Dendritic Trees Revealed by Long-Term in Vivo Imaging in Neonates. Nat. Commun. 2018, 9, 3106. [Google Scholar] [CrossRef] [Green Version]
- Nakazawa, S.; Yoshimura, Y.; Takagi, M.; Mizuno, H.; Iwasato, T. Developmental Phase Transitions in Spatial Organization of Spontaneous Activity in Postnatal Barrel Cortex Layer 4. J. Neurosci. 2020, 40, 7637–7650. [Google Scholar] [CrossRef]
- Zhang, X.; Mennicke, C.V.; Xiao, G.; Beattie, R.; Haider, M.A.; Hippenmeyer, S.; Ghashghaei, H.T. Clonal Analysis of Gliogenesis in the Cerebral Cortex Reveals Stochastic Expansion of Glia and Cell Autonomous Responses to EGFR Dosage. Cells 2020, 9, 2662. [Google Scholar] [CrossRef]
- Han, J.H.; Kushner, S.A.; Yiu, A.P.; Cole, C.J.; Matynia, A.; Brown, R.A.; Neve, R.L.; Guzowski, J.F.; Silva, A.J.; Josselyn, S.A. Neuronal Competition and Selection during Memory Formation. Science 2007, 316, 457–460. [Google Scholar] [CrossRef] [Green Version]
- Brandenburg, C.; Smith, L.A.; Kilander, M.B.C.; Bridi, M.S.; Lin, Y.C.; Huang, S.; Blatt, G.J. Parvalbumin Subtypes of Cerebellar Purkinje Cells Contribute to Differential Intrinsic Firing Properties. Mol. Cell. Neurosci. 2021, 115, 103650. [Google Scholar] [CrossRef]
- Bi, G.Q.; Poo, M.M. Synaptic Modifications in Cultured Hippocampal Neurons: Dependence on Spike Timing, Synaptic Strength, and Postsynaptic Cell Type. J. Neurosci. 1998, 18, 10464–10472. [Google Scholar] [CrossRef] [Green Version]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kohara, K.; Okada, M. Single-Cell Labeling Strategies to Dissect Neuronal Structures and Local Functions. Biology 2023, 12, 321. https://doi.org/10.3390/biology12020321
Kohara K, Okada M. Single-Cell Labeling Strategies to Dissect Neuronal Structures and Local Functions. Biology. 2023; 12(2):321. https://doi.org/10.3390/biology12020321
Chicago/Turabian StyleKohara, Keigo, and Masayoshi Okada. 2023. "Single-Cell Labeling Strategies to Dissect Neuronal Structures and Local Functions" Biology 12, no. 2: 321. https://doi.org/10.3390/biology12020321
APA StyleKohara, K., & Okada, M. (2023). Single-Cell Labeling Strategies to Dissect Neuronal Structures and Local Functions. Biology, 12(2), 321. https://doi.org/10.3390/biology12020321





