Genomic Comparative Analysis of Two Multi-Drug Resistance (MDR) Acinetobacter baumannii Clinical Strains Assigned to International Clonal Lineage II Recovered Pre- and Post-COVID-19 Pandemic
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Isolates
2.2. Genome Sequencing
2.3. Comparative Genome Analysis
2.4. RNA Extraction and qRT-PCR Analysis
2.5. Antibiotic Susceptibility Assays
3. Results and Discussion
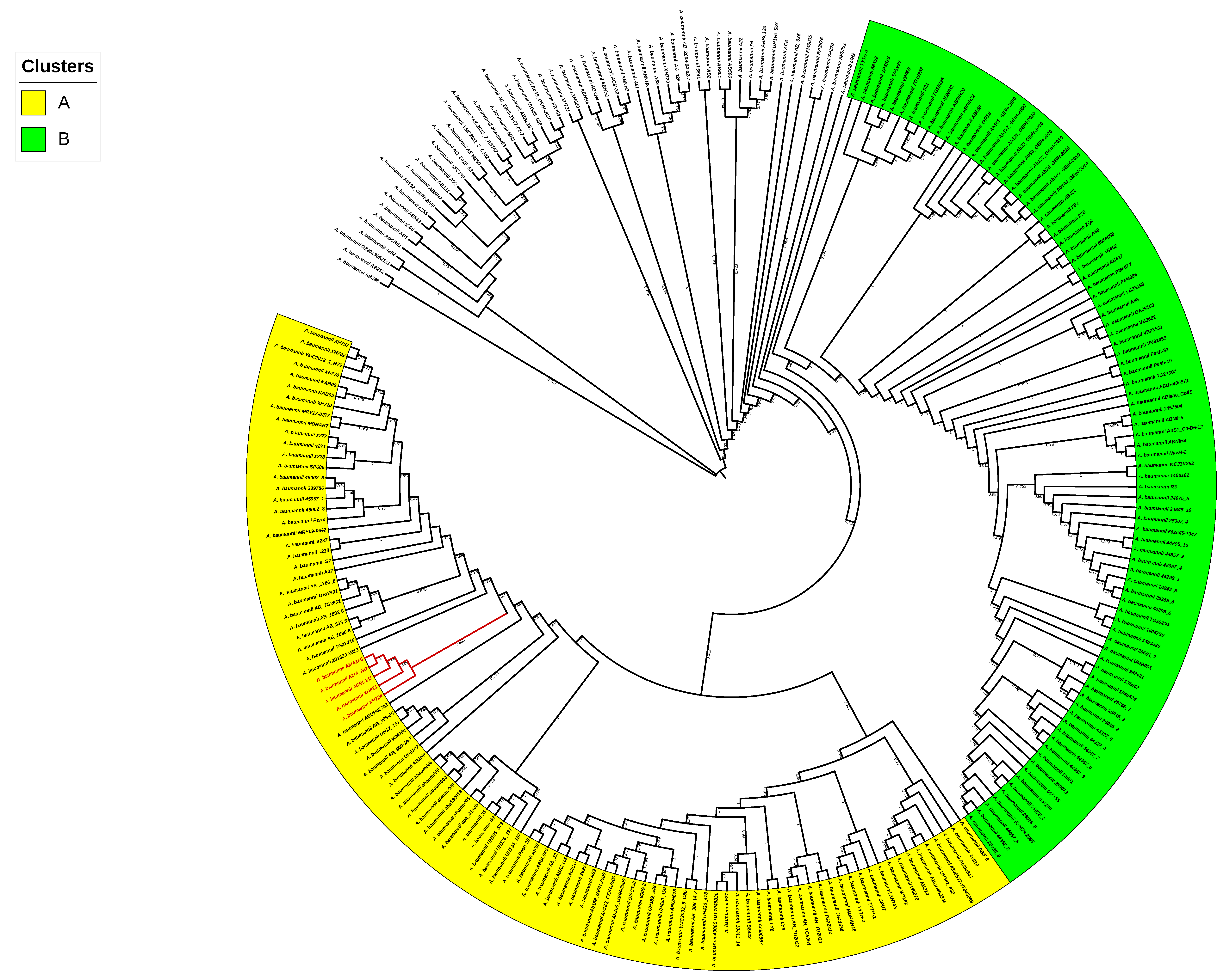
3.1. Sequencing of AMA166 and AMA_NO. Genomic and Phylogenomic Comparative Analyses
3.2. Antibiotic Resistance, Virulence, and Its Association with Horizontal Genetic Transfer (HGT) Elements
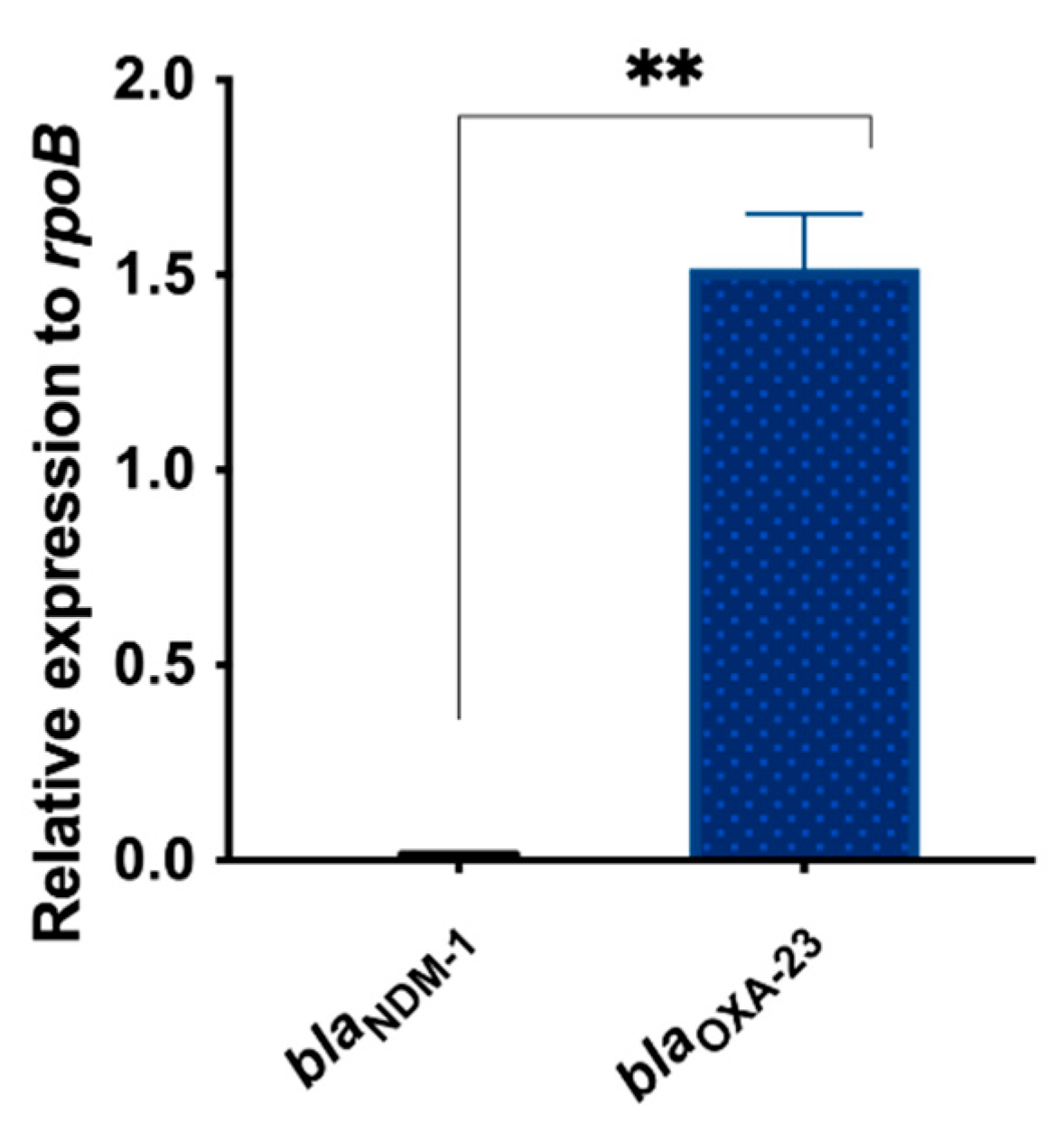
3.3. Differential Expression of blaNDM-1 and blaOXA-23 in AMA_NO
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Karakonstantis, S.; Gikas, A.; Astrinaki, E.; Kritsotakis, E.I. Excess mortality due to pandrug-resistant Acinetobacter baumannii infections in hospitalized patients. J. Hosp. Infect. 2020, 106, 447–453. [Google Scholar] [CrossRef] [PubMed]
- Karakonstantis, S.; Kritsotakis, E.I. Systematic review and meta-analysis of the proportion and associated mortality of polymicrobial (vs monomicrobial) pulmonary and bloodstream infections by Acinetobacter baumannii complex. Infection 2021, 49, 1149–1161. [Google Scholar] [CrossRef] [PubMed]
- Ling, W.; Furuya-Kanamori, L.; Ezure, Y.; Harris, P.N.; Paterson, D.L. Adverse clinical outcomes associated with carbapenem-resistant Acinetobacter (CRA) infections: A systematic review and meta-analysis. JAC Antimicrob. Resist. 2021, 3, dlab157. [Google Scholar] [CrossRef]
- Nguyen, M.; Joshi, S.G. Carbapenem resistance in Acinetobacter baumannii, and their importance in hospital-acquired infections: A scientific review. J. Appl. Microbiol. 2021, 131, 2715–2738. [Google Scholar] [CrossRef] [PubMed]
- WHO. World Health Organisation, Antimicrobial Resistance: Global Report on Surveillance 2014—World | ReliefWeb. 2014. Available online: https://reliefweb.int/report/world/antimicrobial-resistance-global-report-surveillance-2014?gclid=CjwKCAiA68ebBhB-EiwALVC-NsMhCtrt2XXNOS_pNrR_aYSo3JCcAVeGXq5UMxl4RQq7-O35Nzx0LxoCNeQQAvD_BwE (accessed on 30 April 2014).
- CDC. Antibiotic resistance threats in the United States.; US Department of Health and Human Services: Atlanta, GA, USA, 2019.
- Belder JB, D.; Nguyen-Disteche, M.; Houba-Herin, N.; Ghuysen, J.M.; Maruyama, I.N.; Hara, H.; Hirota, Y.; Inouye, M. Overexpression, solubilization and refolding of a genetically engineered derivative of the penicillin-binding protein 3 of Escherichia coli K12. Mol. Microbiol. 1988, 2, 519–525. [Google Scholar] [CrossRef]
- Peleg, A.Y.; Hooper, D.C. Hospital-acquired infections due to gram-negative bacteria. N. Engl. J. Med. 2010, 362, 1804–1813. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.T.; Kuo, S.C.; Lee, Y.T.; Chen, C.P.; Lin, S.W.; Shen, L.J.; Fung, C.-P.; Cho, W.-L.; Chen, T.-L. Sheltering effect and indirect pathogenesis of carbapenem-resistant Acinetobacter baumannii in polymicrobial infection. Antimicrob. Agents Chemother. 2014, 58, 3983–3990. [Google Scholar] [CrossRef] [Green Version]
- Dhamgaye, S.; Qu, Y.; Peleg, A.Y. Polymicrobial infections involving clinically relevant Gram-negative bacteria and fungi. Cell Microbiol. 2016, 18, 1716–1722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safdar, N.; Maki, D.G. The commonality of risk factors for nosocomial colonization and infection with antimicrobial-resistant Staphylococcus aureus, Enterococcus, gram-negative bacilli, Clostridium difficile, and Candida. Ann. Intern. Med. 2002, 136, 834–844. [Google Scholar] [CrossRef]
- Davies, J.C.; Rubin, B.K. Emerging and unusual gram-negative infections in cystic fibrosis. Semin. Respir. Crit. Care Med. 2007, 28, 312–321. [Google Scholar] [CrossRef]
- Cohen, T.S.; Hilliard, J.J.; Jones-Nelson, O.; Keller, A.E.; O’Day, T.; Tkaczyk, C.; DiGiandomenico, A.; Hamilton, M.; Pelletier, M.; Wang, Q.; et al. Staphylococcus aureus α toxin potentiates opportunistic bacterial lung infections. Sci. Transl. Med. 2016, 8, ra31–ra329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moriano, A.; Serra, D.O.; Hoard, A.; Montaña, S.; Degrossi, J.; Bonomo, R.A.; Papp-Wallace, K.M.; Ramirez, M.S. Staphylococcus aureus Potentiates the Hemolytic Activity of Burkholderia cepacia Complex (Bcc) Bacteria. Curr. Microbiol. 2021, 78, 1864–1870. [Google Scholar] [CrossRef] [PubMed]
- Hoenigl, M.; Seidel, D.; Sprute, R.; Cunha, C.; Oliverio, M.; Goldman, G.H.; Ibrahim, A.S.; Carvalho, A. COVID-19-associated fungal infections. Nat. Microbiol. 2022, 7, 1127–1140. [Google Scholar] [CrossRef] [PubMed]
- Fontana, C.; Favaro, M.; Minelli, S.; Bossa, M.C.; Altieri, A. Co-infections observed in SARS-CoV-2 positive patients using a rapid diagnostic test. Sci. Rep. 2021, 11, 16355. [Google Scholar] [CrossRef]
- Scott, H.; Zahra, A.; Fernandes, R.; Fries, B.C.; Thode, H.C.; Singer, A.J. Bacterial infections and death among patients with Covid-19 versus non Covid-19 patients with pneumonia. Am. J. Emerg. Med. 2021, 51, 1–5. [Google Scholar] [CrossRef]
- Swets, M.C.; Russell, C.D.; Harrison, E.M.; Docherty, A.B.; Lone, N.; Girvan, M.; Hardwick, H.; Visser, L.; Openshaw, P.; Groeneveld, G.; et al. SARS-CoV-2 co-infection with influenza viruses, respiratory syncytial virus, or adenoviruses. Lancet 2022, 399, 1463–1464. [Google Scholar] [CrossRef]
- Castellanos, N.; Nakanouchi, J.; Yüzen, D.I.; Fung, S.; Fernandez, J.S.; Barberis, C.; Tuchscherr, L.; Ramirez, M.S. A Study on Acinetobacter baumannii and Staphylococcus aureus Strains Recovered from the Same Infection Site of a Diabetic Patient. Curr. Microbiol. 2019, 76, 842–847. [Google Scholar] [CrossRef]
- Fernandez, J.S.; Tuttobene, M.R.; Montaña, S.; Subils, T.; Cantera, V.; Iriarte, A.; Tuchscherr, L.; Ramirez, M.S. Staphylococcus aureus α-Toxin Effect on Acinetobacter baumannii Behavior. Biology 2022, 11, 570. [Google Scholar] [CrossRef]
- Contou, D.; Claudinon, A.; Pajot, O.; Micaëlo, M.; Longuet Flandre, P.; Dubert, M.; Cally, R.; Logre, E.; Fraissé, M.; Mentec, H.; et al. Bacterial and viral co-infections in patients with severe SARS-CoV-2 pneumonia admitted to a French ICU. Ann. Intensive Care 2020, 10, 119. [Google Scholar] [CrossRef]
- Sharifipour, E.; Shams, S.; Esmkhani, M.; Khodadadi, J.; Fotouhi-Ardakani, R.; Koohpaei, A.; Doosti, Z.; Golzari, S.E. Evaluation of bacterial co-infections of the respiratory tract in COVID-19 patients admitted to ICU. BMC Infect. Dis. 2020, 20, 646. [Google Scholar] [CrossRef]
- Karruli, A.; Boccia, F.; Gagliardi, M.; Patauner, F.; Ursi, M.P.; Sommese, P.; De Rosa, R.; Murino, P.; Ruocco, G.; Corcione, A.; et al. Multidrug-resistant infections and outcome of critically ill patients with coronavirus disease 2019: A single center experience. Microb. Drug Resist. 2021, 27, 1167–1175. [Google Scholar] [CrossRef] [PubMed]
- Nori, P.; Cowman, K.; Chen, V.; Bartash, R.; Szymczak, W.; Madaline, T.; Katiyar, C.P.; Jain, R.; Aldrich, M.; Weston, G.; et al. Bacterial and fungal coinfections in COVID-19 patients hospitalized during the New York City pandemic surge. Infect. Control Hosp. Epidemiol. 2020, 42, 84–88. [Google Scholar] [CrossRef]
- Rangel, K.; Chagas, T.P.G.; De-Simone, S.G. Acinetobacter baumannii infections in times of COVID-19 pandemic. Pathogens 2021, 10, 1006. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Hua, M.; Liu, X.; Du, C.; Pu, L.; Xiang, P.; Wang, L.; Liu, J. Bacterial and fungal co-infections among COVID-19 patients in intensive care unit. Microbes Infect. 2021, 23, 104806. [Google Scholar] [CrossRef] [PubMed]
- Abdollahi, A.; Aliramezani, A.; Salehi, M.; Norouzi Shadehi, M.; Ghourchian, S.; Douraghi, M. Co-infection of ST2IP carbapenem-resistant Acinetobacter baumannii with SARS-CoV-2 in the patients admitted to a Tehran tertiary referral hospital. BMC Infect Dis 2021, 21, 927. [Google Scholar] [CrossRef]
- Almuzara, M.; Barberis, C.; Traglia, G.; Famiglietti, A.; Ramirez, M.S.; Vay, C. Evaluation of matrix-assisted laser desorption ionization-time-of-flight mass spectrometry for species identification of Nonfermenting Gram-Negative Bacilli. J. Microbiol. Methods 2015, 112, 24–27. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Gurevich, A.; Saveliev, V.; Vyahhi, N.; Tesler, G. QUAST: Quality assessment tool for genome assemblies. Bioinformatics 2013, 29, 1072–1075. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [Green Version]
- Cantalapiedra, C.P.; Hernández-Plaza, A.; Letunic, I.; Bork, P.; Huerta-Cepas, J. eggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 2021, 38, 5825–5829. [Google Scholar] [CrossRef]
- Richter, M.; Rosselló-Móra, R.; Oliver Glöckner, F.; Peplies, J. JSpeciesWS: A web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 2016, 32, 929–931. [Google Scholar] [CrossRef] [PubMed]
- Lowe, T.M.; Eddy, S.R. TRNAscan-SE: A program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res. 1996, 25, 955–964. [Google Scholar] [CrossRef]
- Alcock, B.P.; Raphenya, A.R.; Lau, T.T.; Tsang, K.K.; Bouchard, M.; Edalatmand, A.; Huynh, W.; Nguyen, A.-L.; Cheng, A.; Liu, S.; et al. CARD 2020: Antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res. 2020, 48, D517–D525. [Google Scholar] [CrossRef]
- Chen, L.; Xiong, Z.; Sun, L.; Yang, J.; Jin, Q. VFDB 2012 update: Toward the genetic diversity and molecular evolution of bacterial virulence factors. Nucleic Acids Res. 2011, 40, D641–D645. [Google Scholar] [CrossRef] [PubMed]
- Wyres, K.L.; Cahill, S.M.; Holt, K.E.; Hall, R.M.; Kenyon, J.J. Identification of Acinetobacter baumannii loci for capsular polysaccharide (KL) and lipooligosaccharide outer core (OCL) synthesis in genome assemblies using curated reference databases compatible with Kaptive. Microb. Genom. 2020, 6, e000339. [Google Scholar] [CrossRef]
- Antunes, L.C.S.; Imperi, F.; Carattoli, A.; Visca, P. Deciphering the Multifactorial Nature of Acinetobacter baumannii Pathogenicity. PLoS ONE 2011, 6, e22674. [Google Scholar] [CrossRef] [Green Version]
- Siguier, P.; Pérochon, J.; Lestrade, L.; Mahillon, J.; Chandler, M. ISfinder: The reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006, 34, D32–D36. [Google Scholar] [CrossRef] [Green Version]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mindlin, S.; Beletsky, A.; Rakitin, A.; Mardanov, A.; Petrova, M. Acinetobacter Plasmids: Diversity and Development of Classification Strategies. Front. Microbiol. 2020, 11, 588410. [Google Scholar] [CrossRef] [PubMed]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.G.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [Green Version]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wright, E.S.; Yilmaz, L.S.; Noguera, D.R. DECIPHER, a search-based approach to chimera identification for 16S rRNA sequences. Appl. Environ. Microbiol. 2012, 78, 717–725. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Linder, C.R.; Warnow, T. RAxML and FastTree: Comparing two methods for large-scale maximum likelihood phylogeny estimation. PLoS ONE 2011, 6, e27731. [Google Scholar] [CrossRef] [PubMed]
- Darriba, D.; Taboada, G.L.; Doallo, R.; Posada, D. jModelTest 2: More models, new heuristics and parallel computing. Nat. Methods 2012, 9, 772. [Google Scholar] [CrossRef] [Green Version]
- Page, A.J.; Taylor, B.; Delaney, A.J.; Soares, J.; Seemann, T.; Keane, J.A.; Harris, S.R. SNP-sites: Rapid efficient extraction of SNPs from multi-FASTA alignments. Microb. Genom. 2016, 2, e000056. [Google Scholar] [CrossRef] [Green Version]
- Darling, A.E.; Mau, B.; Perna, N.T. Progressivemauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pimentel, C.; Le, C.; Tuttobene, M.; Subils, T.; Martinez, J.; Sieira, R.; Papp-Wallace, K.; Keppetipola, N.; Bonomo, R.; Actis, L.; et al. Human Pleural Fluid and Human Serum Albumin Modulate the Behavior of a Hypervirulent and Multidrug-Resistant (MDR) Acinetobacter baumannii Representative Strain. Pathogens 2021, 10, 471. [Google Scholar] [CrossRef]
- Quinn, B.; Rodman, N.; Jara, E.; Fernandez, J.S.; Martinez, J.; Traglia, G.M.; Montaña, S.; Cantera, V.; Place, K.; Bonomo, R.A.; et al. Human serum albumin alters specific genes that can play a role in survival and persistence in Acinetobacter baumannii. Sci. Rep. 2018, 8, 14741. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCT) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- CLSI M100-ED32; Performance Standards for Antimicrobial Susceptibility Testing, 32nd ed. Clinical and Laboratory Standards Institute: Malvern, PA, USA, 2022.
- Garcia-Cobos, S.; Arroyo, M.; Perez-Vazquez, M.; Aracil, B.; Oteo, J.; Campos, J. Evaluation of the EUCAST disc diffusion susceptibility testing method for Haemophilus influenzae based on the resistance mechanism to β-lactam antibiotics. J. Antimicrob. Chemother. 2013, 68, 159–163. [Google Scholar] [CrossRef] [PubMed]
- Shi, A.; Fan, F.; Broach, J.R. Microbial adaptive evolution. J. Ind. Microbiol. Biotechnol. 2022, 49, 76. [Google Scholar] [CrossRef]
- Langridge, G.C.; Fookes, M.; Connor, T.R.; Feltwell, T.; Feasey, N.; Parsons, B.N.; Seth-Smith, H.M.B.; Barquist, L.; Stedman, A.; Humphrey, T.; et al. Patterns of genome evolution that have accompanied host adaptation in Salmonella. Proc. Natl. Acad. Sci. USA 2015, 112, 863–868. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.-Q.; Charoechai, P.; Khunajakr, N.; Deng, Y.-M.; Widodo, W.; Dunn, N.W. Genetic and transcriptional analysis of a novel plasmid-encoded copper resistance operon from Lactococcus lactis. Gene 2002, 297, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Hall, B.G.; Parker, L.L.; Betts, P.W.; DuBose, R.F.; A Sawyer, S.; Hartl, D.L. IS103, a new insertion element in Escherichia coli: Characterization and distribution in natural populations. Genetics 1989, 121, 423–431. [Google Scholar] [CrossRef]
- Bonnin, R.A.; Poirel, L.; Nordmann, P. A novel and hybrid composite transposon at the origin of acquisition of bla(RTG-5) in Acinetobacter baumannii. Int. J. Antimicrob. Agents 2012, 40, 257–259. [Google Scholar] [CrossRef]
- Gaiarsa, S.; Batisti Biffignandi, G.; Esposito, E.P.; Castelli, M.; Jolley, K.A.; Brisse, S.; Sassera, D.; Zarrilli, R. Comparative analysis of the two Acinetobacter baumannii multilocus sequence typing (MLST) Schemes. Front. Microbiol. 2019, 10, 930. [Google Scholar] [CrossRef] [Green Version]
- Steenwyk, J.L.; Mead, M.E.; de Castro, P.A.; Valero, C.; Damasio, A.; dos Santos, R.A.C.; Labella, A.L.; Li, Y.; Knowles, S.L.; Raja, H.A.; et al. Genomic and Phenotypic Analysis of COVID-19-Associated Pulmonary Aspergillosis Isolates of Aspergillus fumigatus. Microbiol. Spectr. 2021, 9, e0001021. [Google Scholar] [CrossRef] [PubMed]
- Cañada-García, J.E.; Ramírez de Arellano, E.; Jiménez-Orellana, M.; Viedma, E.; Sánchez, A.; Alhambra, A.; Villa, J.; Delgado-Iribarren, A.; Bautista, V.; Lara, N.; et al. Carbapenemase-Producing Klebsiella pneumoniae in COVID-19 intensive care patients: Identification of IncL-VIM-1 plasmid in previously non-predominant sequence types. Antibiotics 2023, 12, 107. [Google Scholar] [CrossRef] [PubMed]
- Yoon, E.-J.; Balloy, V.; Fiette, L.; Chignard, M.; Courvalin, P.; Grillot-Courvalin, C. Contribution of the Ade Resistance-Nodulation-Cell Division-Type Efflux Pumps to Fitness and Pathogenesis of Acinetobacter baumannii. Mbio 2016, 7, e00697-16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moubareck, C.A.; Halat, D.H. Insights into Acinetobacter baumannii: A review of microbiological, virulence, and resistance traits in a threatening nosocomial pathogen. Antibiotics 2020, 9, 119. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, L.-J.; Pan, Y.; Gao, C.-Y.; Hou, P.-F. Distribution of Carbapenemases and Efflux Pump in Carbapenem-resistance Acinetobacter baumannii. Ann. Clin. Lab. Sci. 2020, 50, 241–246. [Google Scholar] [PubMed]
- Smith, M.G.; Gianoulis, T.A.; Pukatzki, S.; Mekalanos, J.J.; Ornston, L.N.; Gerstein, M.; Snyder, M. New insights into Acinetobacter baumannii pathogenesis revealed by high-density pyrosequencing and transposon mutagenesis. Genes Dev. 2007, 21, 601–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, M.D.; Goglin, K.; Molyneaux, N.; Hujer, K.M.; Lavender, H.; Jamison, J.J.; MacDonald, I.J.; Martin, K.M.; Russo, T.; Campagnari, A.A.; et al. Comparative Genome Sequence Analysis of Multidrug-Resistant Acinetobacter baumannii. J. Bacteriol. 2008, 190, 8053–8064. [Google Scholar] [CrossRef] [Green Version]
- Iacono, M.; Villa, L.; Fortini, D.; Bordoni, R.; Imperi, F.; Bonnal, R.J.P.; Sicheritz-Ponten, T.; De Bellis, G.; Visca, P.; Cassone, A.; et al. Whole-Genome Pyrosequencing of an Epidemic Multidrug-Resistant Acinetobacter baumannii Strain Belonging to the European Clone II Group. Antimicrob. Agents Chemother. 2008, 52, 2616–2625. [Google Scholar] [CrossRef] [Green Version]
- Vallenet, D.; Nordmann, P.; Barbe, V.; Poirel, L.; Mangenot, S.; Bataille, E.; Dossat, C.; Gas, S.; Kreimeyer, A.; Lenoble, P.; et al. Comparative Analysis of Acinetobacters: Three Genomes for Three Lifestyles. PLoS ONE 2008, 3, e1805. [Google Scholar] [CrossRef]
- Chen, C.-C.; Lin, Y.-C.; Sheng, W.-H.; Chen, Y.-C.; Chang, S.-C.; Hsia, K.-C.; Liao, M.-H.; Li, S.-Y. Genome Sequence of a Dominant, Multidrug-Resistant Acinetobacter baumannii Strain, TCDC-AB0715. J. Bacteriol. 2011, 193, 2361–2362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.Y.; Kim, S.; Kim, S.M.; Cha, S.H.; Lim, S.K.; Kim, J. Complete genome sequence of Multidrug-Resistant Acinetobacter baumannii strain 1656-2, which forms sturdy biofilm. J. Bacteriol. 2011, 193, 6393. [Google Scholar] [CrossRef] [Green Version]
- Farrugia, D.N.; Elbourne, L.D.; Hassan, K.A.; Eijkelkamp, B.A.; Tetu, S.G.; Brown, M.H.; Shah, B.S.; Peleg, A.Y.; Mabbutt, B.C.; Paulsen, I.T. The complete genome and phenome of a community-acquired Acinetobacter baumannii. PLoS ONE 2013, 8, e58628. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Yan, Z.; Zhang, Z.; Zhou, Q.; Zhou, J.; Wakeland, E.K.; Fang, X.; Xuan, Z.; Shen, D.; Li, Q.-Z. Complete Genome Analysis of Three Acinetobacter baumannii Clinical Isolates in China for Insight into the Diversification of Drug Resistance Elements. PLoS ONE 2013, 8, e66584. [Google Scholar] [CrossRef] [Green Version]
- Russo, T.A.; Luke, N.R.; Beanan, J.M.; Olson, R.; Sauberan, S.L.; MacDonald, U.; Schultz, L.W.; Umland, T.C.; Campagnari, A.A. The K1 Capsular Polysaccharide of Acinetobacter baumannii Strain 307-0294 Is a Major Virulence Factor. Infect. Immun. 2010, 78, 3993–4000. [Google Scholar] [CrossRef] [Green Version]
- Ramirez, M.S.; Penwell, W.F.; Traglia, G.M.; Zimbler, D.L.; Gaddy, J.A.; Nikolaidis, N.; Arivett, B.A.; Adams, M.D.; Bonomo, R.A.; Actis, L.A.; et al. Identification of Potential Virulence Factors in the Model Strain Acinetobacter baumannii A118. Front. Microbiol. 2019, 10, 1599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kenyon, J.J.; Kasimova, A.A.; Sviridova, A.N.; Shpirt, A.M.; Shneider, M.M.; Mikhaylova, Y.V.; Shelenkov, A.A.; Popova, A.V.; Perepelov, A.V.; Shashkov, A.S.; et al. Correlation of Acinetobacter baumannii K144 and K86 capsular polysaccharide structures with genes at the K locus reveals the involvement of a novel multifunctional rhamnosyltransferase for structural synthesis. Int. J. Biol. Macromol. 2021, 193, 1294–1300. [Google Scholar] [CrossRef] [PubMed]
- Weinberg, E.D. Iron withholding: A defense against infection and neoplasia. Physiol. Rev. 1984, 64, 65–102. [Google Scholar] [CrossRef] [PubMed]
- Cuajungco, M.P.; Ramirez, M.S.; Tolmasky, M.E. Zinc: Multidimensional effects on living organisms. Biomedicines 2021, 9, 208. [Google Scholar] [CrossRef] [PubMed]
- Hennigar, S.R.; McClung, J.P. Nutritional immunity: Starving pathogens of trace minerals. Am. J. Lifestyle Med. 2016, 10, 170–173. [Google Scholar] [CrossRef]
- Tolmasky, M.E.; Crosa, J.H. Regulation of plasmid-mediated iron transport and virulence in Vibrio anguillarum. Biol. Met. 1991, 4, 33–35. [Google Scholar] [CrossRef]
- Ratledge, C.; Dover, L.G. Iron metabolism in pathogenic bacteria. Annu. Rev. Microbiol. 2000, 54, 881–941. [Google Scholar] [CrossRef]
- Antunes, L.C.; Imperi, F.; Towner, K.J.; Visca, P. Genome-assisted identification of putative iron-utilization genes in Acinetobacter baumannii and their distribution among a genotypically diverse collection of clinical isolates. Res. Microbiol. 2011, 162, 279–284. [Google Scholar] [CrossRef]
- Chan, A.P.; Sutton, G.; DePew, J.; Krishnakumar, R.; Choi, Y.; Huang, X.-Z.; Beck, E.; Harkins, D.M.; Kim, M.; Lesho, E.P.; et al. A novel method of consensus pan-chromosome assembly and large-scale comparative analysis reveal the highly flexible pan-genome of Acinetobacter baumannii. Genome Biol. 2015, 16, 143. [Google Scholar] [CrossRef] [Green Version]
- Sheldon, J.R.; Skaar, E.P. Acinetobacter baumannii can use multiple siderophores for iron acquisition, but only acinetobactin is required for virulence. PLoS Pathog. 2020, 16, e1008995. [Google Scholar] [CrossRef]
- Evans, B.A.; Amyes, S.G.B. OXA β-lactamases. Clin. Microbiol. Rev. 2014, 27, 241–263. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramirez, M.S.; Bonomo, R.A.; Tolmasky, M.E. Carbapenemases: Transforming Acinetobacter baumannii into a yet more dangerous menace. Biomolecules 2020, 10, 720. [Google Scholar] [CrossRef] [PubMed]
- Joshi, P.R.; Acharya, M.; Kakshapati, T.; Leungtongkam, U.; Thummeepak, R.; Sitthisak, S. Co-existence of bla OXA-23 and bla NDM-1 genes of Acinetobacter baumannii isolated from Nepal: Antimicrobial resistance and clinical significance. Antimicrob. Resist. Infect. Control 2017, 6, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acman, M.; Wang, R.; van Dorp, L.; Shaw, L.P.; Wang, Q.; Luhmann, N.; Yin, Y.; Sun, S.; Chen, H.; Wang, H.; et al. Role of mobile genetic elements in the global dissemination of the carbapenem resistance gene blaNDM. Nat. Commun. 2022, 13, 1131. [Google Scholar] [CrossRef] [PubMed]
- Safavi, M.; Bostanshirin, N.; Hajikhani, B.; Yaslianifard, S.; van Belkum, A.; Goudarzi, M.; Hashemi, A.; Darban-Sarokhalil, D.; Dadashi, M. Global genotype distribution of human clinical isolates of New Delhi metallo-β-lactamase-producing Klebsiella pneumoniae; A systematic review. J. Glob. Antimicrob. Resist. 2020, 23, 420–429. [Google Scholar] [CrossRef]
- Kash, J.C.; Walters, K.-A.; Davis, A.S.; Sandouk, A.; Schwartzman, L.M.; Jagger, B.W.; Chertow, D.S.; Qi, L.; Kuestner, R.E.; Ozinsky, A.; et al. Lethal Synergism of 2009 Pandemic H1N1 Influenza Virus and Streptococcus pneumoniae Coinfection Is Associated with Loss of Murine Lung Repair Responses. Mbio 2011, 2, e00172-11. [Google Scholar] [CrossRef] [Green Version]
- Reinoso-Vizcaíno, N.M.; Cian, M.B.; Cortes, P.R.; Olivero, N.B.; Hernandez-Morfa, M.; Piñas, G.E.; Badapanda, C.; Rathore, A.; Perez, D.R.; Echenique, J. The pneumococcal two-component system SirRH is linked to enhanced intracellular survival of Streptococcus pneumoniae in influenza-infected pulmonary cells. PLOS Pathog. 2020, 16, e1008761. [Google Scholar] [CrossRef]
- McCullers, J.A.; Bartmess, K.C. Role of neuraminidase in lethal synergism between influenza virus and Streptococcus pneumoniae. J. Infect. Dis. 2003, 187, 1000–1009. [Google Scholar] [CrossRef] [Green Version]
- Erickson, A.K.; Jesudhasan, P.R.; Mayer, M.J.; Narbad, A.; Winter, S.E.; Pfeiffer, J.K. Bacteria Facilitate Enteric Virus Co-infection of Mammalian Cells and Promote Genetic Recombination. Cell Host Microbe. 2017, 23, 77–88.e5. [Google Scholar] [CrossRef] [Green Version]


| Strains | Isolates Date | Genome Size (bp) | GC% Content | N50 | N Contig | Coverage Depth | MLST Profile/Clonal Complex | tRNA | ncRNA | NCBI Accession Number |
|---|---|---|---|---|---|---|---|---|---|---|
| AMA166 | 2016 | 3.823.583 | 39 | 220435 | 38 | 108X | ST2/CC2 | 60 | 38 | JANKJZ000000000 |
| AMA_NO | 2021 | 4.078.010 | 39 | 125130 | 79 | 80X | ST2/CC2 | 61 | 41 | JANKKA000000000 |
| Diameters of Inhibition Zones (mm)/Minimum Inhibitory Concentrations (MICs) | ||
|---|---|---|
| AMA 166 | AMA_NO | |
| Ampicillin/sulbactam (AMS) | 6 (R) | 6 (R) |
| Amikacin (AKN) | 24 (S) | 19 (S) |
| Cefepime (FEP) | 6 (R) | 6 (R) |
| Ceftazidime (CAZ) | 19 (R) | 6 (R) |
| Ciprofloxacin (CIP) | 6 (R) | 6 (R) |
| Gentamicin (GEN) | 8 (R) | 7 (R) |
| Imipenem (IMI) | 6 (R) | 6 (R) |
| Meropenem (MEM) | 6 (R) | 6 (R) |
| Minocycline (MIN) | 20 (S) | 22 (S) |
| Tigecycline (TIG) | 20/0.5 mg/L (S) | 20/1 mg/L (S) |
| Colistin (COL) | 14/1 mg/L (S) | 14/1 mg/L (S) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Traglia, G.M.; Pasteran, F.; Escalante, J.; Nishimura, B.; Tuttobene, M.R.; Subils, T.; Nuñez, M.R.; Rivollier, M.G.; Corso, A.; Tolmasky, M.E.; et al. Genomic Comparative Analysis of Two Multi-Drug Resistance (MDR) Acinetobacter baumannii Clinical Strains Assigned to International Clonal Lineage II Recovered Pre- and Post-COVID-19 Pandemic. Biology 2023, 12, 358. https://doi.org/10.3390/biology12030358
Traglia GM, Pasteran F, Escalante J, Nishimura B, Tuttobene MR, Subils T, Nuñez MR, Rivollier MG, Corso A, Tolmasky ME, et al. Genomic Comparative Analysis of Two Multi-Drug Resistance (MDR) Acinetobacter baumannii Clinical Strains Assigned to International Clonal Lineage II Recovered Pre- and Post-COVID-19 Pandemic. Biology. 2023; 12(3):358. https://doi.org/10.3390/biology12030358
Chicago/Turabian StyleTraglia, German Matias, Fernando Pasteran, Jenny Escalante, Brent Nishimura, Marisel R. Tuttobene, Tomás Subils, Maria Rosa Nuñez, María Gabriela Rivollier, Alejandra Corso, Marcelo E. Tolmasky, and et al. 2023. "Genomic Comparative Analysis of Two Multi-Drug Resistance (MDR) Acinetobacter baumannii Clinical Strains Assigned to International Clonal Lineage II Recovered Pre- and Post-COVID-19 Pandemic" Biology 12, no. 3: 358. https://doi.org/10.3390/biology12030358
APA StyleTraglia, G. M., Pasteran, F., Escalante, J., Nishimura, B., Tuttobene, M. R., Subils, T., Nuñez, M. R., Rivollier, M. G., Corso, A., Tolmasky, M. E., & Ramirez, M. S. (2023). Genomic Comparative Analysis of Two Multi-Drug Resistance (MDR) Acinetobacter baumannii Clinical Strains Assigned to International Clonal Lineage II Recovered Pre- and Post-COVID-19 Pandemic. Biology, 12(3), 358. https://doi.org/10.3390/biology12030358









