Effects of a Nanonetwork-Structured Soil Conditioner on Microbial Community Structure
Abstract
:Simple Summary
Abstract
1. Introduction
2. Materials and Methods
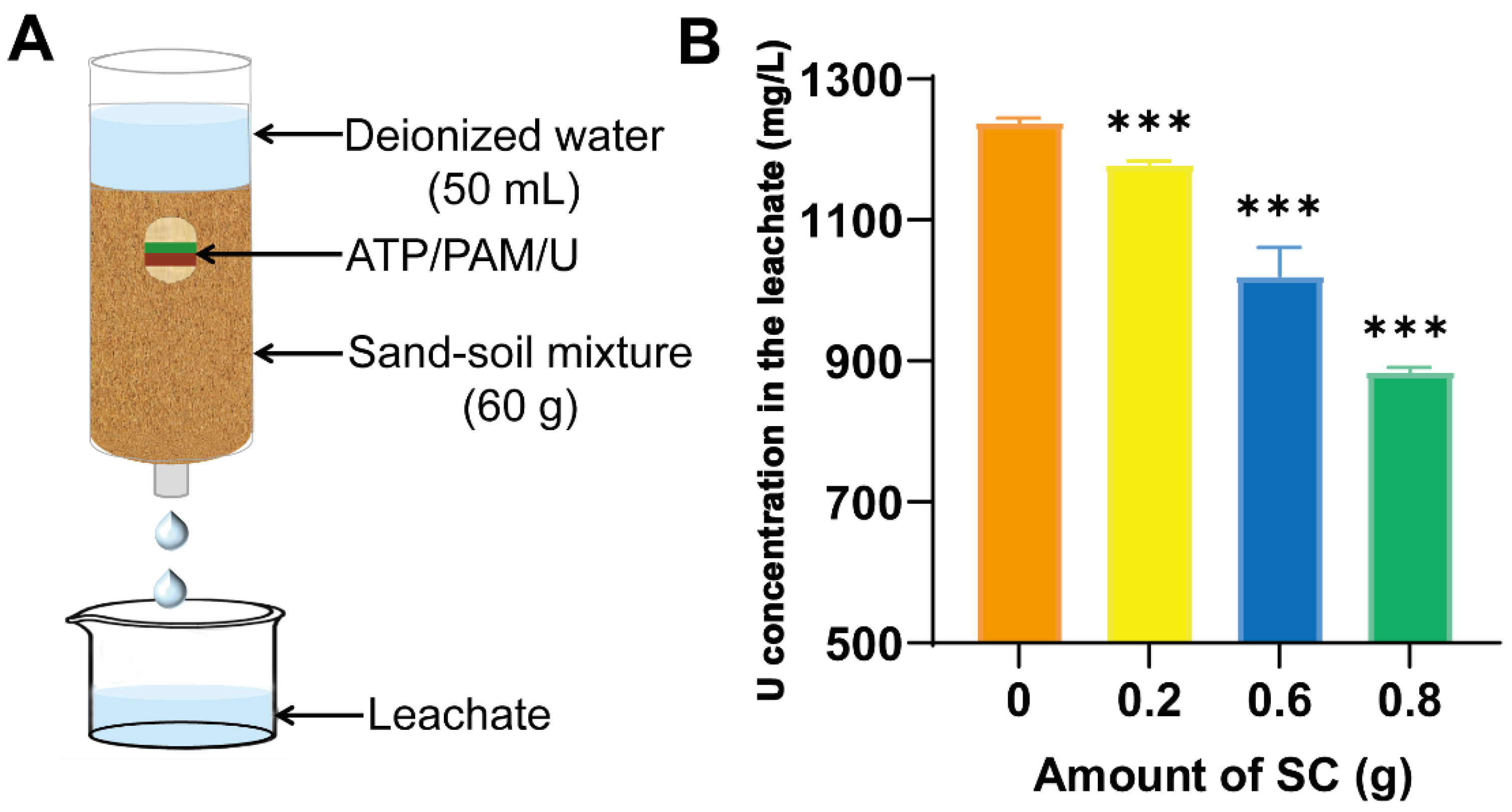
2.1. Preparation of Soil Conditioner and Leaching Assay
2.2. Interaction Analyses of Soil Conditioner
2.3. Greenhouse Experiment and Design
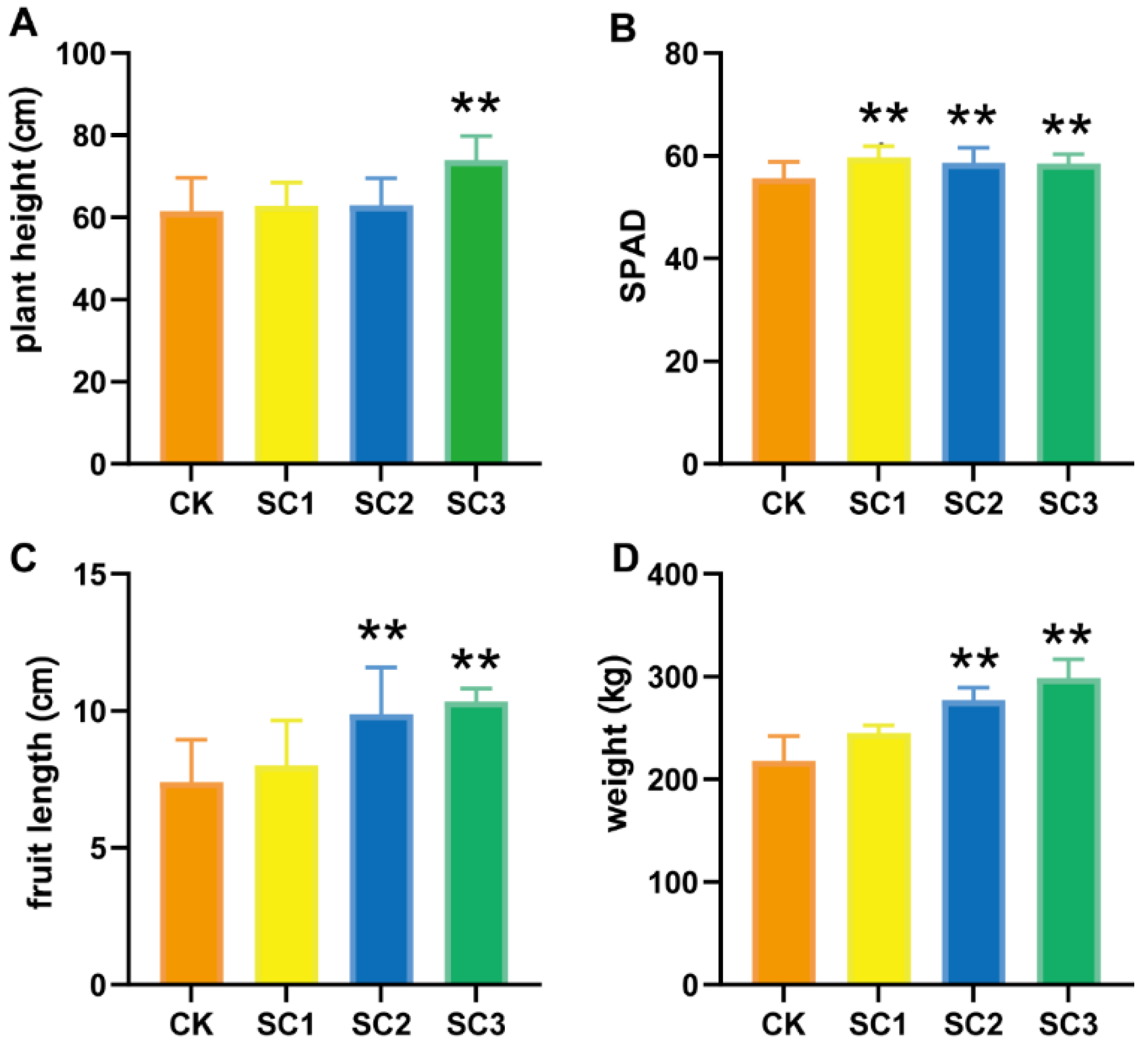
2.4. Plant Growth, Soil Collection, and Properties
2.5. High-Throughput Sequencing of the Microbe Communities and Data Processing
2.6. Statistical Analysis
3. Results
3.1. Effect of Controlling Urea Leaching Using the Soil Conditioner
3.2. Interaction Characteristics of Soil Conditioner Components
3.3. Effects of Soil Conditioner on Plant Growth
3.4. Effect of Soil Conditioner on Soil Characteristics
3.5. The Microbe Communities Identified Using High-Throughput Sequencing
3.5.1. Alpha Diversity of the Microbial Community
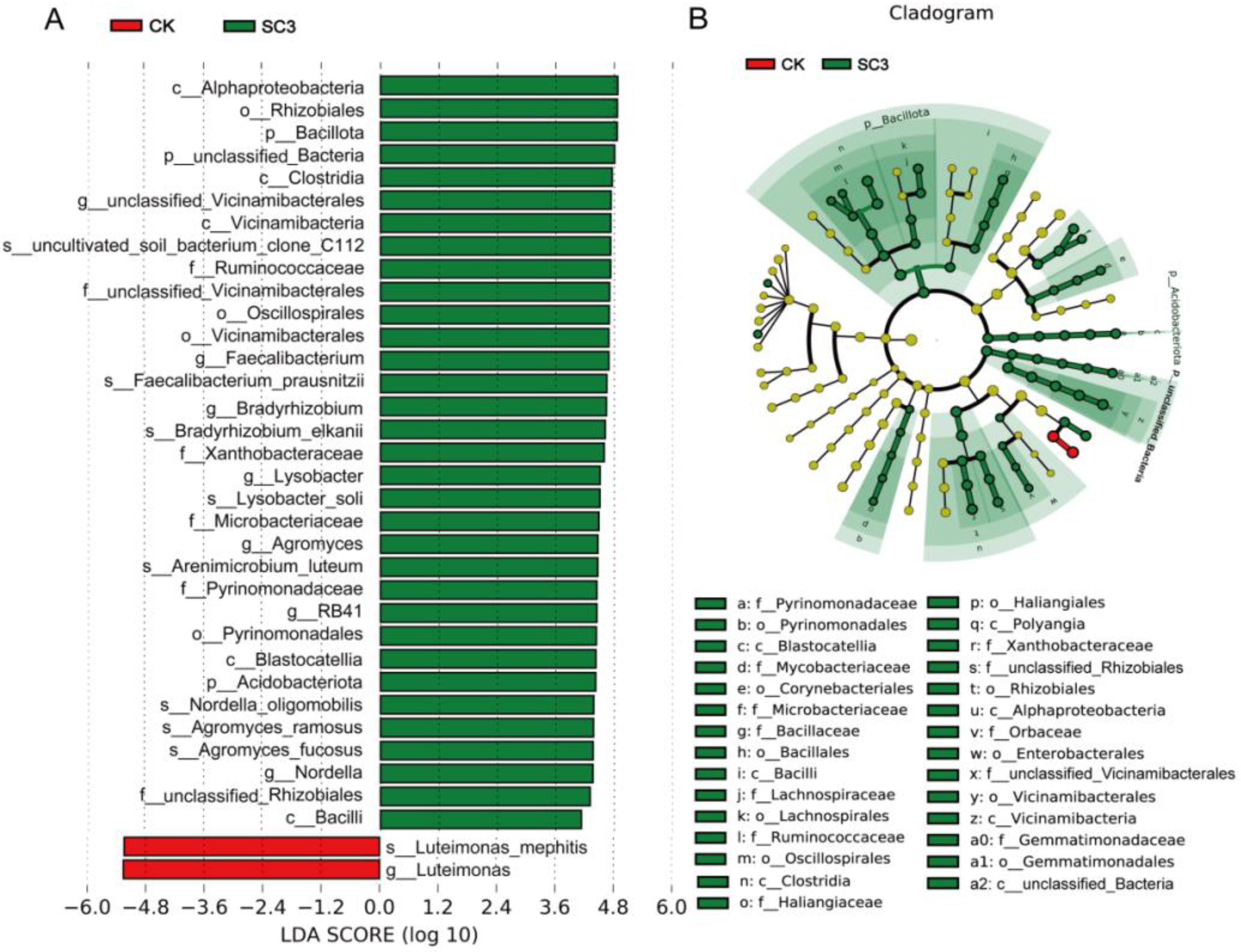
3.5.2. Comparison of Bacterial Community Composition Based on 16S rRNA Sequencing
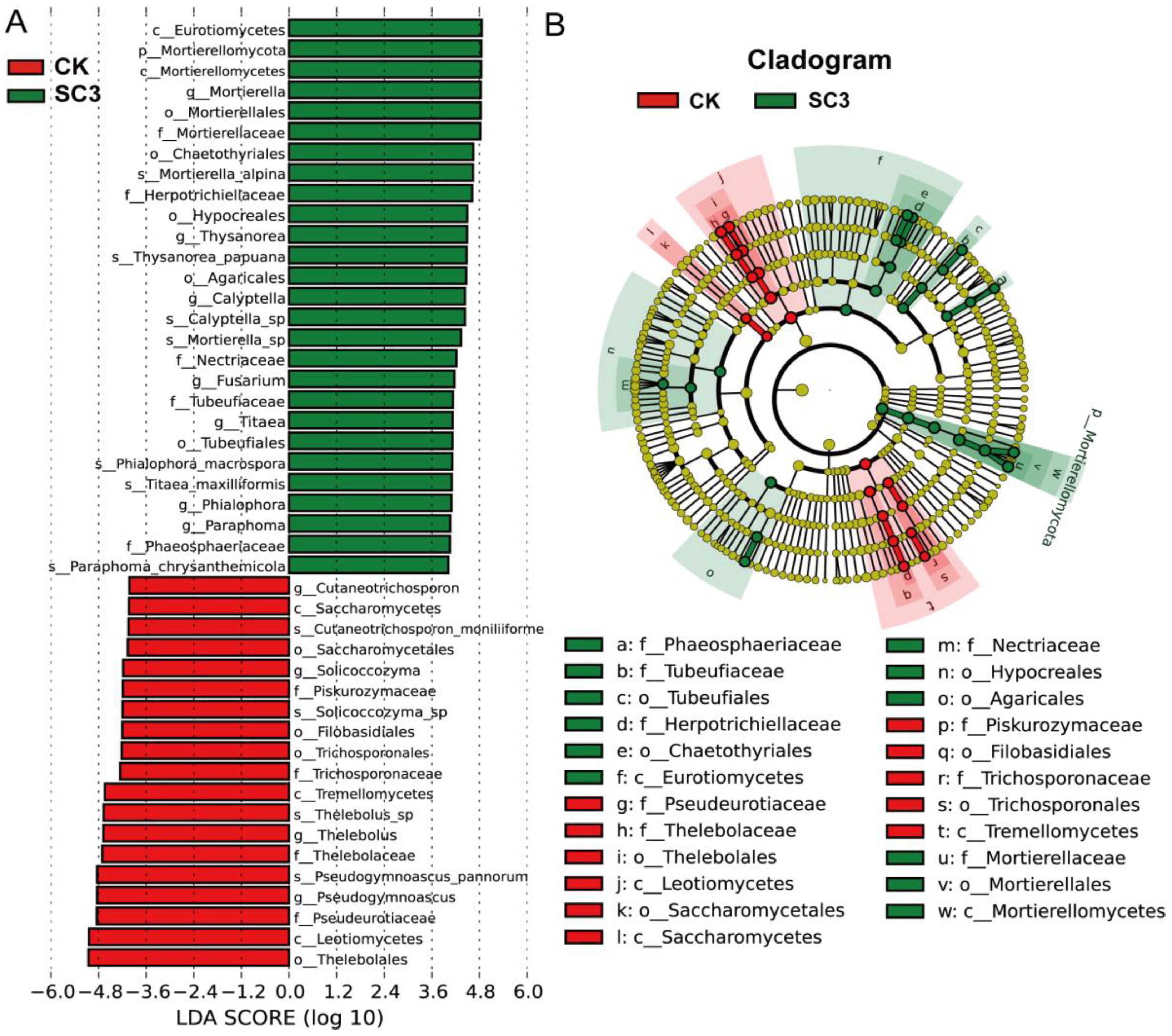
3.5.3. Comparison of Fungal Community Composition Based on ITS Sequencing
3.5.4. Correlation Analysis between Microbes and Environmental Parameters
3.5.5. Predicted Functional Gene Analysis for Microbial Communities
4. Discussion
4.1. Material Characteristics of Nanostructured Soil Conditioner and Characteristics of Reducing Nitrogen Loss
4.2. Effect of SC on Soil Physicochemical Properties
4.3. Effect of SC on Microbial Community Structure
4.3.1. Effect of SC on Bacterial Microorganisms
4.3.2. Effect of SC on Fungal Microorganisms
4.4. Relationship between Soil Physicochemical Properties and Microbial Community after Application of SC
4.5. Predicted Functional Analysis for Microbial Communities
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fierer, N.; Wood, S.A.; Bueno de Mesquita, C.P. How microbes can, and cannot, be used to assess soil health. Soil. Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Cai, D.; Wu, Z.; Jiang, J.; Wu, Y.; Feng, H.; Brown, I.G.; Chu, P.K.; Yu, Z. Controlling nitrogen migration through micro-nano networks. Sci. Rep. 2014, 4, 3665. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.; Ji, H.; Sheng, J.; Zhang, Y.; Feng, Y.; Guo, Z.; Chen, L. Combining Azolla and urease inhibitor to reduce ammonia volatilization and increase nitrogen use efficiency and grain yield of rice. Sci. Total Environ. 2020, 743, 140799. [Google Scholar] [CrossRef] [PubMed]
- Miransari, M.; Mackenzie, A. Development of a Soil N Test for Fertilizer Requirements for Corn Production in Quebec. Commun. Soil Sci. Plant Anal. 2011, 42, 50–65. [Google Scholar] [CrossRef]
- Miransari, M.; Mackenzie, A. Development of a soil n test for fertilizer requirements for wheat. J. Plant Nutr. 2011, 34, 762–777. [Google Scholar] [CrossRef]
- Chai, R.; Ye, X.; Ma, C.; Wang, Q.; Tu, R.; Zhang, L.; Gao, H. Greenhouse gas emissions from synthetic nitrogen manufacture and fertilization for main upland crops in China. Carbon Balance Manag. 2019, 14, 20. [Google Scholar] [CrossRef]
- Nimmo, J. Encyclopedia of Soil in the Environment. Fertil. Fertil. 2004, 3, 295–303. [Google Scholar]
- Singh, B.; Craswell, E. Fertilizers and nitrate pollution of surface and ground water: An increasingly pervasive global problem. SN Appl. Sci. 2021, 3, 518. [Google Scholar] [CrossRef]
- Miransari, M. Soil microbes and plant fertilization. Appl. Microbiol. Biotechnol. 2011, 92, 875–885. [Google Scholar] [CrossRef]
- Liu, R.; Lal, R. Potentials of engineered nanoparticles as fertilizers for increasing agronomic productions. Sci. Total Environ. 2015, 514, 131–139. [Google Scholar] [CrossRef]
- Diacono, M.; Montemurro, F. Long-Term Effects of Organic Amendments on Soil Fertility. A Review. Agron. Sustain. Dev. 2010, 30, 401–422. [Google Scholar] [CrossRef]
- Sohi, S.; Krull, E.; Lopez-Capel, E.; Bol, R. A Review of Biochar and Its Use and Function in Soil. Adv. Agron. 2010, 105, 47–82. [Google Scholar] [CrossRef]
- Jagadamma, S.; Mayes, M.; Steinweg, J.M.; Schaeffer, S. Substrate quality alters microbial mineralization of added substrate and soil organic carbon. Biogeosci. Discuss. 2014, 11, 4451–4482. [Google Scholar] [CrossRef]
- Byrne, M.; Tobin, J.; Forrestal, P.; Danaher, M.; Nkwonta, C.; Richards, K.; Cummins, E.; Hogan, S.; O’Callaghan, T. Urease and Nitrification Inhibitors—As Mitigation Tools for Greenhouse Gas Emissions in Sustainable Dairy Systems: A Review. Sustainability 2020, 12, 6018. [Google Scholar] [CrossRef]
- Lawrencia, D.; Wong, S.K.; Low, D.Y.S.; Goh, B.H.; Goh, J.K.; Ruktanonchai, U.R.; Soottitantawat, A.; Lee, L.H.; Tang, S.Y. Controlled Release Fertilizers: A Review on Coating Materials and Mechanism of Release. Plants 2021, 10, 238. [Google Scholar] [CrossRef]
- Dinca, L.; Grenni, P.; Onet, A.; Onet, C. Fertilization and Soil Microbial Community: A Review. Appl. Sci. 2022, 12, 1198. [Google Scholar] [CrossRef]
- Zhou, L.; Zhao, P.; Chi, Y.; Wang, D.; Wang, P.; Liu, N.; Cai, D.; Wu, Z.; Zhong, N. Controlling the Hydrolysis and Loss of Nitrogen Fertilizer (Urea) by using a Nanocomposite Favors Plant Growth. ChemSusChem 2017, 10, 2068–2079. [Google Scholar] [CrossRef]
- Mercado-Blanco, J.; Abrantes, I.; Barra Caracciolo, A.; Bevivino, A.; Ciancio, A.; Grenni, P.; Hrynkiewicz, K.; Kredics, L.; Proença, D.N. Belowground Microbiota and the Health of Tree Crops. Front Microbiol. 2018, 9, 1006. [Google Scholar] [CrossRef]
- Saleem, M.; Hu, J.; Jousset, A. More Than the Sum of Its Parts: Microbiome Biodiversity as a Driver of Plant Growth and Soil Health. Annu. Rev. Ecol. Evol. Syst. 2019, 50, 145–168. [Google Scholar] [CrossRef]
- Mohammad, M. Soil microbes and the availability of soil nutrients. Acta. Physiol. Plant. 2013, 35, 3075–3084. [Google Scholar] [CrossRef]
- Berendsen, R.L.; Pieterse, C.M.; Bakker, P.A. The rhizosphere microbiome and plant health. Trends. Plant Sci. 2012, 17, 478–486. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Bending, G.D.; Turner, M.K.; Rayns, F.; Marx, M.-C.; Wood, M. Microbial and biochemical soil quality indicators and their potential for differentiating areas under contrasting agricultural management regimes. Soil. Biol. Biochem. 2004, 36, 1785–1792. [Google Scholar] [CrossRef]
- Singh, J.S.; Gupta, V.K. Soil microbial biomass: A key soil driver in management of ecosystem functioning. Sci. Total Environ. 2018, 634, 497–500. [Google Scholar] [CrossRef] [PubMed]
- Wani, G.A.; Khan, M.A.; Dar, M.A.; Shah, M.A.; Reshi, Z.A. Next Generation High Throughput Sequencing to Assess Microbial Communities: An Application Based on Water Quality. Bull. Environ. Contam. Toxicol. 2021, 106, 727–733. [Google Scholar] [CrossRef]
- Wang, J.X.; Liu, S.S.; Han, S.Y.; Wang, A.Y. High-throughput sequencing reveals soil bacterial community structure and their interactions with environmental factors of the grassland fairy ring. Environ. Microbiol. Rep. 2022, 14, 479–493. [Google Scholar] [CrossRef]
- Wegner, C.E.; Richter-Heitmann, T.; Klindworth, A.; Klockow, C.; Richter, M.; Achstetter, T.; Glöckner, F.O.; Harder, J. Expression of sulfatases in Rhodopirellula baltica and the diversity of sulfatases in the genus Rhodopirellula. Mar. Genom. 2013, 9, 51–61. [Google Scholar] [CrossRef]
- Schloter, M.; Nannipieri, P.; Sørensen, S.; van Elsas, J. Microbial indicators for soil quality. Biol. Fertil. Soils. 2018, 54, 1–10. [Google Scholar] [CrossRef]
- Gao, Z.; Hu, Y.; Han, M.; Xu, J.; Wang, X.; Liu, L.; Tang, Z.; Jiao, W.; Jin, R.; Liu, M.; et al. Effects of continuous cropping of sweet potatoes on the bacterial community structure in rhizospheric soil. BMC Microbiol. 2021, 21, 102. [Google Scholar] [CrossRef]
- Yao, Q.; Liu, J.; Yu, Z.; Li, Y.; Jin, J.; Liu, X. Three years of biochar amendment alters soil physiochemical properties and fungal community composition in a black soil of northeast China. Soil. Biol. Biochem. 2017, 110, 56–67. [Google Scholar] [CrossRef]
- Lavecchia, A.; Curci, M.; Jangid, K.; Whitman, W.; Ricciuti, P.; Pascazio, S.; Crecchio, C. Microbial 16S gene-based composition of a sorghum cropped rhizosphere soil under different fertilization managements. Biol. Fertil Soils 2015, 51, 661–672. [Google Scholar] [CrossRef]
- Smalla, K.; Wieland, G.; Buchner, A.; Zock, A.; Parzy, J.; Kaiser, S.; Roskot, N.; Heuer, H.; Berg, G. Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: Plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 2001, 67, 4742–4751. [Google Scholar] [CrossRef]
- Berg, J.; Brandt, K.; Abu Al-Soud, W.; Holm, P.; Hansen, L.; Sørensen, S.; Nybroe, O. Selection for Cu-Tolerant Bacterial Communities with Altered Composition, but Unaltered Richness, via Long-Term Cu Exposure. Appl. Environ. Microbiol. 2012, 78, 7438–7446. [Google Scholar] [CrossRef] [PubMed]
- Zhao, S.; Liu, D.; Ling, N.; Chen, F.; Fang, W.; Shen, Q. Bio-organic fertilizer application significantly reduces the Fusarium oxysporum population and alters the composition of fungi communities of watermelon Fusarium wilt rhizosphere soil. Biol. Fertil. Soils 2014, 50, 765–774. [Google Scholar] [CrossRef]
- Magoč, T.; Salzberg, S.L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 2011, 27, 2957–2963. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Haas, B.J.; Gevers, D.; Earl, A.M.; Feldgarden, M.; Ward, D.V.; Giannoukos, G.; Ciulla, D.; Tabbaa, D.; Highlander, S.K.; Sodergren, E.; et al. Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome. Res. 2011, 21, 494–504. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic. Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Kõljalg, U.; Nilsson, R.H.; Abarenkov, K.; Tedersoo, L.; Taylor, A.F.; Bahram, M.; Bates, S.T.; Bruns, T.D.; Bengtsson-Palme, J.; Callaghan, T.M.; et al. Towards a unified paradigm for sequence-based identification of fungi. Mol. Ecol. 2013, 22, 5271–5277. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef] [PubMed]
- Langille, M.G.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814–821. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.; Song, Z.; Bates, S.; Branco, S.; Tedersoo, L.; Menke, J.; Schilling, J.; Kennedy, P. FUNGuild: An open annotation tool for parsing fungal community datasets by ecological guild. Fungal. Ecol. 2015, 20, 241–248. [Google Scholar] [CrossRef]
- Liang, Z.; Liu, F.; Wang, W.; Zhang, P.; Sun, X.; Wang, F.; Kell, H. High-throughput sequencing revealed differences of microbial community structure and diversity between healthy and diseased Caulerpa lentillifera. BMC Microbiol. 2019, 19, 225. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Zhang, G.; Zhou, L.; Wang, M.; Cai, D.; Wu, Z. Synthesis of a Multifunctional Graphene Oxide-Based Magnetic Nanocomposite for Efficient Removal of Cr(VI). Langmuir 2017, 33, 7007–7014. [Google Scholar] [CrossRef] [PubMed]
- Buckerfield, J.C.; Lee, K.E.; Davoren, C.W.; Hannay, J.N. Earthworms as indicators of sustainable production in dryland cropping in southern Australia. Soil. Biol. Biochem. 1997, 29, 547–554. [Google Scholar] [CrossRef]
- Shi, Z.; Tang, Z.; Wang, C. Effect of phenanthrene on the physicochemical properties of earthworm casts in soil. Ecotoxicol. Environ. Saf. 2019, 168, 348–355. [Google Scholar] [CrossRef]
- Singh, N.; Bhuker, A.; Jeevanadam, J. Effects of metal nanoparticle-mediated treatment on seed quality parameters of different crops. Naunyn. Schmiedebergs. Arch Pharm. 2021, 394, 1067–1089. [Google Scholar] [CrossRef]
- Tilman, D.; Cassman, K.G.; Matson, P.A.; Naylor, R.; Polasky, S. Agricultural sustainability and intensive production practices. Nature 2002, 418, 671–677. [Google Scholar] [CrossRef]
- Sajjadi, M.; Ahmadpoor, F.; Nasrollahzadeh, M.; Ghafuri, H. Lignin-derived (nano)materials for environmental pollution remediation: Current challenges and future perspectives. Int. J. Biol. Macromol. 2021, 178, 394–423. [Google Scholar] [CrossRef]
- Ma, C.; White, J.C.; Zhao, J.; Zhao, Q.; Xing, B. Uptake of Engineered Nanoparticles by Food Crops: Characterization, Mechanisms, and Implications. Annu. Rev. Food Sci. Technol. 2018, 9, 129–153. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; White, J.C.; Wang, Z.; Xing, B. Nano-enabled fertilizers to control the release and use efficiency of nutrients. Curr. Opin. Environ. Sci. Health 2018, 6, 77–83. [Google Scholar] [CrossRef]
- Sohi, S.P.; Krull, E.; Lopez-Capel, E.; Bol, R. Chapter 2—A Review of Biochar and Its Use and Function in Soil. In Advances in Agronomy; Academic Press: Cambridge, MA, USA, 2010; Volume 105, pp. 47–82. [Google Scholar]
- Ehaliotis, C.; Cadisch, G.; Giller, K.E. Substrate amendments can alter microbial dynamics and N availability from maize residues to subsequent crops. Soil. Biol. Biochem. 1998, 30, 1281–1292. [Google Scholar] [CrossRef]
- Kah, M. Nanopesticides and Nanofertilizers: Emerging Contaminants or Opportunities for Risk Mitigation? Front Chem. 2015, 3, 64. [Google Scholar] [CrossRef]
- Huang, D.; Wang, W.; Xu, J.; Wang, A. Mechanical and water resistance properties of chitosan/poly(vinyl alcohol) films reinforced with attapulgite dispersed by high-pressure homogenization. Chem. Eng. J. 2012, 210, 166–172. [Google Scholar] [CrossRef]
- Xiang, Y.; Wang, M.; Sun, X.; Cai, D.; Wu, Z. Controlling Pesticide Loss through Nanonetworks. ACS Sustain. Chem. Eng. 2014, 2, 918–924. [Google Scholar] [CrossRef]
- Kallenbach, C.M.; Conant, R.T.; Calderón, F.; Wallenstein, M.D. A novel soil amendment for enhancing soil moisture retention and soil carbon in drought-prone soils. Geoderma 2019, 337, 256–265. [Google Scholar] [CrossRef]
- Liang, B.; Ma, C.; Fan, L.; Wang, Y.; Yuan, Y. Soil amendment alters soil physicochemical properties and bacterial community structure of a replanted apple orchard. Microbiol. Res. 2018, 216, 1–11. [Google Scholar] [CrossRef]
- Raymond, J.; Siefert, J.L.; Staples, C.R.; Blankenship, R.E. The natural history of nitrogen fixation. Mol. Biol. Evol. 2004, 21, 541–554. [Google Scholar] [CrossRef] [PubMed]
- Debruyn, J.M.; Mead, T.J.; Wilhelm, S.W.; Sayler, G.S. PAH biodegradative genotypes in Lake Erie sediments: Evidence for broad geographical distribution of pyrene-degrading mycobacteria. Environ. Sci. Technol. 2009, 43, 3467–3473. [Google Scholar] [CrossRef]
- Bhatti, A.A.; Haq, S.; Bhat, R.A. Actinomycetes benefaction role in soil and plant health. Microb. Pathog. 2017, 111, 458–467. [Google Scholar] [CrossRef]
- Thomas, F.; Hehemann, J.H.; Rebuffet, E.; Czjzek, M.; Michel, G. Environmental and gut bacteroidetes: The food connection. Front Microbiol. 2011, 2, 93. [Google Scholar] [CrossRef] [PubMed]
- Mujakić, I.; Piwosz, K.; Koblížek, M. Phylum Gemmatimonadota and Its Role in the Environment. Microorganisms 2022, 10, 151. [Google Scholar] [CrossRef] [PubMed]
- Tkacz, A.; Cheema, J.; Chandra, G.; Grant, A.; Poole, P.S. Stability and succession of the rhizosphere microbiota depends upon plant type and soil composition. ISME J. 2015, 9, 2349–2359. [Google Scholar] [CrossRef]
- Bodenhausen, N.; Horton, M.W.; Bergelson, J. Bacterial communities associated with the leaves and the roots of Arabidopsis thaliana. PLoS ONE 2013, 8, e56329. [Google Scholar] [CrossRef]
- Halo, B.; Khan, A.; Waqas, M.; Al-Harrasi, A.; Hussain, J.; Ali, L.; Adnan, M.; Lee, I.-J. Endophytic bacteria (Sphingomonas sp. LK11) and Gibberellin can improve Solanum lycopersicum growth and oxidative stress under salinity. J. Plant Interact. 2015, 10, 117–125. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef]
- Yoon, J.H.; Lee, C.H.; Oh, T.K. Aeromicrobium alkaliterrae sp. nov., isolated from an alkaline soil, and emended description of the genus Aeromicrobium. Int. J. Syst. Evol. Microbiol. 2005, 55, 2171–2175. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Le, W.; Zhang, Y.; Jiang, Y.; Wu, W.; Zu, L.; Jiang, C. A study on polyphasic taxonomy of one antifungal actinomycete strain YIM31530~(T). J. Yunnan Univ. (Nat. Sci.) 2004, 26, 265–269. [Google Scholar]
- Fengyan, Z.; Zhang, Y.; Dong, W.; Zhang, Y.; Zhang, G.; Sun, Z.; Yang, L. Vermicompost can suppress Fusarium oxysporum f. sp. lycopersici via generation of beneficial bacteria in a long-term tomato monoculture soil. Plant Soil 2019, 440, 491–505. [Google Scholar] [CrossRef]
- Franke-Whittle, I.; Manici, L.; Insam, H.; Stres, B. Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted apple orchards. Plant Soil 2015, 395, 317–333. [Google Scholar] [CrossRef]
- Wolff, D.; Krah, D.; Dötsch, A.; Ghattas, A.K.; Wick, A.; Ternes, T.A. Insights into the variability of microbial community composition and micropollutant degradation in diverse biological wastewater treatment systems. Water Res. 2018, 143, 313–324. [Google Scholar] [CrossRef] [PubMed]
- Frąc, M.; Hannula, S.E.; Bełka, M.; Jędryczka, M. Fungal Biodiversity and Their Role in Soil Health. Front Microbiol. 2018, 9, 707. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Cheng, J.; Li, T.; Liao, Y. Response of soil fungal communities to continuous cropping of flue-cured tobacco. Sci. Rep. 2020, 10, 19911. [Google Scholar] [CrossRef] [PubMed]
- Ma, A.; Zhuang, X.; Wu, J.; Cui, M.; Lv, D.; Liu, C.; Zhuang, G. Ascomycota members dominate fungal communities during straw residue decomposition in arable soil. PLoS ONE 2013, 8, e66146. [Google Scholar] [CrossRef] [PubMed]
- Zhao, R.-L.; Li, G.-J.; Sanchez-Ramirez, S.; Stata, M.; Yang, Z.-L.; Wu, G.; Dai, Y.-C.; He, S.-H.; Cui, B.-K.; Zhou, J.-L.; et al. A six-gene phylogenetic overview of Basidiomycota and allied phyla with estimated divergence times of higher taxa and a phyloproteomics perspective. Fungal. Divers. 2017, 84, 43–74. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Spatafora, J.W.; Chang, Y.; Benny, G.L.; Lazarus, K.; Smith, M.E.; Berbee, M.L.; Bonito, G.; Corradi, N.; Grigoriev, I.; Gryganskyi, A.; et al. A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 2016, 108, 1028–1046. [Google Scholar] [CrossRef]
- Žifčáková, L.; Větrovský, T.; Howe, A.; Baldrian, P. Microbial activity in forest soil reflects the changes in ecosystem properties between summer and winter. Environ. Microbiol. 2016, 18, 288–301. [Google Scholar] [CrossRef]
- Phosri, C.; Põlme, S.; Taylor, A.; Kõljalg, U.; Suwannasai, N.; Tedersoo, L. Diversity and community composition of ectomycorrhizal fungi in a dry deciduous dipterocarp forest in Thailand. Biodivers. Conserv. 2012, 21, 2287–2298. [Google Scholar] [CrossRef]
- Schreiter, S.; Ding, G.C.; Heuer, H.; Neumann, G.; Sandmann, M.; Grosch, R.; Kropf, S.; Smalla, K. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front Microbiol. 2014, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Chu, H.; Fierer, N.; Lauber, C.L.; Caporaso, J.G.; Knight, R.; Grogan, P. Soil bacterial diversity in the Arctic is not fundamentally different from that found in other biomes. Environ. Microbiol 2010, 12, 2998–3006. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.G.; Miller, R.M. Carbon cycling by arbuscular mycorrhizal fungi in soil-plant systems. Trends Plant Sci. 2003, 8, 407–409. [Google Scholar] [CrossRef] [PubMed]
- Deng, Q.; Cheng, X.; Hui, D.; Zhang, Q.; Li, M.; Zhang, Q. Soil microbial community and its interaction with soil carbon and nitrogen dynamics following afforestation in central China. Sci. Total Environ. 2016, 541, 230–237. [Google Scholar] [CrossRef]
- Fierer, N.; Strickland, M.S.; Liptzin, D.; Bradford, M.A.; Cleveland, C.C. Global patterns in belowground communities. Ecol. Lett. 2009, 12, 1238–1249. [Google Scholar] [CrossRef]
- Burke, C.; Thomas, T.; Lewis, M.; Steinberg, P.; Kjelleberg, S. Composition, uniqueness and variability of the epiphytic bacterial community of the green alga Ulva australis. ISME J. 2011, 5, 590–600. [Google Scholar] [CrossRef]
- Burke, C.; Steinberg, P.; Rusch, D.; Kjelleberg, S.; Thomas, T. Bacterial community assembly based on functional genes rather than species. Proc. Natl. Acad Sci. USA 2011, 108, 14288–14293. [Google Scholar] [CrossRef]
- Chen, L.; Xiang, W.; Wu, H.; Ouyang, S.; Lei, P.; Hu, Y.; Ge, T.; Ye, J.; Kuzyakov, Y. Contrasting patterns and drivers of soil fungal communities in subtropical deciduous and evergreen broadleaved forests. Appl. Microbiol. Biotechnol. 2019, 103, 5421–5433. [Google Scholar] [CrossRef]








| Treatment | pH Value | Organic Matter (g/kg) | Total Salt (%) | Available Nitrogen (mg/kg) | Available Phosphorus (mg/kg) | Available Potassium (mg/kg) | Soil Capacity (g/cm3) | Porosity (%) | Water Retention Rate (%) |
|---|---|---|---|---|---|---|---|---|---|
| CK | 7.86 ± 0.13 | 26 ± 3 | 0.10 ± 0.02 | 80 ± 7 | 95 ± 2 | 426 ± 10 | 1 ± 0.01 | 17.5 ± 0.34 | 23 ± 1 |
| SC3 | 7.87 ± 0.06 | 26 ± 2 | 0.11 ± 0.02 | 97 ± 8 | 82 ± 4 | 438 ± 14 | 1 ± 0.01 | 19.7 ± 0.97 | 25 ± 2 |
| Treatment | Oberved_Species | chao1 | ACE | Shannon | Simpson | PD_Whole_Tree | Good’s_Coverage | |
|---|---|---|---|---|---|---|---|---|
| 16S rRNA | CK | 1240 ± 100 | 1336 ± 101 | 1346 ± 102 | 7.01 ± 0.19 | 0.96 ± 0.00 | 104 ± 4 | 0.996 |
| SC3 | 1550 ± 8 | 1614 ± 10 * | 1604 ± 18 * | 8.85 ± 0.01 *** | 0.99 ± 0.00 *** | 115 ± 2 | 0.997 | |
| ITS1-5F | CK | 435 ± 22 | 459 ± 21 | 464 ± 20 | 4.93 ± 0.67 | 0.89 ± 0.07 | 125 ± 6 | 0.999 |
| SC3 | 521 ± 8 ** | 548 ± 11 ** | 549 ± 15 ** | 6.59 ± 0.10 * | 0.97 ± 0.00 | 145 ± 5 ** | 0.999 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, J.; Zhao, P.; Wang, D.; Zhao, Y.; Wang, Z.; Zhong, N. Effects of a Nanonetwork-Structured Soil Conditioner on Microbial Community Structure. Biology 2023, 12, 668. https://doi.org/10.3390/biology12050668
Cao J, Zhao P, Wang D, Zhao Y, Wang Z, Zhong N. Effects of a Nanonetwork-Structured Soil Conditioner on Microbial Community Structure. Biology. 2023; 12(5):668. https://doi.org/10.3390/biology12050668
Chicago/Turabian StyleCao, Jingjing, Pan Zhao, Dongfang Wang, Yonglong Zhao, Zhiqin Wang, and Naiqin Zhong. 2023. "Effects of a Nanonetwork-Structured Soil Conditioner on Microbial Community Structure" Biology 12, no. 5: 668. https://doi.org/10.3390/biology12050668
APA StyleCao, J., Zhao, P., Wang, D., Zhao, Y., Wang, Z., & Zhong, N. (2023). Effects of a Nanonetwork-Structured Soil Conditioner on Microbial Community Structure. Biology, 12(5), 668. https://doi.org/10.3390/biology12050668






