Population Structure and Selection Signatures of Domestication in Geese
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
2.1. Sampling and Sequencing
2.2. Sequence Mapping and SNP Calling
2.3. Population Genetic Analysis
2.4. Identification of Divergent Regions
2.5. Genotype Validation of Candidate Variations
3. Results
3.1. Genetic Variation from Genome Resequencing
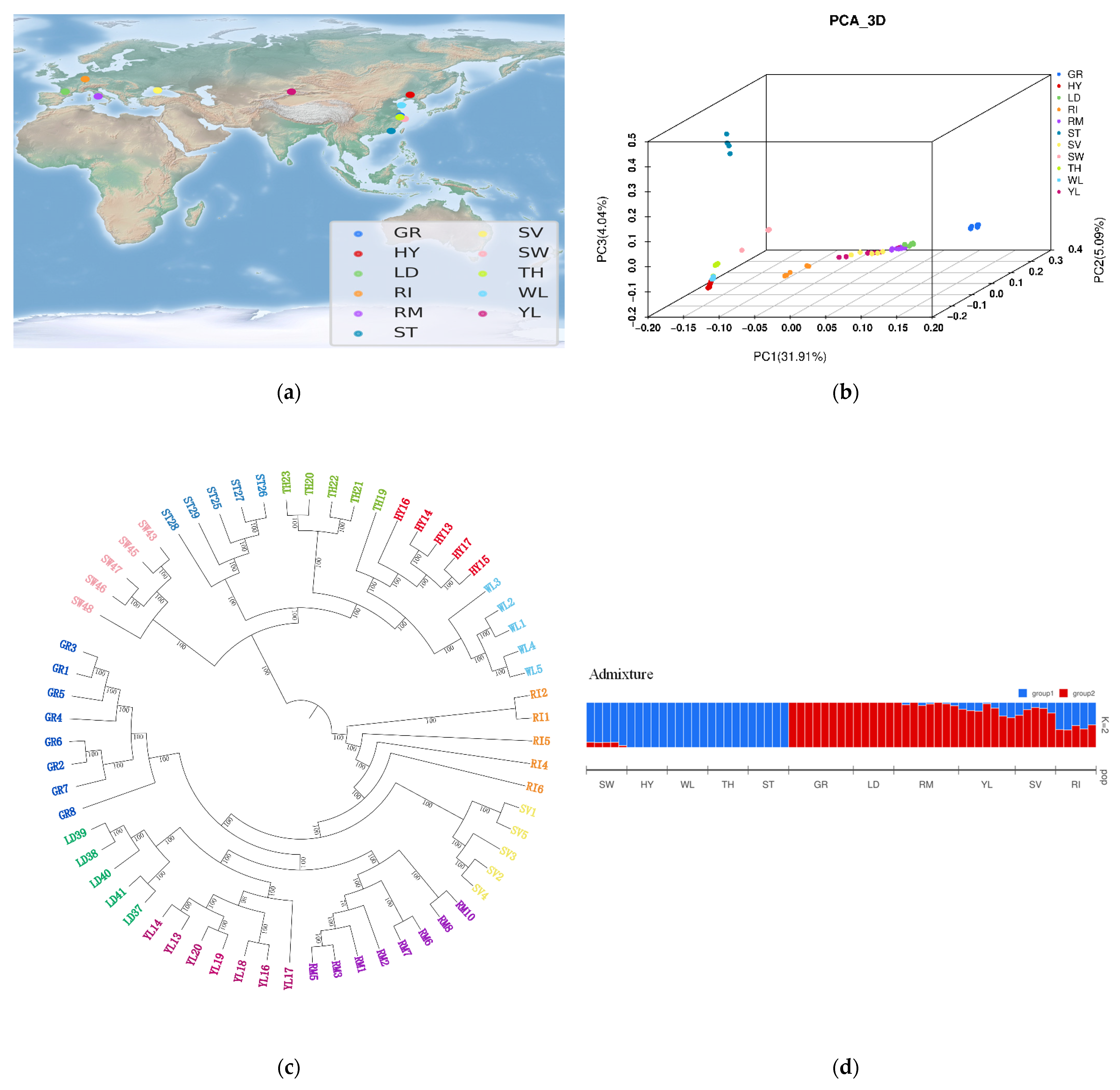
3.2. Independent Origins of Chinese and European Domestic Geese
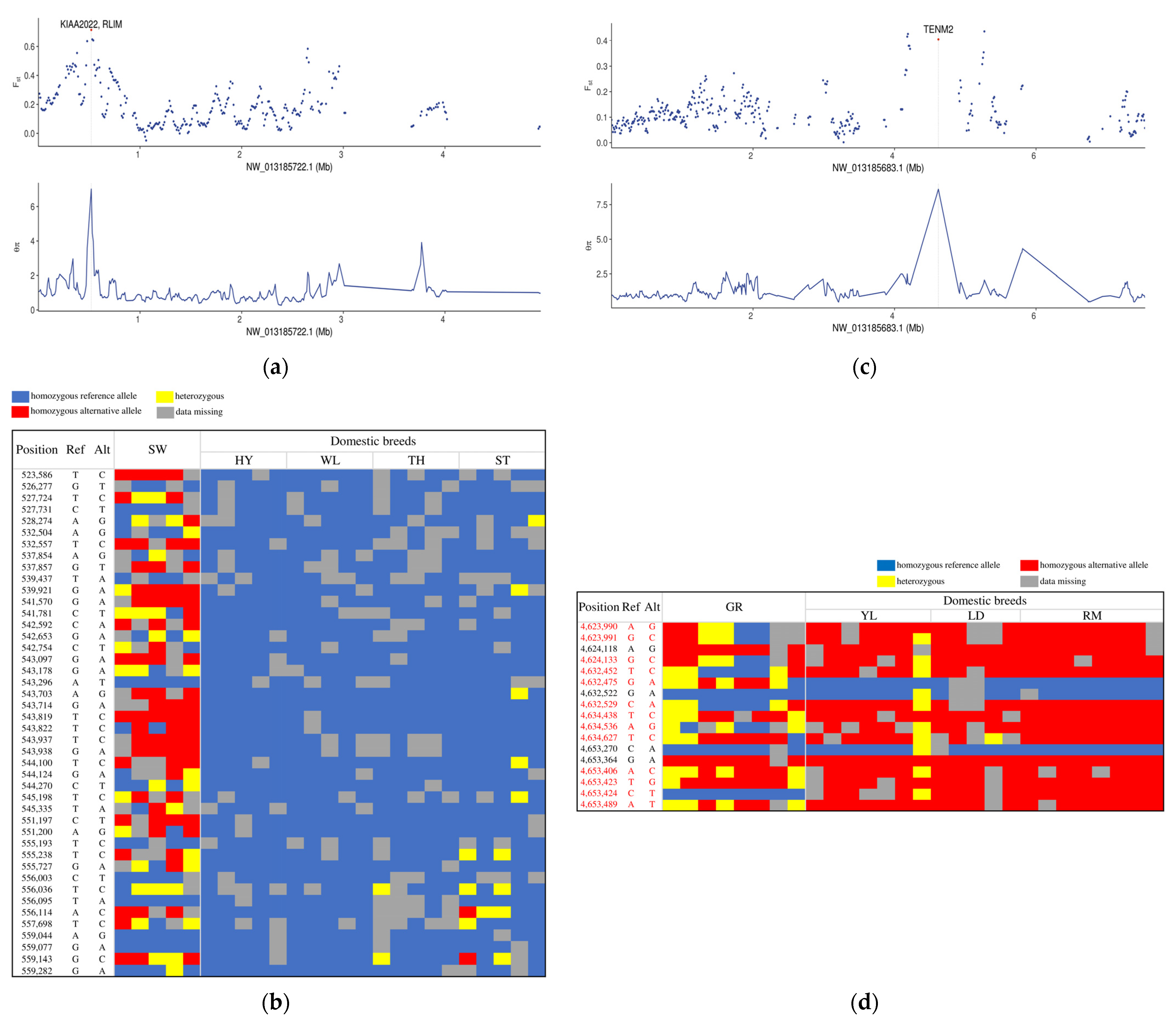
3.3. Independent Selection Signatures in Chinese and European Domestic Geese
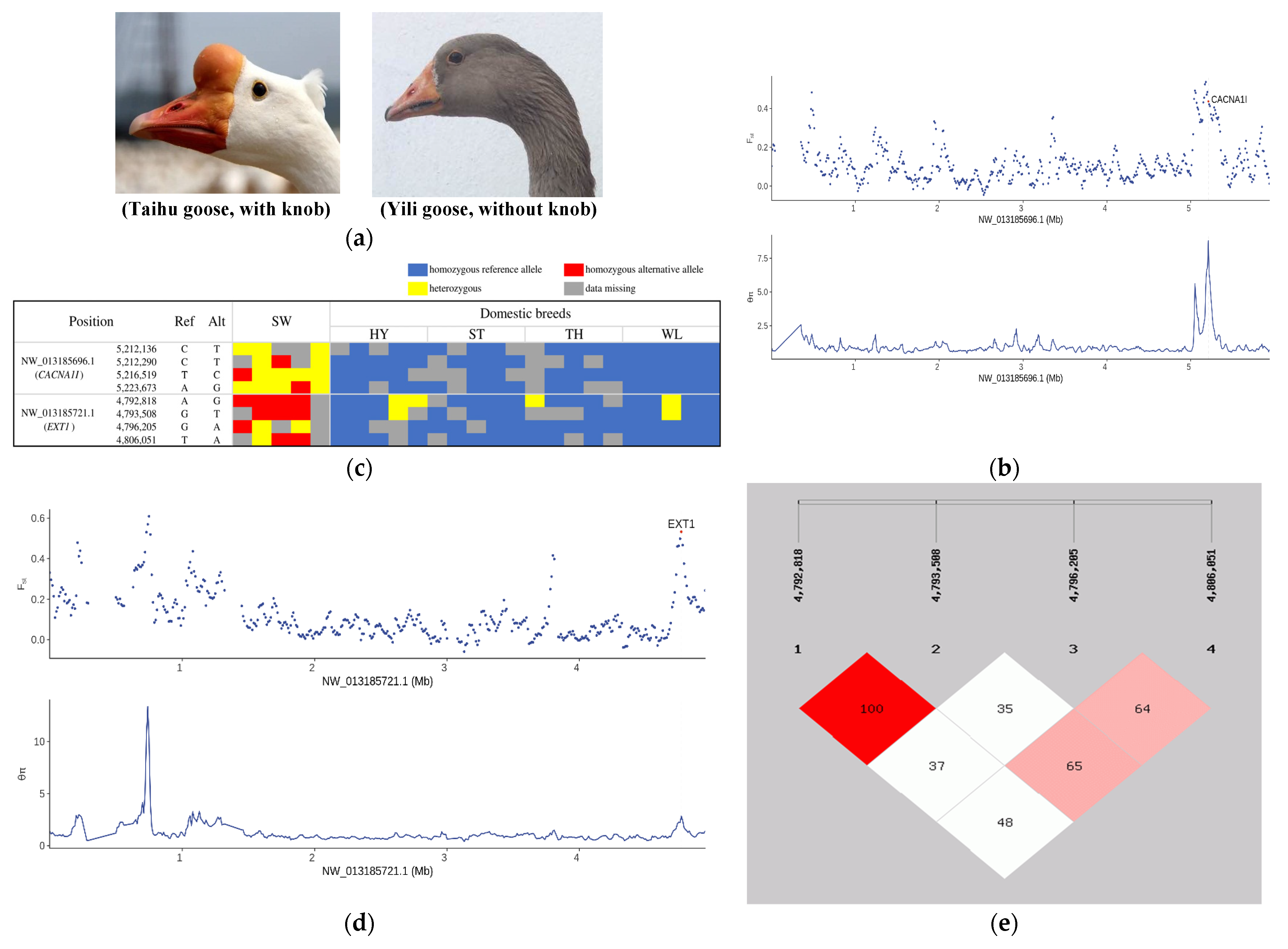
3.4. Selection Signatures Controlling Protuberant Knob
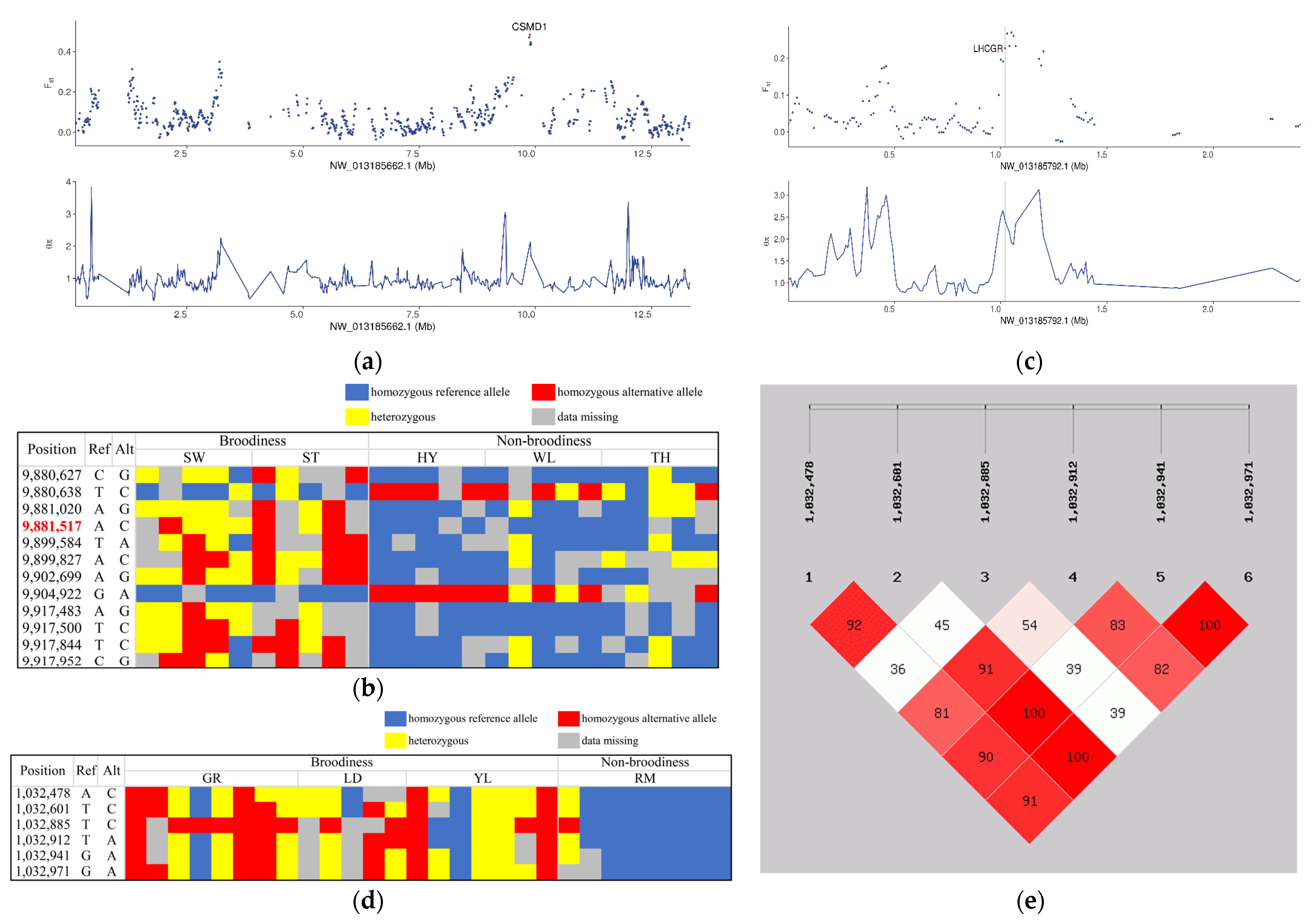
3.5. Genetic Signatures Related to Broodiness Behavior
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Zhang, Z.; Jia, Y.; Almeida, P.; Mank, J.E.; van Tuinen, M.; Wang, Q.; Jiang, Z.; Chen, Y.; Zhan, K.; Hou, S.; et al. Whole-genome resequencing reveals signatures of selection and timing of duck domestication. Gigascience 2018, 7, giy027. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Z.; Li, M.; Cheng, H.; Fan, W.; Yuan, Z.; Gao, Q.; Xu, Y.; Guo, Z.; Zhang, Y.; Hu, J.; et al. An intercross population study reveals genes associated with body size and plumage color in ducks. Nat. Commun. 2018, 9, 3974. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, X.; Yang, J.; Shen, M.; Xie, X.L.; Liu, G.J.; Xu, Y.X.; Lv, F.H.; Yang, H.; Yang, Y.L.; Liu, C.B.; et al. Whole-genome resequencing of wild and domestic sheep identifies genes associated with morphological and agronomic traits. Nat. Commun. 2020, 1, 2815. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.J.; Wang, G.D.; Ma, P.; Zhang, L.L.; Yin, T.T.; Liu, Y.H.; Otecko, N.O.; Wang, M.; Ma, Y.P.; Wang, L.; et al. Genomic regions under selection in the feralization of the dingoes. Nat. Commun. 2020, 11, 671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rubin, C.J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Chen, Y.; Wang, Z.; Li, X.; Chen, W.; Tao, Z.; Shen, J.; Tian, Y.; Wang, D.; Li, G.; et al. The goose genome sequence leads to insights into the evolution of waterfowl and susceptibility to fatty liver. Genome Biol. 2015, 16, 89. [Google Scholar] [CrossRef] [Green Version]
- Eda, M.; Itahashi, Y.; Kikuchi, H.; Sun, G.; Hsu, K.H.; Gakuhari, T.; Yoneda, M.; Jiang, L.; Yang, G.; Nakamura, S. Multiple lines of evidence of early goose domestication in a 7,000-y-old rice cultivation village in the lower Yangtze River, China. Proc. Natl. Acad. Sci. USA 2022, 119, e2117064119. [Google Scholar] [CrossRef]
- Albarella, U. Alternate fortunes? The role of domestic ducks and geese from Roman to Medieval times in Britain. In Documenta Archaeobiologiae III. Feathers, Grit and Symbolism; Grupe, G., Peters, J., Eds.; VML Verlag Marie Leidorf GmbH: Rahden, Germany, 2005; pp. 249–258. [Google Scholar]
- Ouyang, J.; Zheng, S.; Huang, M.; Tang, H.; Qiu, X.; Chen, S.; Wang, Z.; Zhou, Z.; Gao, Y.; Xiong, Y.; et al. Chromosome-level genome and population genomics reveal evolutionary characteristics and conservation status of Chinese indigenous geese. Commun. Biol. 2022, 5, 1191. [Google Scholar] [CrossRef]
- Li, H.F.; Zhu, W.Q.; Chen, K.W.; H, Y.; Xu, W.J.; Song, W. Two maternal origins of Chinese domestic goose. Poult. Sci. 2011, 90, 2705–2710. [Google Scholar] [CrossRef]
- Ren, T.; Liang, S.; Zhao, A.; He, K. Analysis of the complete mitochondrial genome of the Zhedong White goose and characterization of NUMTs: Reveal domestication history of goose in China and Euro. Gene 2016, 577, 75–81. [Google Scholar] [CrossRef]
- Heikkinen, M.E.; Ruokonen, M.; White, T.A.; Alexander, M.M.; Gündüz, İ.; Dobney, K.M.; Aspi, J.; Searle, J.B.; Pyhäjärvi, T. Long-Term Reciprocal Gene Flow in Wild and Domestic Geese Reveals Complex Domestication History. G3 Genes Genomes Genet. 2020, 10, 3061–3070. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:1303.3997v2. [Google Scholar]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Lee, S.H.; Goddard, M.E.; Visscher, P.M. GCTA: A tool for genome-wide complex trait analysis. Am. J. Hum. Genet. 2011, 88, 76–82. [Google Scholar] [CrossRef] [Green Version]
- Stamatakis, A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 2006, 22, 2688–2690. [Google Scholar] [CrossRef] [Green Version]
- Alexander, D.H.; Novembre, J.; Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009, 19, 1655–1664. [Google Scholar] [CrossRef] [Green Version]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. 1000 Genomes Project Analysis Group. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Magome, T.; Hattori, T.; Taniguchi, M.; Ishikawa, T.; Miyata, S.; Yamada, K.; Takamura, H.; Matsuzaki, S.; Ito, A.; Tohyama, M.; et al. XLMR protein related to neurite extension (Xpn/KIAA2022) regulates cell-cell and cell-matrix adhesion and migration. Neurochem. Int. 2013, 63, 561–569. [Google Scholar] [CrossRef]
- Del Toro, D.; Carrasquero-Ordaz, M.A.; Chu, A.; Ruff, T.; Shahin, M.; Jackson, V.A.; Chavent, M.; Berbeira-Santana, M.; Seyit-Bremer, G.; Brignani, S.; et al. Structural Basis of Teneurin-Latrophilin Interaction in Repulsive Guidance of Migrating Neurons. Cell 2020, 180, 323–339.e19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, W.; Hou, L.E.; Yuan, X.; Gu, T.; Chen, Z.; Zhang, Y.; Zhang, Y.; Chen, G.; Xu, Q.; Zhao, W. Identifying molecular pathways and candidate genes associated with knob traits by transcriptome analysis in the goose (Anser cygnoides). Sci. Rep. 2021, 11, 11978. [Google Scholar] [CrossRef] [PubMed]
- Deng, Y.; Hu, S.; Luo, C.; Ouyang, Q.; Li, L.; Ma, J.; Lin, Z.; Chen, J.; Liu, H.; Hu, J.; et al. Integrative analysis of histomorphology, transcriptome and whole genome resequencing identified DIO2 gene as a crucial gene for the protuberant knob located on forehead in geese. BMC Genom. 2021, 22, 487. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Sun, X.; Chimbaka, I.M.; Qin, N.; Xu, X.; Liswaniso, S.; Xu, R.; Gonzalez, J.M. Transcriptome Analysis of Ovarian Follicles Reveals Potential Pivotal Genes Associated with Increased and Decreased Rates of Chicken Egg Production. Front. Genet. 2021, 12, 622751. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.D.; Zhai, W.; Yang, H.C.; Fan, R.X.; Cao, X.; Zhong, L.; Wang, L.; Liu, F.; Wu, H.; Cheng, L.G.; et al. The genomics of selection in dogs and the parallel evolution between dogs and humans. Nat. Commun. 2013, 4, 1860. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ward, S.; Bishop, C.M.; Woakes, A.J.; Butler, P.J. Heart rate and the rate of oxygen consumption of flying and walking barnacle geese (Branta leucopsis) and bar-headed geese (Anser indicus). J. Exp. Biol. 2002, 205 Pt 21, 3347–3356. [Google Scholar] [CrossRef]
- Wang, M.S.; Li, Y.; Peng, M.S.; Zhong, L.; Wang, Z.J.; Li, Q.Y.; Tu, X.L.; Dong, Y.; Zhu, C.L.; Wang, L.; et al. Genomic Analyses Reveal Potential Independent Adaptation to High Altitude in Tibetan Chickens. Mol. Biol. Evol. 2015, 32, 1880–1889. [Google Scholar] [CrossRef]
- Projecto-Garcia, J.; Natarajan, C.; Moriyama, H.; Weber, R.E.; Fago, A.; Cheviron, Z.A.; Dudley, R.; McGuire, J.A.; Witt, C.C.; Storz, J.F. Repeated elevational transitions in hemoglobin function during the evolution of Andean hummingbirds. Proc. Natl. Acad. Sci. USA 2013, 110, 20669–20674. [Google Scholar] [CrossRef] [Green Version]
- Zeder, M.A.; Hesse, B. The initial domestication of goats (Capra hircus) in the Zagros Mountains 10,000 years ago. Science 2000, 287, 2254–2257. [Google Scholar] [CrossRef]
- Mannermaa, K. Goose: Domestication. In Encyclopedia of Global Archaeology; Smith, C., Ed.; Springer: New York, NY, USA, 2014; pp. 3096–3098. [Google Scholar]
- Wang, M.S.; Zhang, R.W.; Su, L.Y.; Li, Y.; Peng, M.S.; Liu, H.Q.; Zeng, L.; Irwin, D.M.; Du, J.L.; Yao, Y.G.; et al. Positive selection rather than relaxation of functional constraint drives the evolution of vision during chicken domestication. Cell Res. 2016, 26, 556–573. [Google Scholar] [CrossRef]
- Peichl, L. Topography of ganglion cells in the dog and wolf retina. J. Comp. Neurol. 1992, 324, 603–620. [Google Scholar] [CrossRef]
- Evans, K.E.; McGreevy, P.D. The distribution of ganglion cells in the equine retina and its relationship to skull morphology. Anat. Histol. Embryol. 2007, 36, 151–156. [Google Scholar] [CrossRef]
- Ji, W.Y. Anatomical, histological characteristic and genetic basis of different size knobs in geese (Anser Cygnoides). Master’s Thesis, Yangzhou University, Yangzhou, China, 2022. [Google Scholar]
- Imsland, F.; Feng, C.; Boije, H.; Bed’hom, B.; Fillon, V.; Dorshorst, B.; Rubin, C.J.; Liu, R.; Gao, Y.; Gu, X.; et al. The Rose-comb mutation in chickens constitutes a structural rearrangement causing both altered comb morphology and defective sperm motility. PLoS Genet. 2012, 8, e1002775. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vanpé, C.; Gaillard, J.M.; Kjellander, P.; Mysterud, A.; Magnien, P.; Delorme, D.; Van Laere, G.; Klein, F.; Liberg, O.; Hewison, A.J. Antler size provides an honest signal of male phenotypic quality in roe deer. Am. Nat. 2007, 169, 481–493. [Google Scholar] [CrossRef]
- Ferns, P.N.; Reed, J.P.; Gregg, E.D.; O’Hara, C.; Lang, A.; Sinkowski, G.R. Bill knob size and reproductive effort in common shelducks tadorna tadorna. Wildfowl 2005, 55, 49–60. [Google Scholar]
- Al-Zayed, Z.; Al-Rijjal, R.A.; Al-Ghofaili, L.; BinEssa, H.A.; Pant, R.; Alrabiah, A.; Al-Hussainan, T.; Zou, M.; Meyer, B.F.; Shi, Y. Mutation spectrum of EXT1 and EXT2 in the Saudi patients with hereditary multiple exostoses. Orphanet. J. Rare Dis. 2021, 16, 100. [Google Scholar] [CrossRef] [PubMed]
- Maas, S.M.; Shaw, A.C.; Bikker, H.; Lüdecke, H.J.; van der Tuin, K.; Badura-Stronka, M.; Belligni, E.; Biamino, E.; Bonati, M.T.; Carvalho, D.R.; et al. Phenotype and genotype in 103 patients with tricho-rhino-phalangeal syndrome. Eur. J. Med. Genet. 2015, 58, 279–292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, A.S.; Rusch, J.; Lima, A.C.; Usmani, A.; Huang, N.; Lepamets, M.; Vigh-Conrad, K.A.; Worthington, R.E.; Mägi, R.; Wu, X.; et al. Rare Mutations in the Complement Regulatory Gene CSMD1 Are Associated with Male and Female Infertility. Nat. Commun. 2019, 10, 4626. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Bai, X.; Liu, H.; Zhao, B.; Yan, Z.; Hou, Y.; Chu, Q. Population Genomic Sequencing Delineates Global Landscape of Copy Number Variations that Drive Domestication and Breed Formation of in Chicken. Front. Genet. 2022, 13, 830393. [Google Scholar] [CrossRef]
- Oduwole, O.O.; Huhtaniemi, I.T.; Misrahi, M. The Roles of Luteinizing Hormone, Follicle-Stimulating Hormone and Testosterone in Spermatogenesis and Folliculogenesis Revisited. Int. J. Mol. Sci. 2021, 22, 12735. [Google Scholar] [CrossRef]
- Prabhudesai, K.S.; Raje, S.; Dhamanaskar, A.; Modi, D.; Dighe, V.; Contini, A.; Idicula-Thomas, S. Identification and in vivo validation of a 9-mer peptide derived from FSHβ with FSHR antagonist activity. Peptides 2020, 132, 170367. [Google Scholar] [CrossRef]
| Scaffold | Start (bp) | End (bp) | θπ Ratio (Wild/Domestic) | FST | Gene Name | Functions |
|---|---|---|---|---|---|---|
| NW_013185722.1 | 520,001 | 560,001 | 7.02084 | 0.714427 | RLIM | Innate immune system pathway |
| NW_013185722.1 | 520,001 | 560,001 | 7.02084 | 0.714427 | KIAA2022 | Involved in neurite outgrowth |
| NW_013185659.1 | 5,240,001 | 5,280,001 | 5.69171 | 0.555266 | AKT3 | Hippocampal neurogenesis |
| NW_013185696.1 | 5,050,001 | 5,090,001 | 5.6146 | 0.492042 | RPS19BP1 | Cellular responses to stimuli |
| NW_013185696.1 | 5,180,001 | 5,220,001 | 5.57682 | 0.536577 | CACNA1I | Involved in sensory processing, sleep, and hormone and neurotransmitter release |
| NW_013185664.1 | 5,530,001 | 5,570,001 | 5.48137 | 0.47688 | LPIN1 | Involved in lipid metabolism |
| NW_013185657.1 | 7,890,001 | 7,930,001 | 4.82417 | 0.480043 | PTGR2 | Arachidonic acid metabolism |
| NW_013185662.1 | 12,000,001 | 12,040,001 | 4.62785 | 0.48803 | DLGAP2 | Plays a role in synapse organization and signaling in neuronal cells |
| NW_013185657.1 | 7,910,001 | 7,950,001 | 4.4634 | 0.493049 | UBR1 | Class I MHC mediated antigen processing and presentation pathway |
| NW_013185706.1 | 4,210,001 | 4,250,001 | 4.30241 | 0.644791 | ANKS1B | Brain development |
| NW_013185706.1 | 4,210,001 | 4,250,001 | 4.30241 | 0.644791 | APAF1 | Visual system |
| NW_013185654.1 | 22,740,001 | 22,780,001 | 4.0863 | 0.572681 | KAT6B | Involved in cerebral cortex development |
| NW_013185779.1 | 1,580,001 | 1,620,001 | 4.04274 | 0.362452 | PAPPA2 | Regulates bone structure and mass |
| NW_013185885.1 | 180,001 | 220,001 | 4.00233 | 0.444173 | HTR1D | Affects neural activity |
| NW_013185716.1 | 70,001 | 110,001 | 3.98051 | 0.441914 | LDLRAP1 | Lipid metabolism |
| NW_013185722.1 | 480,001 | 520,001 | 3.58249 | 0.636686 | SLC16A2 | Development of central nervous system |
| NW_013185930.1 | 710,001 | 750,001 | 3.4615 | 0.45944 | BEGAIN | Regulates postsynaptic neurotransmitter receptor activity |
| NW_013185657.1 | 8,100,001 | 8,140,001 | 3.31629 | 0.63377 | TTBK2 | Involved in atrophy of the cerebellum and brainstem |
| NW_013185657.1 | 8,130,001 | 8,170,001 | 3.13448 | 0.695921 | STARD9 | Lipid binding |
| NW_013185657.1 | 8,130,001 | 8,170,001 | 3.13448 | 0.695921 | CDAN1 | Essential for primitive erythropoiesis |
| NW_013185662.1 | 11,990,001 | 12,030,001 | 3.06677 | 0.490442 | CLN8 | Involved in neuronal differentiation |
| NW_013185654.1 | 6,470,001 | 6,510,001 | 2.99721 | 0.610978 | PAX2 | Participates in optic nerve development |
| NW_013185885.1 | 130,001 | 170,001 | 2.93483 | 0.510357 | LUZP1 | Affects neural tube |
| NW_013185657.1 | 8,230,001 | 8,270,001 | 2.92731 | 0.597566 | SNAP23 | Class I MHC mediated antigen processing and presentation pathway |
| NW_013185662.1 | 11,980,001 | 12,020,001 | 2.92009 | 0.469008 | ARHGEF10 | Neural morphogenesis |
| NW_013185663.1 | 10,900,001 | 10,940,001 | 2.83154 | 0.520603 | NCOA2 | Circadian clock pathway; Glucose metabolism regulation |
| NW_013185779.1 | 1,550,001 | 1,590,001 | 2.61207 | 0.494237 | RFWD2 | Class I MHC mediated antigen processing and presentation pathway |
| NW_013185657.1 | 9,070,001 | 9,110,001 | 2.55479 | 0.512538 | TYRO3 | Innate immune response; Neuron protection |
| NW_013185799.1 | 2,270,001 | 2,310,001 | 2.45909 | 0.493535 | BACH1 | Heme binding |
| NW_013185672.1 | 7,580,001 | 7,620,001 | 2.35532 | 0.475914 | HBS1L | Controls fetal hemoglobin level |
| NW_013185672.1 | 7,580,001 | 7,620,001 | 2.35532 | 0.475914 | ALDH8A1 | Visual system |
| NW_013185666.1 | 8,860,001 | 8,900,001 | 2.30898 | 0.484818 | PI4KA | Neurodevelopment |
| NW_013185741.1 | 2,060,001 | 2,100,001 | 2.22717 | 0.450407 | DHRS3 | Visual phototransduction pathway |
| NW_013185799.1 | 2,230,001 | 2,270,001 | 2.19869 | 0.566629 | GRIK1 | Involved in transmission of light information |
| NW_013185722.1 | 2,650,001 | 2,690,001 | 2.1901 | 0.584116 | SMARCA1 | Promotes brain development |
| NW_013185657.1 | 9,010,001 | 9,050,001 | 1.92798 | 0.56243 | MAPKBP1 | Immune function |
| NW_013185885.1 | 80,001 | 120,001 | 1.84796 | 0.657325 | KDM1A | Involved in blood cell development |
| NW_013185661.1 | 4,790,001 | 4,830,001 | 1.81422 | 0.484537 | JAZF1 | Glucose and lipid metabolism |
| NW_013185657.1 | 8,260,001 | 8,300,001 | 1.72594 | 0.577736 | ZNF106 | Essential for skeletal muscle function |
| NW_013185769.1 | 770,001 | 810,001 | 1.69181 | 0.517866 | FOXP2 | Neurodevelopment |
| NW_013185657.1 | 7,400,001 | 7,440,001 | 1.68652 | 0.481765 | DPF3 | Plays an essential role in heart and skeletal muscle development |
| NW_013185722.1 | 700,001 | 740,001 | 1.68606 | 0.446956 | ABCB7 | Involved in the transport of heme |
| NW_013185682.1 | 300,001 | 340,001 | 1.5411 | 0.478458 | SHPRH | Metabolism of proteins pathway |
| NW_013185746.1 | 1,540,001 | 1,580,001 | 1.52872 | 0.478827 | NLGN4X | Remodels central nervous system synapses |
| Scaffold | Start (bp) | End (bp) | θπ Ratio (Wild/Domestic) | FST | Gene Name | Functions |
|---|---|---|---|---|---|---|
| NW_013185673.1 | 20,001 | 60,001 | 1.7753 | 0.573003 | TRAPPC3L | Bone development |
| NW_013185676.1 | 6,660,001 | 6,700,001 | 7.19466 | 0.456279 | WWOX | Bone development |
| NW_013185673.1 | 200,001 | 240,001 | 3.73375 | 0.230746 | NT5DC1 | Bone development |
| NW_013185673.1 | 230,001 | 270,001 | 3.52744 | 0.380911 | COL10A1 | Bone development |
| NW_013185655.1 | 5,790,001 | 5,830,001 | 2.06836 | 0.416861 | HMX1 | Development |
| NW_013185859.1 | 1,260,001 | 1,300,001 | 1.77307 | 0.431281 | BLMH | Immunity |
| NW_013185855.1 | 1,150,001 | 1,190,001 | 4.47664 | 0.322131 | EDA2R | Immunity |
| NW_013185882.1 | 440,001 | 480,001 | 1.70704 | 0.44064 | BANP | Immunity |
| NW_013185655.1 | 16,110,001 | 16,150,001 | 3.93205 | 0.381584 | BCL11B | Immunity |
| NW_013185714.1 | 560,001 | 600,001 | 1.84461 | 0.413279 | SLC25A5 | Metabolism |
| NW_013185676.1 | 3,500,001 | 3,540,001 | 3.6863 | 0.236796 | FAM96B | Metabolism |
| NW_013185718.1 | 2,100,001 | 2,140,001 | 3.60565 | 0.280045 | MCAT | Metabolism |
| NW_013185718.1 | 2,100,001 | 2,140,001 | 3.60565 | 0.280045 | TSPO | Metabolism |
| NW_013185718.1 | 2,100,001 | 2,140,001 | 3.60565 | 0.280045 | TTLL12 | Metabolism |
| NW_013185673.1 | 30,001 | 70,001 | 2.01444 | 0.48736 | DSE | Metabolism |
| NW_013185667.1 | 9,680,001 | 9,720,001 | 4.9104 | 0.391882 | XDH | Metabolism |
| NW_013185667.1 | 9,870,001 | 9,910,001 | 3.73004 | 0.253108 | GALNT14 | Metabolism |
| NW_013185656.1 | 15,270,001 | 15,310,001 | 2.0786 | 0.410677 | GBE1 | Metabolism |
| NW_013185683.1 | 4,620,001 | 4,660,001 | 8.61059 | 0.405009 | TENM2 | Nervous system |
| NW_013185656.1 | 17,300,001 | 17,340,001 | 7.02891 | 0.241569 | ROBO2 | Nervous system |
| NW_013185659.1 | 14,670,001 | 14,710,001 | 4.59723 | 0.278432 | PRKCE | Nervous system |
| NW_013185792.1 | 190,001 | 230,001 | 4.44657 | 0.35268 | ALK | Nervous system |
| NW_013185660.1 | 4,120,001 | 4,160,001 | 3.69867 | 0.261144 | SLC4A10 | Nervous system |
| NW_013186039.1 | 90,001 | 130,001 | 3.64631 | 0.340177 | FLOT2 | Nervous system |
| NW_013185810.1 | 920,001 | 960,001 | 3.50133 | 0.403726 | EXOC2 | Nervous system |
| NW_013185725.1 | 2,390,001 | 2,430,001 | 3.37828 | 0.281384 | TRAPPC6B | Nervous system |
| NW_013185725.1 | 2,390,001 | 2,430,001 | 3.37828 | 0.281384 | GEMIN2 | Nervous system |
| NW_013185656.1 | 10,860,001 | 10,900,001 | 0.459251 | 0.459251 | EPHA6 | Nervous system |
| NW_013185745.1 | 430,001 | 470,001 | 1.74703 | 0.433555 | GRM8 | Nervous system |
| NW_013185859.1 | 1,260,001 | 1,300,001 | 1.77307 | 0.431281 | SLC6A4 | Nervous system |
| NW_013185702.1 | 3,680,001 | 3,720,001 | 2.04116 | 0.42798 | SEMA5A | Nervous system |
| NW_013185677.1 | 5,000,001 | 5,040,001 | 4.49648 | 0.375977 | DNAH3 | Reproduction |
| NW_013185676.1 | 6,370,001 | 6,410,001 | 4.84849 | 0.467322 | ADAMTS18 | Visual system |
| NW_013185868.1 | 780,001 | 820,001 | 4.031 | 0.220905 | OPTN | Visual system |
| NW_013186039.1 | 90,001 | 130,001 | 3.64631 | 0.340177 | ERAL1 | Growth |
| NW_013185664.1 | 7,050,001 | 7,090,001 | 7.54069 | 0.219905 | NBAS | Growth; Visual system |
| NW_013185703.1 | 1,950,001 | 1,990,001 | 2.17876 | 0.42861 | JAG1 | Hematopoiesis |
| Breed/Species | N | Segregation of Genotype with Phenotype (Yes/No) | |||
|---|---|---|---|---|---|
| 4,792,818 | 4,793,508 | 4,796,205 | 4,806,051 | ||
| Zhedong goose | 17 | 17/0 | 17/0 | 17/0 | 13/4 |
| Panshi grey goose | 18 | 18/0 | 18/0 | 18/0 | 18/0 |
| Yongkang grey goose | 20 | 20/0 | 20/0 | 20/0 | 10/10 |
| Swan goose | 7 | 7/0 | 7/0 | 5/2 | 2/5 |
| Total | 62 | 62/0 | 62/0 | 60/2 | 43/19 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, L.; Cao, Y.; Li, G.; Tian, Y.; Zeng, T.; Gu, T.; Xu, W.; Konoval, O.; Lu, L. Population Structure and Selection Signatures of Domestication in Geese. Biology 2023, 12, 532. https://doi.org/10.3390/biology12040532
Chen L, Cao Y, Li G, Tian Y, Zeng T, Gu T, Xu W, Konoval O, Lu L. Population Structure and Selection Signatures of Domestication in Geese. Biology. 2023; 12(4):532. https://doi.org/10.3390/biology12040532
Chicago/Turabian StyleChen, Li, Yongqing Cao, Guoqin Li, Yong Tian, Tao Zeng, Tiantian Gu, Wenwu Xu, Oksana Konoval, and Lizhi Lu. 2023. "Population Structure and Selection Signatures of Domestication in Geese" Biology 12, no. 4: 532. https://doi.org/10.3390/biology12040532





